Abstract
Complex functional coupling exists between transcriptional elongation and pre-mRNA alternative splicing. Pausing sites and changes in the rate of transcription by RNA polymerase II (RNAPII) may therefore have fundamental impacts in the regulation of alternative splicing. Here, we show that the elongation and splicing-related factor TCERG1 regulates alternative splicing of the apoptosis gene Bcl-x in a promoter-dependent manner. TCERG1 promotes the splicing of the short isoform of Bcl-x (Bcl-xs) through the SB1 regulatory element located in the first half of exon 2. Consistent with these results, we show that TCERG1 associates with the Bcl-x pre-mRNA. A transcription profile analysis revealed that the RNA sequences required for the effect of TCERG1 on Bcl-x alternative splicing coincide with a putative polymerase pause site. Furthermore, TCERG1 modifies the impact of a slow polymerase on Bcl-x alternative splicing. In support of a role for an elongation mechanism in the transcriptional control of Bcl-x alternative splicing, we found that TCERG1 modifies the amount of pre-mRNAs generated at distal regions of the endogenous Bcl-x. Most importantly, TCERG1 affects the rate of RNAPII transcription of endogenous human Bcl-x. We propose that TCERG1 modulates the elongation rate of RNAPII to relieve pausing, thereby activating the proapoptotic Bcl-xS 5′ splice site.
INTRODUCTION
The expression of protein-coding genes in eukaryotes is a highly orchestrated process that involves multiple coordinated events. Genomic DNA must be transcribed into precursor mRNAs (pre-mRNA) by RNA polymerase II (RNAPII) and processed through subsequent steps to yield a mature mRNA that is exported from the nucleus to the cytoplasm and used by the translational machinery. The pre-mRNA undergoes several processing steps, including capping, splicing, and cleavage/polyadenylation, which appear to be precisely coordinated with nascent transcript formation (41, 44, 49). Of these RNA processing mechanisms, alternative splicing occurs as a widespread means to achieve proteomic diversity. Results of deep sequencing-based expression analyses estimate that more than 90% of multiexon human genes undergo alternative splicing (50, 66). The misregulation of alternative splicing underlies multiple diseases, including neurological disorders and cancer (5, 19, 32, 67).
Although transcription and alternative splicing can occur independently, both processes are physically and functionally interconnected (44, 49), and this coupling and coordination may be important for the regulation of gene expression. To date, two models have been proposed to explain the link between transcription and splicing. In the recruitment model, the unique carboxyl-terminal domain (CTD) of RNAPII functions as a “landing pad” for factors involved in pre-mRNA splicing in a manner that is dependent on the phosphorylation of RNAPII and the resulting functional state of the transcriptional complex (4, 7, 28, 38, 40, 42, 43, 71). In the kinetic model, an alternative but not exclusive model, the transcript elongation rate determines the outcome of competing splicing reactions that occur cotranscriptionally (5). There is extensive evidence indicating that the rate of transcription elongation could be used to control alternative splicing (16, 17, 26, 27, 35, 48, 56). Conversely, cis-acting elements and splicing factors have been shown to affect RNAPII processivity (13, 18, 34). The physiological relevance of the coupling between transcriptional elongation and alternative splicing was demonstrated in a recent report that reported that DNA damage affected specific alternative splicing events through changes in the RNAPII elongation rate (45). Recently, a global analysis of the nascent RNA in yeast revealed that cotranscriptional splicing is associated with RNAPII pausing at specific sites (1, 6, 69). Thus, it is proposed that transcriptional pausing is imposed by a regulatory checkpoint that is associated with cotranscriptional splicing (1). One hypothesis suggests that proteins acting at the interface of these processes serve as checkpoint factors to regulate cotranscriptional splicing.
TCERG1 (previously designated CA150) is a human nuclear factor that has been implicated in transcriptional elongation and pre-mRNA splicing. TCERG1 is composed of multiple protein domains, most notable of which are three WW domains in the amino-terminal half and six FF repeat motifs in the carboxyl-terminal half (63). Transcription and splicing components bind to both domains (23, 33, 57, 61), and TCERG1 has been identified in highly purified spliceosomes in multiple studies (14, 37, 47, 52). The subnuclear distribution of TCERG1 resembles that of an RNA metabolism factor, with enrichment at the interface of splicing factor-rich nuclear speckles and what are presumably nearby transcription sites (57). TCERG1 can affect pre-mRNA splicing of β-globin, β-tropomyosin, CD44, and fibronectin splicing reporters (11, 33, 51, 58) and of putative cellular targets identified by microarray analysis following TCERG1 knockdown (51). In the manuscript, we report that TCERG1 regulates the alternative splicing of Bcl-x exon 2 by modulating the rate of RNAPII transcriptional elongation. We speculate that TCERG1 relieves pausing of RNAPII and therefore acts as a checkpoint regulator to promote cotranscriptional splicing.
MATERIALS AND METHODS
Plasmids.
The pEFBOST7-TCERG1(1–1098), pEFBOST7-TCERG1(1–662), and pEFBOST7-TCERG1(591–1098) plasmids have been previously described (57, 62). Alberto Kornblihtt (Universidad de Buenos Aires, Buenos Aires, Argentina) and David Bentley (University of Colorado School of Medicine) kindly provided the expression vectors for the α-amanitin-resistant variants of the human RNAPII (hRpb1) large subunit.
Alternative splicing reporter minigenes.
The CMV-X2, HIV-X2, and HIV-X2.13 reporter minigenes have been previously described (20, 59). Bcl-x inserts of plasmids X2 and X2.13 were produced by PCR amplification using plasmids CMV-X2 and CMV-X2.13 (27) as templates, the Pfu-Turbo polymerase,and primers AscI-X-Fwd and X-Age-Rev. The derived PCR products were cleaved with AscI and AgeI and ligated into the SVEDA-HIV-2 vector (a kind gift of Alberto Kornblihtt, Buenos Aires, Argentina) cut with the same enzymes. Overlap PCR mutagenesis was used to generate deletions in the SB1 element (Δ9, Δ11, Δ13, Δ16, and Δ23).
Transfections.
HEK293T and HeLa cells were used for the transfection assays. The transfections were performed in 35-mm plates (Falcon). Each plate was seeded with approximately 2 × 105 cells 20 h prior to transfection. The cells were grown to approximately 60 to 70% confluence and transfected with the appropriate amounts of the indicated constructs by using calcium phosphate. Approximately 48 h after transfection, the cells were harvested and processed for Western blotting and reverse transcriptase PCR (RT-PCR) analysis.
For the RNA interference (RNAi) knockdown experiments, the cells were transfected using JetPrime (Polyplus) or Lipofectamine 2000 reagent (Invitrogen) according to the manufacturers' protocols with 60 nM (final concentration) of either one of the following small interfering RNA (siRNA) duplexes: siEGFP, 5′-CUACAACAGCCACAACGE-3′, or siTCERG1, 5′-GGAGUUGCACAAGAUAGUU-3′ (51). The cells were harvested 48 h after transfection. For double transfections, the cells were transfected with the reporter minigene 48 h after siRNA transfection. The cells were harvested 24 h later. Under these conditions, we consistently achieved ∼6-fold overexpression and at least ∼80% knockdown of TCERG1. α-Amanitin (Sigma) was added to the cells at 10 μg/ml 18 h after transfection, and cells were collected 24 h later. A total of 5 μM MG132 (Calbiochem) was administered at 3 h after transfection for 18 h.
RNA extraction and RT-PCR analysis.
Total RNA was extracted from transfected cells with TRIzol (Invitrogen) using the manufacturer's protocol. The RNA was digested with 10 U RNase-free DNase I (Roche) for 30 min at 37°C. After DNase inactivation at 70°C for 5 min, 1 μg of endogenous RNA or 400 ng plasmid-derived RNA was reverse transcribed by the Moloney murine leukemia virus RT (Invitrogen) for 1 h at 37°C using oligo(dT)15 for endogenous RNA and primer RT-Sveda or RT3 for HIV-X2 or CMV-X2 reporter minigenes, respectively. One-tenth of the resulting cDNA was amplified by PCR using the oligonucleotides X3 and X2 for the endogenous gene or X34 and XAgeIR for the plasmid-derived transcripts. After 30 cycles, the PCR products were analyzed on a 1.5% agarose gel. The intensity of the bands was quantified using Quantity One software (Bio-Rad). For PCR of the endogenous Bcl-x, [α-32P]dCTP (PerkinElmer Life Sciences) was added to the PCR mixtures, and the amplification products were fractionated onto a 4% native polyacrylamide gel. The gels were exposed on screens that were scanned on a Storm PhosphorImager 860 (GE Healthcare). The intensity of the bands was quantified using ImageQuant software.
For the pre-mRNA quantification, RNA was extracted from cells grown in 60-mm plates (Falcon) as described before. Approximately 5 μg digested RNA was reverse transcribed using random hexamers. The quantification of the Bcl-x transcripts was carried out by real-time PCR using the Perfecta SYBR green supermix for iQ (Quanta Biosciences) and the iCycler thermal cycler station (Bio-Rad) with oligonucleotides D-fwd and D-rev for the distal regions of the Bcl-x gene. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal gene control. The values are represented as (E Bcl-X)ΔCT Bcl-x/(E GAPDH)ΔCT GAPDH, where E is the PCR efficiency and ΔCT = (the cycle threshold [CT] for mock treatment) − (CT for TCERG1 knockdown/overexpression).
The statistical analysis was performed using Prism 5.0 software (GraphPad). Two-tailed Student's t tests were used to compare the means between the samples and their respective controls. The P values are represented in the figures by asterisks (*, P < 0.05; **, P < 0.01). The absence of an asterisk indicates that the change relative to control was not statistically significant.
Chromatin immunoprecipitation assay.
HEK293T cells were seeded in 100-mm-diameter plates at 60 to 70% confluence and transfected with 6 μg splicing reporter minigene HIV-X2 or CMV-X2 using the calcium phosphate precipitation method. After 48 h, the cells were fixed with 1% formaldehyde to cross-link the chromatin and were incubated at room temperature for 10 min. For the experiment shown in Fig. 3E, below, we used 20 min of cross-linking. The cross-linking was arrested by adding glycine (0.125 M) for an additional 5 min at room temperature. Subsequently, the cells were pelleted, washed three times with phosphate-buffered saline (PBS), and lysed in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], protease inhibitor mixture [Complete; Roche], and 1 mM phenylmethylsulfonyl fluoride [PMSF]) for 10 min on ice. The lysates were sonicated 10 times for 15 s on ice and centrifuged at maximum speed. The sheared chromatin was diluted by the addition of 10 volumes of ChIP buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl, protease inhibitor mixture, and 1 mM PMSF) and precleared with a salmon sperm DNA/protein A-agarose fast-flow slurry (Millipore) for 2 h. The beads were removed by centrifugation. A 5% sample of the precleared chromatin supernatant was removed to serve as the preimmunoprecipitation (pre-IP; input) control, and the remaining precleared chromatin was incubated overnight with 10 μg anti-RNAPII (NP-20; Santa Cruz Biotechnology), anti-TCERG1 (57), or nonspecific rabbit IgG. The chromatin-antibody complexes were collected by incubation with salmon sperm DNA/protein-A agarose (50% slurry) and centrifugation. The bead pellets were washed in low or high salt conditions using a buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 150 mM NaCl) containing 20 mM and 500 mM NaCl, respectively. The beads were then washed once with LiCl buffer (0.25 M LiCl, 1% NP-40, 1% Na-deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl [pH 8.0]) followed by two washes with Tris-EDTA buffer. The antibody-chromatin complexes were eluted from the beads by incubation with elution buffer (0.1% SDS, 0.1 M NaHCO3). A final concentration of 0.2 M NaCl was added to eluates and incubated at 65°C for 4 to 6 h. The samples were treated with RNase A and proteinase K, and the DNA was purified using phenol-chloroform extraction. The pre-IP input sample was purified in a manner similar to the bound chromatin immunoprecipitation (ChIP) fraction described above. The DNA obtained was amplified by quantitative PCR (qPCR) using Perfecta SYBR green supermix for iQ (Quanta Biosciences). The following primers were used: LTR-fwd and LTR-rev for the HIV-2 long terminal repeat (LTR) promoter; CMV-fwd and CMV-rev for the cytomegalovirus (CMV) promoter; P-fwd, P-rev, E2-fwd, E2-rev, D-fwd, and D-rev for the endogenous Bcl-x gene. Dilutions of the input were used to normalize the obtained values. The statistical analysis of the data was performed using Prism 5.0 software (GraphPad) as described above.
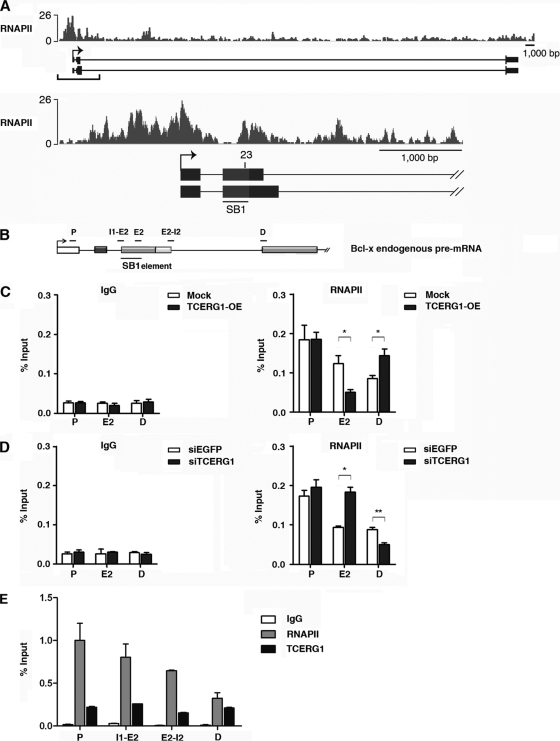
Fig 3.
TCERG1 modulates RNAPII distribution on Bcl-x exon 2. (A) An RNAPII-paused region coincided with region 23 in the Bcl-x gene. The densities of sequence reads from the RNAPII chromatin-immunopurified samples (bars) are displayed above the Bcl-x promoter region (−1,000 bp upstream of the start site) and the first 4,000 bp of the transcribed region. The alternative splicing regulatory region SB1 (gray box) and region 23, required for TCERG1 activity, are indicated. (B) Schematic representation of the structure of the Bcl-x gene, drawn with exons (boxes) and introns (lines). The positions of the SB1 element and of the primers used to amplify mRNA products by qPCR are indicated (P, promoter region; E2, exon 2; I1-E2, intron 1-exon 2 junction; E2-I2, exon 2-intron 2 junction; D, distal region). (C) The polymerase distribution at different positions of the gene was detected by ChIP followed by qPCR of cells transfected with an empty vector (mock) or a TCERG1 overexpression (OE) expression vector. (D) The same experiment described for panel B was carried out with siRNAs against enhanced green fluorescent protein (EGFP) or TCERG1. (E) Distributions of RNAPII and TCERG1 at different positions of the gene were detected by ChIP followed by qPCR of cells. IgG was used as a control in all the ChIP experiments. Data from three independent experiments are presented as percentages of the bound input (means ± standard deviations). *, P < 0.05; **, P < 0.01.
RNAPII ChIP-Seq.
HCT116 cells were seeded in 100-mm-diameter plates and grown under normal conditions (10% fetal bovine serum in Dulbecco's modified Eagle's medium). When cells reached 70 to 80% confluence they were fixed with 1% formaldehyde to cross-link the chromatin and were incubated at room temperature for 10 min. Subsequently, the cells were washed two times with ice-cold PBS. ChIP was performed with antibody H-224 against RNAPII (sc-9001; Santa Cruz Biotechnology) using the procedure described by Svotelis et al. in 2009 (64). The DNA samples from the RNAPII chromatin-immunopurified sample were processed into an Illumina compatible library using a ChIP-Seq kit following the manufacturer's protocol (Illumina). The library was then sequenced for 40 cycles on the Illumina GAIIx platform following the manufacturer's recommendations (Illumina). The 40-nucleotide (nt) reads from the sequencer Fastq files were then aligned to the human genome (hg19) with bowtie, using the options -S -a -p6 -m1 -n2 -best -strata, and the resulting alignment were then converted to binary sorted alignments in BAM format using SamTools (31). A total of 9.2 M and 9.7 M 40-nt reads were generated for the control and 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside (DRB; Sigma)-treated samples, respectively, and 64.3% (5.9 M) and 69.1% (6.7 M) of the reads were uniquely aligned to the human genome for the respective library.
In vitro RNA immunoprecipitation.
Templates for the RNA transcripts were amplified from the pS2 and pS2.13 plasmids (20) by PCR (PfuTurbo; Stratagene) and were gel purified (Qiagen). Transcription was performed using T3 RNA polymerase (Promega) in the presence of a cap analog and [α-32P]UTP (PerkinElmer Life Sciences). Total RNA was purified from a 4.5% denaturing acrylamide gel as previously described (70).
Sepharose-protein A beads (CL-4B; GE Healthcare) were incubated in NET2 buffer (150 mM NaCl, 50 mM Tris HCl [pH 7.5], and 0.05% NP-40) for 1 h at room temperature and then incubated with 1 μl anti-hnRNPK (a kind gifte of Gideon Dreyfuss, University of Pennsylvania School of Medicine), 4 μl anti-TCERG1, or 4 μl IgG antibodies per 50 μl of 50% slurry and incubated for 1 h at 4°C. After three washes with cold NET2 buffer, equivalent quantities of the transcripts were incubated in 12.5 μl splicing mix without ATP for 30 min at 4°C as previously described (46). Total counts were used for the input measurements, and the radioactivity was measured again after five washes with 1 ml NET2 cold buffer.
In vivo RNA immunoprecipitation.
HEK293T cells were used to investigate the association of TCERG1 with Bcl-x RNA. For the detection of Bcl-x transcripts derived from the minigenes, HEK293T cells grown in 100-mm-diameter plates were transfected with 6 μg of splicing reporter HIV-X2 by using calcium phosphate. Cells were washed twice in cold PBS and fixed with 1% formaldehyde for 10 min at room temperature with slow mixing. The cross-linking was arrested by adding glycine at 0.125 M for 5 min at room temperature. Cells were pelleted and washed twice with ice-cold PBS. Subsequently, cells were lysed in 2 ml of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% SDS, 1 mM EDTA, 150 mM NaCl, and protease inhibitor mixture [Complete; Roche]) and sonicated 3 times for 20 s on ice. The cell extracts were centrifuged at maximum speed for 10 min. Supernatants were cleared with a protein A-agarose fast-flow slurry (Millipore) and tRNA at 100 μg/μl with end-over-end rotation for 2 h at 4°C. Beads were removed by centrifugation, and the supernatant was incubated with anti-TCERG1 antibodies and nonspecific rabbit IgG overnight. A fraction of the supernatant was saved for RNA extraction as the input value. The RNA-antibody complexes were collected with protein A-agarose beads for 2 h at 4°C. (Beads were previously incubated with RNaseOUT [Invitrogen] for 10 min at room temperature.) The beads were collected and washed six times in high-stringency RIPA buffer (50 mM Tris-HCl [pH 7.5], 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 1 M NaCl, 1 M urea, and 0.2 mM PMSF) with end-over-end rotation for 10 min at room temperature. The beads were resuspended in a buffer containing 100 μl of 50 mM Tris-HCl (pH 7.0), 5 mM EDTA, 10 mM dithiothreitol, and 1% SDS, incubated at 70°C for 45 min, and then treated with proteinase K (Ambion) for 30 min at 50°C. RNA was extracted with phenol-chloroform-isoamyl alcohol and precipitated with ethanol. RNA precipitates were resuspended in 15 μl of water, treated with DNase I (Roche) for 30 min at 37°C, and reversed transcribed using random hexaprimers and RT-Sveda for the endogenous Bcl-x and the Bcl-x minigene, respectively. qPCR was performed using a LightCycler 2.0 and Fast Start DNA Master Plus SYBR green (Roche) with D-fwd and D-rev primers for the endogenous Bcl-x and for the minigene-derived products.
RNAPII processivity assay.
The rates of RNAPII transcription were measured as previously described (60) with minor modifications. Briefly, HEK293T cells were grown to approximately 70 to 80% confluence and transfected with 60 nM concentrations of the indicated siRNAs. The next day, the cells were treated with 100 μM DRB in culture medium for 3 h. The cells were washed with PBS to remove the DRB and incubated in fresh medium for various periods of time as indicated in the legend for Fig. 5, below. Following the incubation period, the cells were lysed, and the total RNA was isolated using TRIzol reagent. The cDNA was amplified by qPCR using the Perfecta SYBR green supermix for iQ (Quanta Biosciences) and D-fwd and D-rev primers. GAPDH was used as a reference gene control. The CT was calculated as follows: (E Bcl-X)ΔCT Bcl-X/(E GAPDH)ΔCT GAPDH, where E is the PCR efficiency and ΔCT = CT for the control − CT for DRB-treated cultures. The reactions were performed in triplicate. The results show the averages and standard deviations of two independent experiments.
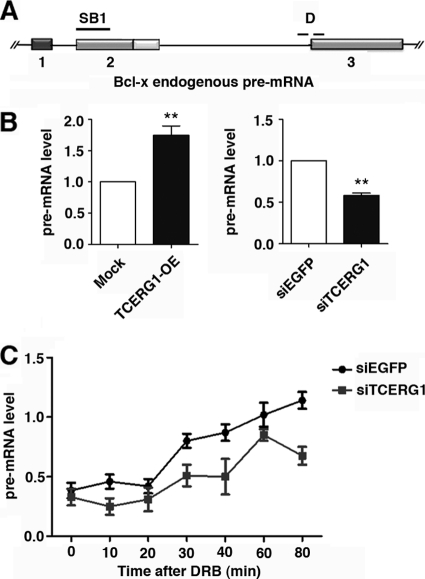
Fig 5.
TCERG1 increases the rate of RNAPII transcription in vivo. (A) Schematic representation of the structure of the Bcl-x gene, drawn with exons (boxes) and introns (lines). The position of the SB1 element and of the primers spanning the intron-exon 3 junction (D) used to amplify transcripts by qRT-PCR are indicated. (B) Quantitative analysis of the amount of nascent Bcl-x transcripts. (Left) HEK293T cells were transfected with an empty vector (mock) or the TCERG1 overexpression (OE) plasmid. (Right) HEK293T cells were transfected with siRNAs against EGFP or TCERG1. The quantification of the experimental data from four independent experiments performed in triplicates is shown in graphic form (means ± standard deviations [SD]). **, P < 0.01. (C) Kinetics of RNAPII-dependent transcription elongation in the presence or absence of TCERG1. HEK293T cells were transfected with siEGFP or siTCERG1 and treated 48 h later with 100 μM DRB for 3 h. After DRB removal, fresh medium was added and samples were taken at the times indicated. qRT-PCR was performed using primer set D to measure the levels of pre-mRNA expression. The graph shows pre-mRNA levels at different times from the control (siEGFP) or TCERG1-depleted cells (siTCERG1). The data shown are the averages from triplicates of two independent experiments (means ± SD). *, P < 0.05; **, P < 0.01.
Western blot analysis.
A fraction of the transfected cells were lysed in cold T7 buffer (20 mM HEPES [pH 7.9], 150 mM NaCl, 5 mM EDTA, 1% NP-40, 1 mM dithiothreitol, protease inhibitor mixture [Complete, Roche], and 1 mM PMSF). The proteins were separated by 10% SDS-PAGE, transferred to a nitrocellulose membrane (Amersham Biosciences), and then incubated with specific antibodies against TCERG1 (57), CDK9 (Santa Cruz Biotechnology) at a 1:500 dilution, and anti-T7 (Bethyl) at a 1:20,000 dilution. Peroxidase-conjugated secondary antibodies (PerkinElmer Life Science) were used at a 1:500 dilution, and the bound antibodies were detected by enhanced chemiluminescence (PerkinElmer Life Science).
Primers.
The following primers were used in the study: LTR-fwd, 5′-CTGCAGGTCGGAGTACTGTC-3′; LTR-rev, 5′-GGAACACCCAGGCTCTACCT-3′; CMV-fwd, 5′-CACCAAAATCAACGGGACTT-3′; CMV-rev, 5′-TTCTGAAGCTCGAGACTGACC-3′; P-fwd, 5′-GGACGGATGAAATAGGCTGA-3′; P-rev, 5′-GAAGAGACAGGGGAACTTGC-3′; E2-fwd, 5′-CCCAGAAAGGATACAGCTGG-3′; E2-rev, 5′-GCGATCCGACTCACCAATAC-3′; D-fwd, 5′-TGGAGGGTCAAGAAAGAGGA-3′; D-rev, 5′-TGCTGCATTGTTCCCATAGA-3′; RT-Sveda, 5′-GGGAAGCTAGAGTAAGTAG-3′; RT3, 5′-GAAGGCACAGTCGAGGCTG-3′; X3, 5′-ATGGCAGCAGTAAAGCAAGCG-3′; X2, 5′-TCATTTCCGACTGAAGAGTGA-3′; X34, 5′-AGGGAGGCAGGCGACGGCGACGAGTTT-3′; XAgeIR, 5′-GTGGATCCCCCGGGCTGCAGGAATTCGAT-3′; SB1-fwd, 5′-GCTGGTGGTTGACTTTCTCTC-3′; SB1-rev, 5′-GGTCTCCATCTCCGATTCAG-3′; E2-fwd, 5′-CAGCTTGGATGGCCACTTAC-3′; I2-rev, 5′-TCTCCAACAATCACCCAACA-3′; I1-fwd, 5′-CACTGGTGCTTTCGATTTGA-3′; E2-rev, 5′-CCAAAACACCTGCTCACTCA-3′; hGH-fwd, 5′-CAACAGAAATCCAACCTAGAGCTGCT-3′; hGH-rev, 5′-TCTTCCAGCCTCCCATCAGCGTTTGG-3′; GAPDH fwd, 5′-ATGGGGAAGGTGAAGGTCG-3′; GAPDH rev, 5′-GGGTCATTGATGGCAACAATATC-3′; AscI-X-Fwd, 5′-GGCGCGCCTCACTATAGGGAGACCCAAGCTGGCTAG-3′; X-Age-Rev, 5′-CTTACCGGTGGATCCCCCGGGCTGCAGGAATTCGAT-3′.
Microarray data accession number.
We deposited the ChIP-Seq data in the Gene Expression Omnibus (GEO) database (accession number GSE 33716).
RESULTS
TCERG1 promotes the splicing of the short isoform of Bcl-x (Bcl-xs) in a promoter-dependent manner.
Several studies have implicated TCERG1 in the regulation of apoptosis (61, 68). For this reason, we conducted in vivo splicing assays to test whether TCERG1 regulated the alternative splicing of genes involved in apoptosis. Among others, we tested the effect of TCERG1 on the alternative splicing of the Bcl-x gene. Human Bcl-x pre-mRNA undergoes alternative splicing at 5′ splice sites to produce the antiapoptotic long (Bcl-xL) and the proapoptotic short (Bcl-xS) isoforms; the use of the downstream site produces Bcl-xL, and the use of the upstream site produces Bcl-xS (Fig. 1A) (3). Endogenous Bcl-x expression predominantly produces the Bcl-xL mRNA variant in HEK293T cells (Fig. 1B) (20). Exogenous TCERG1 expression stimulated the endogenous production of the Bcl-xS splice variant (Fig. 1B). Given the already-high level of Bcl-xL isoforms, changes in the splicing pattern were barely detected upon TCERG1 depletion using siRNAs (unpublished observations). The impact of TCERG1 on Bcl-x splicing may be through its ability to coordinate transcription elongation and alternative splicing (24, 57). To assess this possibility, we tested the effect of TCERG1 on the splicing of Bcl-x reporter minigenes programmed by two different promoters, the human immunodeficiency virus type 2 promoter and the cytomegalovirus promoter (HIV-X2 and CMV-X2, respectively). Although the Bcl-x splicing profile from these minigenes did not exactly mimic the pattern of the endogenous transcripts, the minigenes have been shown to be useful in attemps to unravel the complex regulation of the alternative splicing of Bcl-x (20). Total RNA was isolated from HEK293T cells transfected with either HIV-X2 or CMV-X2 minigenes under conditions of TCERG1 overexpression or gene knockdown, and splicing was analyzed by RT-PCR using specific primers to detect the Bcl-xL and Bcl-xS isoforms. In the case of HIV-X2, increased levels of TCERG1 produced a more efficient utilization of the Bcl-xS 5′ splice site, resulting in a decreased ratio of Bcl-xL to Bcl-xS, while the converse effect was observed upon TCERG1 depletion using siRNA (Fig. 1C). An intact carboxyl region in TCERG1 is necessary and sufficient for promoting the expression of the Bcl-xs splice isoform (unpublished observations). However, no effects of TCERG1 overexpression/knockdown on Bcl-x alternative splicing were observed when the CMV-X2 minigene was used (Fig. 1D). Taken together, these data indicate that TCERG1 regulates the alternative splicing of Bcl-x in a promoter-dependent manner.
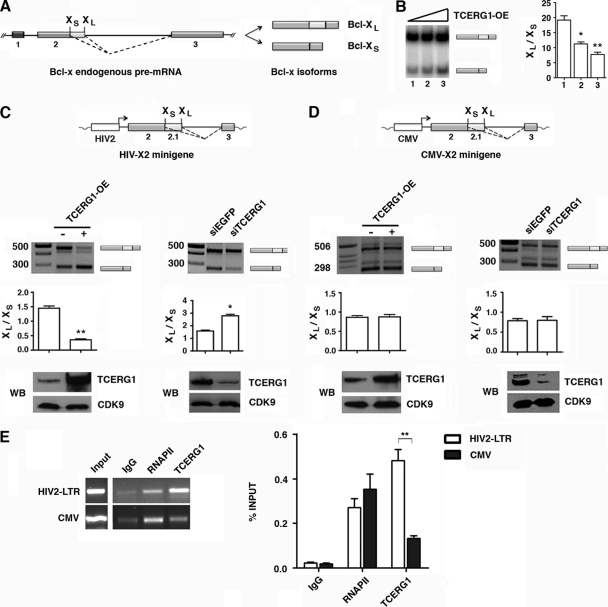
Fig 1.
Regulation of Bcl-x alternative splicing by TCERG1. (A) Schematic representation of the structure of the Bcl-x gene, with exons (boxes) and introns (lines). Two splice variants derived from the Bcl-x gene, antiapoptotic Bcl-xL and proapoptotic Bcl-xs, were generated via alternative 5′ splice site selection within exon 2. The positions of the 5′ splice sites of Bcl-xs and Bcl-xL are indicated. The dotted lines indicate the alternative splicing events. (B) Overexpression (OE) of TCERG1 increased the levels of proapoptotic Bcl-xs in a dose-dependent manner. HEK293T cells were transfected with an empty vector (lane 1) or plasmids carrying TCERG1 (lanes 2 and 3, respectively). After total RNA extraction, the RNA splicing variants were amplified by radioactive RT-PCR, and the products were separated on a native 4% polyacrylamide gel. The graph on the left shows the densitometric analyses results as the ratio of Bcl-xL to Bcl-xS isoforms from three independent experiments (means ± standard deviations [SD]). *, P < 0.05; **, P < 0.01. (C and D) Effects of TCERG1 overexpression/knockdown on alternative splicing of Bcl-x minigenes carrying the HIV-2 LTR (C) and CMV (D) promoters. A schematic of the minigenes is shown. HEK293T cells were cotransfected with the Bcl-x minigene together with empty plasmid (-) or a TCERG1 expression plasmid (+). For the RNAi experiments, HEK293T cells were cotransfected with the Bcl-x minigene together with siTCERG1 or the control (siEGFP). RT-PCR was performed to analyze alternatively spliced forms of Bcl-x. The graphs show the densitometric analysis results as the ratio of Bcl-xL to Bcl-xS isoforms from four independent experiments (means ± SD). *, P < 0.05; **, P < 0.01. A fraction of the cell lysates was analyzed by immunoblotting with the indicated antibodies to detect the TCERG1 and CDK9 proteins. (E) ChIP analysis of the recruitment of TCERG1 to the HIV-2 and CMV promoters. TCERG1 binds preferentially to the HIV-2 promoter in vivo. HEK293T cells were cotransfected with either HIV2-X2 or CMV-X2 minigenes together with a TCERG1 expression plasmid. The cross-linked and shared chromatin was immunoprecipitated with the indicated antibodies. After reversal of the cross-linking and purification of the DNA, PCR was used to detect the sequences corresponding to the HIV-2 and CMV promoters. The input shows the signal from the chromatin before immunoprecipitation. The primers used in the ChIP assays are described in Materials and Methods. The bar graph presents the quantification of the data by qPCR from four independent experiments (means ± SD). **, P < 0.01.
The differences observed in the TCERG1 splicing effects with HIV-X2 or CMV-X2 minigenes prompted us to measure TCERG1 occupancy at the promoter region in ChIP experiments. The ChIP experiments revealed an increased amount of TCERG1 bound to the HIV-2 promoter in transfected HEK293T cells (Fig. 1E), consistent with the observation that the HIV-X2-dependent alternative splicing is affected by TCERG1 in these cells. In contrast, the ChIP assays detected low levels of TCERG1 at the CMV promoter region (Fig. 1E). Thus, TCERG1 associates specifically with the HIV-2 gene promoter. These results suggest that complexes with different protein compositions assemble at each promoter to regulate Bcl-x alternative splicing.
TCERG1 promotes use of the Bcl-xs 5′ splice site through the SB1 regulatory element in exon 2.
It has been reported that a cis regulatory element, called SB1, is located in the first half of Bcl-x exon 2 (55). This 361-nucleotide-long region behaves as a splicing silencer, and its deletion stimulates the use of the Bcl-xS 5′ splice site, thus promoting the expression of the Bcl-xS isoform in transfection assays performed with HEK293 and HeLa cells (55). We sought to determine whether TCERG1 affects Bcl-x alternative splicing through the SB1 element. To this end, the splicing ratios of transcripts derived from the HIV2-X2 and HIV2-X2.13 minigenes (Fig. 2A, X2 and X2.13, respectively) were compared. The X2.13 and X2 minigenes were identical; however, X2.13 lacks the SB1 region. Consistent with previous reports, transcripts from X2 were spliced more efficiently at the Bcl-xL splice site than those expressed from X2.13 (Fig. 2B and C) (55). Increased levels of TCERG1 resulted in a shift toward the Bcl-xS isoform for transcripts derived from X2, thereby reproducing our previous results. In contrast, we observed no effect of increased levels of TCERG1 on the ratio of isoforms produced from X2.13 (Fig. 2B). Similar results were obtained with a TCERG1 knockdown; the amount of the Bcl-xL isoform was increased for transcripts derived from X2, whereas there was no change in the ratio of isoforms produced from X2.13 (Fig. 2C). These results indicate that TCERG1 activates the Bcl-xS 5′ splice site in an SB1-dependent manner.
Fig 2.
Effects of TCERG1 on Bcl-x splicing depend on the SB1 regulatory element. (A) Structure of the Bcl-x gene and X2 and X2.13 (ΔSB1) minigenes. (B and C) Analysis of the effects of SB1 deletion on Bcl-x splice site selection in response to TCERG1 overexpression (OE) (B) and knockdown (C). HEK293T cells were cotransfected with Bcl-X2 or Bcl-X2.13 together with empty vector (-) or TCERG1 expression plasmid. For the RNAi experiments, HEK293T cells were cotransfected with the Bcl-x minigene together with siTCERG1 or control (siEGFP). RT-PCR was performed to analyze the alternatively spliced forms of Bcl-x. The graphs show the densitometric analysis results as the ratio of Bcl-xL to Bcl-xS isoforms from three independent experiments (means ± standard deviations [SD]). *, P < 0.05; **, P < 0.01. A fraction of the cell lysates was analyzed by immunoblotting with the indicated antibodies to detect the TCERG1 and CDK9 proteins. (D) RNA coimmunoprecipitation of the Bcl-x pre-mRNA with antibodies against TCERG1 is dependent on the SB1 region. The X2 or X2.13 transcripts were incubated in HeLa nuclear extracts under splicing conditions and then immunoprecipitated using the indicated antibodies. After five washes, the RNA was precipitated and quantified. The data are presented as the percentage of the bound input (means ± SD). **, P < 0.01. (E) TCERG1 associates with transcripts derived from the endogenous Bcl-x gene. In vivo immunoprecipitation assays were carried out with specific antibodies against TCERG1. Transcripts derived from the endogenous Bcl-x gene were detected by qPCR (see Materials and Methods). The data are presented as percentages of the bound input (means ± SD). **, P < 0.01. (F) Diagrammatic representation of the deleted sequences (in bold) in the Bcl-x minigene tested in the experiments. The numbers below the lines indicate the deleted regions. (G) HEK293T cells were cotransfected with 0.5 μg HIV-2 reporter minigene (X2, wild type; X2.13, carrying a complete deletion of the SB1 element; Δ9, Δ11, Δ13, Δ16, Δ17, Δ23, carrying 10-nucleotide deletions in the SB1 element) together with 0.5 μg empty vector (-) or 0.5 μg TCERG1 expression vector (+). RT-PCR was performed to analyze the alternatively spliced forms of Bcl-x. The bar graph shows the densitometric analysis results as the ratio of Bcl-xL to Bcl-xS isoforms from three independent experiments. (H) HEK293T cells were cotransfected with 0.5 μg of the indicated Bcl-x minigenes together with siRNAs against enhanced green fluorescent protein (siEGFP) or TCERG1 (siTCERG1). RT-PCR was performed to determine alternatively spliced forms of Bcl-x. The bar graph shows the densitometric analysis results as the ratio of Bcl-xL to Bcl-xS isoforms from three independent experiments. (I) RNA coimmunoprecipitation was carried out using the Δ12 and Δ23 transcripts as described in the legend for panel D. Data are presented as percentages of the bound input (means ± SD). *, P < 0.05; **, P < 0.01.
To analyze the interaction of TCERG1 with Bcl-x pre-mRNA, we performed RNA immunoprecipitation assays using S2 and S2.13 as templates for the synthesis of transcripts in vitro (see Materials and Methods). The radiolabeled transcripts were incubated in nuclear extracts under in vitro splicing conditions, and specific antibodies were added. The results of this assay revealed that TCERG1 binds to S2 but not to S2.13 (Fig. 2D), which is consistent with the data presented above. The ribonucleoprotein hnRNP K, which regulates Bcl-x splicing and is known to interact in vitro with S2 and S2.13 transcripts (54), was used as a positive control in the experiment (Fig. 2D). To lend additional support to our data, we investigated whether TCERG1 associated with Bcl-x transcripts derived from the endogenous pre-mRNA and the minigenes. We performed in vivo RNA immunoprecipitation assays with anti-TCERG1-specific antibodies and assessed TCERG1 binding by RT-PCR. We found a specific association between TCERG1 and transcripts derived from the endogenous Bcl-x pre-mRNA (Fig. 2E) and transcripts derived from the minigenes (see Fig. S1 in the supplemental material).
To further delineate the TCERG1-responsive sequences within the SB1 regulatory element, we used Bcl-x constructs that each contained 10-nucleotide deletions along the SB1 element. The splicing patterns of all of the mutant constructs, except the Δ23 mutant, were affected by TCERG1 overexpression (Fig. 2G). However, the reduction of endogenous TCERG1 expression by siRNA only slightly increased the expression of the Bcl-xL isoform for transcripts derived from the Δ23 mutant construct (Fig. 2H). In the RNAi experiments, the change in the alternative splicing pattern of Δ23 was less evident than for the X2 and Δ17 mutant constructs, probably because the deletion of region 23 greatly diminishes the use of the Bcl-xL 5′ splice site. RNA immunoprecipitation assays showed reduced binding of TCERG1 to the Δ23 transcript (Fig. 2I), and these results are consistent with the previous RT-PCR data. Collectively, these results suggest that TCERG1 regulates the alternative splicing of Bcl-x through region 23 in the SB1 element.
Recently, the existence of an SB1 splicing repressor that regulates the alternative splicing of Bcl-x was suggested. In the proposed model, a balance between protein synthesis and proteasome-mediated protein degradation controls the level of this putative Bcl-x splicing repressor (59). Blocking proteasome-mediated protein degradation by using proteasome inhibitors (such as MG132 or bortezomib), which alone have no impact on Bcl-x splicing, might shift the equilibrium toward the accumulation of the repressor that binds to SB1 and downregulate the production of Bcl-xS (59). Interestingly, treatment with MG132 (see Fig. S2 in the supplemental material) or bortezomib (unpublished observations) suppressed the TCERG1-mediated increase in the expression of the Bcl-xS isoform. The shift in the ratio of Bcl-xS to Bcl-xL was not due to inadequate expression of TCERG1, because the amount of protein was nearly identical under all conditions (see Fig. S2).
TCERG1 modulates the distribution of RNAPII on Bcl-x exon 2.
An analysis of the sequences present in region 23 of the SB1 regulatory element revealed a high GC content. Transcripts with GC-rich regions can potentially form stem-and-loop structures or stabilize nucleosomes that can attenuate RNAPII elongation (25, 53). Thus, a high GC content may make RNAPII elongation difficult in vivo (9, 65). To assess whether the sequences within SB1 that confer TCERG1 responsiveness contained a higher level of polymerase molecules, we analyzed RNAPII occupancy by ChIP-sequences (ChIP-Seq). The clear accumulation of an RNAPII signal within region 23 (Fig. 3A) is consistent with the existence of an RNAPII pausing site at this position.
To test whether TCERG1 modified the distribution of polymerase at the SB1 site, therefore linking splice site selection with transcriptional elongation, we measured the density of RNAPII at three regions of the endogenous Bcl-x gene by quantitative ChIP analysis. We designed specific primers to detect polymerase recruitment at the promoter (P), SB1 (E2), and distal (D) regions of the gene (Fig. 3B) under conditions of TCERG1 overexpression/knockdown. Increased levels of TCERG1 resulted in a decreased accumulation of polymerase molecules at the E2 position (Fig. 3C). Conversely, TCERG1 depletion resulted in an accumulation of polymerases at the same position (Fig. 3D). The recruitment of RNAPII at the promoter region remained unaltered, whereas a shift in the amount of polymerase at the distal site was observed (Fig. 3C and D). We also measured the density of TCERG1 along the endogenous Bcl-x gene by using specific primers to detect recruitment at the promoter (P), intron-exon junctions (I1-E2 and E2-I2 [Fig. 3B]), and distal (D) regions of the gene. The recruitment of RNAPII was also assessed. The ChIP experiments revealed a specific association of TCERG1 at these regions of the endogenous Bcl-x gene (Fig. 3E). These results indicate that TCERG1 is able to modify RNAPII distribution at the SB1 element and suggest that this modification affects the dynamics of transcript elongation.
TCERG1 modifies the behavior of a slow RNAPII for alternative splicing of Bcl-x.
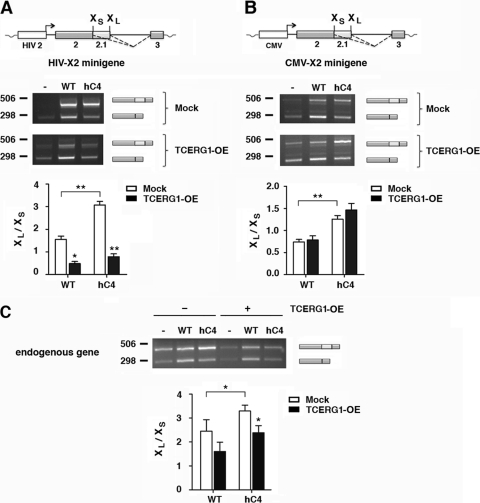
The RNAPII elongation rate controls alternative splicing in human cells. Previous studies (16) have provided direct proof for the involvement of the elongation mechanism in the transcriptional control of alternative splicing by using a mutant form of RNAPII (hC4), which has a decreased elongation rate (10, 12). To further understand the control of alternative splicing by TCERG1, we carried out in vivo splicing assays with human cells cotransfected with a Bcl-x reporter minigene and vectors expressing the α-amanitin-resistant wild-type polymerase and the α-amanitin-resistant hC4 variant. After α-amanitin treatment, most of the endogenous RNAPII was inhibited, and its transcript was made by the resistant mutant variant (21) (see Fig. S3 in the supplemental material). The slow polymerase increased the formation of the Bcl-xL isoform in comparison with the wild-type mutant form of RNAPII, thus favoring the use of the downstream 5′ splice site (Fig. 4A). Remarkably, increasing the level of TCERG1 resulted in a shift toward the Bcl-xS isoform for transcripts derived from HIV-X2 (Fig. 4A), but it did not cause changes for transcripts derived from CMV-X2 (Fig. 4B). The effects on Bcl-x alternative splicing of transcription by the slow hC4 and TCERG1 recapitulated those for the endogenous gene (Fig. 4C). These results support the hypothesis of a transcriptional effect in the control of Bcl-x alternative splicing by TCERG1.
Fig 4.
A slow polymerase (hC4) favors the use of the Bcl-xL splice site, and overexpression of TCERG1 promotes activation of the Bcl-xS splice site. (A) The HIV-X2 minigene was cotransfected into HEK293T cells with empty vector (-) or plasmids containing the α-amanitin-resistant wild-type (WT) or slow (hC4) RNAPII genes. In each case, the cells were also transfected with either the empty vector (mock) or the TCERG1 expression vector and treated with α-amanitin. The alternative splicing was assessed by RT-PCR as described in Materials and Methods. The data are presented as the ratios of Bcl-xL to Bcl-xS from three independent experiments (means ± standard deviations [SD]). *, P < 0.05; **, P < 0.01. (B) The same experiment described for panel A was carried out with the CMV-X2 minigene. The data are presented as the ratio of Bcl-xL to Bcl-xS from three independent experiments (means ± SD). **, P < 0.01. (C) The same experiment described for panel A was carried out in the absence of minigenes. The effect on the Bcl-x endogenous gene was assessed by RT-PCR as described in Materials and Methods. The data are presented as the ratio of Bcl-xL to Bcl-xS from three independent experiments (means ± SD). *, P = 0.0399 (WT-mock versus hC4-mock); P = 0.038 for hC4-mock versus hC4-TCERG1; P = 0.054 for WT-mock versus WT-TCERG1.
The CTD of the largest subunit of RNAPII may contribute to transcription-splicing coupling (38). For example, it has been reported that the serine/arginine-rich (SR) protein SRp20 requires the CTD to inhibit fibronectin EDI exon inclusion (15). Given that TCERG1 binds to hyperphosphorylated CTD (7) and that our results suggest a link between transcriptional elongation and the effect of TCERG1 on alternative splicing, we sought to assess the role of the RNAPII CTD on TCERG1-mediated alternative splicing. We transfected plasmids expressing α-amanitin-resistant polymerases containing a wild-type CTD, a completely deleted CTD (ΔCTD), or either the amino- or carboxyl-terminal half of the CTD (amino acids 1 to 25 and 27 to 52, respectively), which contains the 10-residue carboxyl-terminal motif (Cter) that is crucial for RNAPII stability (8). The CTD is necessary to transcribe endogenous genes (39); however, reporter gene constructs have proven to be useful for study of the role of the CTD and the mechanism by which transcription controls alternative splicing (15). An inappropriate assembly of chromatin onto these expressed reporters seems to be the reason why these transiently transfected constructs are transcribed by ΔCTD polymerases. Interestingly, the alternative splicing of Bcl-x occurred in a similar pattern as the α-amanitin-resistant RNAPII wild type (see Fig. S4 in the supplemental material), under conditions in which the endogenous polymerase was mostly inhibited (see Fig. S3). Next, we tested the effect of those mutants on splicing upon TCERG1 overexpression. The analysis revealed that both halves of the CTD were equally effective in mediating the usage of the Bcl-xS 5′ splice site (see Fig. S4). The complete deletion of the CTD seemed to abrogate the effect of TCERG1 on Bcl-x splicing (see Fig. S4), although formally we cannot exclude that the mRNAs present under these conditions are stable mRNAs transcribed by the endogenous polymerase during the first 24 h in the absence of the drug. These results suggest that the effect of TCERG1 on Bcl-x alternative splicing is independent of the length and the heptad composition of the CTD. These results further suggest the involvement of elongation control in the mechanism of splice site selection elicited by TCERG1.
TCERG1 increases the rate of RNAPII transcription in vivo.
Our results implicate an elongation mechanism in the transcriptional control of Bcl-x alternative splicing and suggest a role for TCERG1 in this process. One possible explanation for the influence of TCERG1 on Bcl-x alternative splicing is that TCERG1 directly affects the elongation rate of RNAPII. To test this possibility, we calculated the amount of pre-mRNAs generated at distal regions of the endogenous Bcl-x gene with respect to the transcription start site under conditions of TCERG1 overexpression/knockdown. The cDNA was synthesized using random hexamers followed by quantitative PCR using primers that span the junction between intron/exon 3, thus corresponding to unspliced pre-mRNAs (Fig. 5A). We observed increased levels of distal transcripts upon TCERG1 overexpression and a diminished accumulation of distal transcripts upon TCERG1 knockdown (Fig. 5B). If TCERG1 increases elongation, a prediction is that the increase in transcription rates would be more noticeable beyond the region where TCERG1 releases the RNAPII pause than before the pause. We tested this by measuring the amount of RNAs generated at a region encompassing the SB1 element. We observed similar levels of transcripts upon TCERG1 overexpression/knockdown (see Fig. S5 in the supplemental material). These results imply that TCERG1 affects the transcription of Bcl-x, and they further suggest that TCERG1 acts through an elongation mechanism.
To directly test whether TCERG1 affects the rate of RNAPII transcription, we used Padgett's protocol (60). In this elegant procedure, DRB, which reversibly blocks gene transcription in vivo, is used in combination with quantitative RT-PCR to analyze the transcription and splicing of endogenous human genes (60). Control and TCERG1-depleted cells were treated with DRB, and samples were collected at different time points after removal of DRB. We performed quantitative RT-PCR using primers spanning the intron-exon 3 junction to detect pre-mRNA expression. The DRB-treated control cells were able to recover transcription of the exon 3 region of the Bcl-x gene within 30 to 80 min after drug release (Fig. 5C), which is consistent with a transcriptional lag due to the genomic distance from the start site of transcription (60). In contrast, the recovery of transcription was significantly slower in cells in which the expression of TCERG1 had been knocked down. These results demonstrate an active role for TCERG1 in RNAPII elongation in vivo.
DISCUSSION
The results presented here show that TCERG1 modulates the alternative splicing of Bcl-x, a key gene in the control of apoptosis, a process to which TCERG1 has been previously linked (61, 68). TCERG1 regulates the alternative splicing of Bcl-x exon 2 by favoring the use of the most upstream of the two alternative 5′ splice sites that compete for a common 3′ splice site. The proposed mechanism for this effect is based on the elongation rate of RNAPII. In this study, we provided direct proof for the elongation mechanism in the transcriptional control of alternative splicing in human cells (29).
Our results support the kinetic coupling model that was first suggested by Eperon and colleagues to explain the mechanism of transcriptional control of alternative splicing; they found that the rate of RNA synthesis could affect its secondary structure and, in turn, affect alternative splicing (17). This type of regulation was also suggested from experiments in which the insertion of a polymerase pause site downstream of weak alternative splice sites induced alternative splicing by delaying the synthesis of an inhibitory element (56). More recently, seminal work by Kornblihtt and coworkers provided evidence for the functional link between RNAPII elongation and the control of alternative splicing (15, 16, 27, 48). In the experiments by Kornblihtt and coworkers, the dynamics of transcription elongation affected downstream splicing decisions by regulating the accessibility of cis elements that control alternative splice sites. Thus, low RNAPII processivity or internal pause signals for elongation would favor the inclusion of an alternative exon, although its 3′ splice site is weak. A highly active elongating polymerase or the absence of pause sites would favor the simultaneous presentation of competing weak/strong 3′ splicing sites, leading to exon exclusion (29).
Our results offer a new kinetic mechanism to explain the transcriptional control of the alternative splice site selection in the apoptotic regulator Bcl-x. Shkreta et al. (59) recently reported that a splicing repressor present at a nearly limiting concentration bound to SB1 and downregulated the production of Bcl-xS. The identity of this putative repressor remains unknown. We hypothesize that SB1-mediated repression of Bcl-xS splicing occurs because the polymerase pauses or slows down near region 23. A slow elongation rate or pause would allow more time for recruitment of the putative repressor to the SB1 element, favoring the production of the Bcl-xL isoform. Based on our results, we propose the existence of a transcriptional checkpoint near region 23 at the SB1 element where RNAPII transiently pauses. The deletion of this region would change the splicing pattern in favor of Bcl-xS due to the absence of the polymerase pausing site; failure of RNAPII to stall would give less time for the repressor to bind to SB1 and block the use of the Bcl-xS 5′ splice site. TCERG1, by stimulating the transcriptional elongation of the Bcl-x gene, would also offer less time for the assembly and activity of a functional repressor at SB1, thus favoring the production of the Bcl-xS isoform. Using high-resolution analysis of transcription and splicing, the recent discovery of cotranscriptional splicing regulated by RNAPII pausing (1, 6) is consistent with our findings. Whether the transcriptional checkpoint at the SB1 element is associated with the phosphorylation of Ser5 (1) and whether phosphorylation of Ser2 in the RNAPII CTD occurs by the action of TCERG1 remain to be investigated.
Using fluorescence recovery after photobleaching (FRAP), Muñoz et al. (45) showed that UV irradiation inhibited transcriptional elongation in vivo and in real time. UV irradiation affected the splicing pattern of endogenous Bcl-x by increasing the formation of the Bcl-xS isoform, a result that we have reproduced (unpublished data). This discrepancy with the results described here using the endogenous and Bcl-x minigenes might be associated with UV treatment. UV irradiation might affect the stability or nuclear distribution of other splicing regulators involved in Bcl-x alternative splicing. Although no change in the localization pattern of some splicing factors was observed, the size and behavior of nuclear speckles were clearly affected by UV treatment (45). The molecular responses elicited by UV irradiation may also vary depending on the type of cells. Moreover, although UV may affect Bcl-x alternative splicing by inhibiting transcription elongation, the contribution of other splicing regulatory pathways linked to DNA damage (59) and different pausing sites may explain why UV increases the production of Bcl-xS.
Transcription by RNAPII, modification of histones, and processing of pre-mRNAs are thought to be integrated processes for the generation of mature mRNAs (36). It will be of interest to address whether TCERG1 affects Bcl-x alternative splicing through changes in histone modification patterns that affect transcriptional elongation rates. In a recent report, siRNAs triggered histone silencing and RNAPII stalling through a mechanism called transcriptional gene silencing (TGS) (2). Microarray analyses (reference 51 and our unpublished data) identified genes that are affected by changes in TCERG1 levels and that are also regulated by microRNAs. Based on those observations, it is an exciting possibility that TCERG1 affects RNAPII processivity and alternative splicing via a mechanism that may also be involving microRNA processing.
Our results show that TCERG1 stimulates the rate of RNAPII elongation and that its overexpression elicits a shift toward the Bcl-xS proapoptotic splice variant. Recent work suggests that pausing could act as checkpoint to ensure optimal activity of RNAPII during development, especially in large highly regulated genes (22, 30). Insufficient time at the pausing site could therefore result in an RNAPII more prone to make mistakes. Thus, a pausing site in Bcl-x may provide the cell with an opportunity to test its pausing machinery; an incapacity to pause efficiently would trigger the production of Bcl-xS and induce apoptosis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Spanish Ministry of Science and Innovation (BFU2008-01599), the Fundación para la Investigación y Prevención del SIDA en España (36768), the Junta de Andalucía (Proyecto de Excelencia 2009/CVI-4626) to C.S., and the Spanish Ministry of Science and Innovation (BFU2009-08796) to C.H.M. Support from the Fondo Europeo de Desarrollo Regional (FEDER) is also acknowledged. Work in the laboratory of B.C. was supported by a grant from the Canadian Institutes of Health Research. B.C. is the Canada Research Chair in Functional Genomics. M.M. was supported by a fellowship from the Spanish Ministry of Education (FPU program). N.S.-H. was supported by a fellowship from the CSIC (JAE program). A.C. holds a scholarship from the FRSQ.
We are grateful to the members of the laboratory for their helpful suggestions, critical discussions, and comments. We thank A. R. Kornblihtt, D. Bentley, and G. Dreyfuss for providing reagents.
Footnotes
Published ahead of print 12 December 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Alexander RD, Innocente SA, Barrass JD, Beggs JD. 2010. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell 40:582–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allo M, et al. 2009. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 16:717–724 [DOI] [PubMed] [Google Scholar]
- 3. Boise LH, et al. 1993. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74:597–608 [DOI] [PubMed] [Google Scholar]
- 4. Bourquin JP, et al. 1997. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 25:2055–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caceres JF, Kornblihtt AR. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18:186–193 [DOI] [PubMed] [Google Scholar]
- 6. Carrillo Oesterreich F, Preibisch S, Neugebauer KM. 2010. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell 40:571–581 [DOI] [PubMed] [Google Scholar]
- 7. Carty SM, Goldstrohm AC, Suñé C, Garcia-Blanco MA, Greenleaf AL. 2000. Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 97:9015–9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chapman RD, Palancade B, Lang A, Bensaude O, Eick D. 2004. The last CTD repeat of the mammalian RNA polymerase II large subunit is important for its stability. Nucleic Acids Res. 32:35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chavez S, Garcia-Rubio M, Prado F, Aguilera A. 2001. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:7054–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y, Chafin D, Price DH, Greenleaf AL. 1996. Drosophila RNA polymerase II mutants that affect transcription elongation. J. Biol. Chem. 271:5993–5999 [PubMed] [Google Scholar]
- 11. Cheng D, Cote J, Shaaban S, Bedford MT. 2007. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25:71–83 [DOI] [PubMed] [Google Scholar]
- 12. Coulter DE, Greenleaf AL. 1985. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J. Biol. Chem. 260:13190–13198 [PubMed] [Google Scholar]
- 13. Damgaard CK, et al. 2008. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol. Cell 29:271–278 [DOI] [PubMed] [Google Scholar]
- 14. Deckert J, et al. 2006. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26:5528–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de la Mata M, Kornblihtt AR. 2006. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat. Struct. Mol. Biol. 13:973–980 [DOI] [PubMed] [Google Scholar]
- 16. de la Mata M, et al. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12:525–532 [DOI] [PubMed] [Google Scholar]
- 17. Eperon LP, Graham IR, Griffiths AD, Eperon IC. 1988. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell 54:393–401 [DOI] [PubMed] [Google Scholar]
- 18. Furger A, O'Sullivan JM, Binnie A, Lee BA, Proudfoot NJ. 2002. Promoter proximal splice sites enhance transcription. Genes Dev. 16:2792–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garcia-Blanco MA, Baraniak AP, Lasda EL. 2004. Alternative splicing in disease and therapy. Nat. Biotechnol. 22:535–546 [DOI] [PubMed] [Google Scholar]
- 20. Garneau D, Revil T, Fisette JF, Chabot B. 2005. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 280:22641–22650 [DOI] [PubMed] [Google Scholar]
- 21. Gerber HP, et al. 1995. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature 374:660–662 [DOI] [PubMed] [Google Scholar]
- 22. Gilchrist DA, et al. 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143:540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goldstrohm AC, Albrecht TR, Suñé C, Bedford MT, Garcia-Blanco MA. 2001. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 21:7617–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldstrohm AC, Greenleaf AL, Garcia-Blanco MA. 2001. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene 277:31–47 [DOI] [PubMed] [Google Scholar]
- 25. Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. 2008. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A. 105:7762–7767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Howe KJ, Kane CM, Ares M., Jr 2003. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA 9:993–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kadener S, et al. 2001. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J. 20:5759–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim E, Du L, Bregman DB, Warren SL. 1997. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell Biol. 136:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kornblihtt AR. 2007. Coupling transcription and alternative splicing. Adv. Exp. Med. Biol. 623:175–189 [DOI] [PubMed] [Google Scholar]
- 30. Levine M. 2011. Paused RNA polymerase II as a developmental checkpoint. Cell 145:502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, et al. 2009. The sequence alignment/map format and SAM tools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Licatalosi DD, Darnell RB. 2006. Splicing regulation in neurologic disease. Neuron 52:93–101 [DOI] [PubMed] [Google Scholar]
- 33. Lin KT, Lu RM, Tarn WY. 2004. The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol. Cell. Biol. 24:9176–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 15:819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Listerman I, Sapra AK, Neugebauer KM. 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 13:815–822 [DOI] [PubMed] [Google Scholar]
- 36. Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144:16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Makarov EM, et al. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205–2208 [DOI] [PubMed] [Google Scholar]
- 38. McCracken S, et al. 1997a. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357–361 [DOI] [PubMed] [Google Scholar]
- 39. Meininghaus M, Chapman RD, Horndasch M, Eick D. 2000. Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxyl-terminal domain of the large subunit affects early steps in transcription. J. Biol. Chem. 275:24375–24382 [DOI] [PubMed] [Google Scholar]
- 40. Misteli T, Spector DL. 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3:697–705 [DOI] [PubMed] [Google Scholar]
- 41. Moore MJ, Proudfoot NJ. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136:688–700 [DOI] [PubMed] [Google Scholar]
- 42. Morris DP, Greenleaf AL. 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275:39935–39943 [DOI] [PubMed] [Google Scholar]
- 43. Mortillaro MJ, et al. 1996. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. U. S. A. 93:8253–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Munoz MJ, de la Mata M, Kornblihtt AR. 2010. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem. Sci. 35:497–504 [DOI] [PubMed] [Google Scholar]
- 45. Munoz MJ, et al. 2009. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 137:708–720 [DOI] [PubMed] [Google Scholar]
- 46. Nasim FU, Hutchison S, Cordeau M, Chabot B. 2002. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA. 8:1078–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Neubauer G, et al. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20:46–50 [DOI] [PubMed] [Google Scholar]
- 48. Nogues G, Kadener S, Cramer P, Bentley D, Kornblihtt AR. 2002. Transcriptional activators differ in their abilities to control alternative splicing. J. Biol. Chem. 277:43110–43114 [DOI] [PubMed] [Google Scholar]
- 49. Oesterreich FC, Bieberstein N, Neugebauer KM. 2011. Pause locally, splice globally. Trends Cell. Biol. 21:328–335 [DOI] [PubMed] [Google Scholar]
- 50. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40:1413–1415 [DOI] [PubMed] [Google Scholar]
- 51. Pearson JL, Robinson TJ, Munoz MJ, Kornblihtt AR, Garcia-Blanco MA. 2008. Identification of the cellular targets of the transcription factor TCERG1 reveals a prevalent role in mRNA processing. J. Biol. Chem. 283:7949–7961 [DOI] [PubMed] [Google Scholar]
- 52. Rappsilber J, Ryder U, Lamond AI, Mann M. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12:1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Resnekov O, Kessler M, Aloni Y. 1989. RNA secondary structure is an integral part of the in vitro mechanism of attenuation in simian virus 40. J. Biol. Chem. 264:9953–9959 [PubMed] [Google Scholar]
- 54. Revil T, Pelletier J, Toutant J, Cloutier A, Chabot B. 2009. Heterogeneous nuclear ribonucleoprotein K represses the production of pro-apoptotic Bcl-xS splice isoform. J. Biol. Chem. 284:21458–21467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Revil T, et al. 2007. Protein kinase C-dependent control of Bcl-x alternative splicing. Mol. Cell. Biol. 27:8431–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW. 1998. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 26:5568–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sánchez-Alvarez M, Goldstrohm AC, Garcia-Blanco MA, Suñé C. 2006. Human transcription elongation factor CA150 localizes to splicing factor-rich nuclear speckles and assembles transcription and splicing components into complexes through its amino and carboxyl regions. Mol. Cell. Biol. 26:4998–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sánchez-Alvarez M, Montes M, Sánchez-Hernández N, Hernández-Munain C, SuÑé C. 2010. Differential effects of sumoylation on transcription and alternative splicing by transcription elongation regulator 1 (TCERG1). J. Biol. Chem. 285:15220–15233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shkreta L, Michelle L, Toutant J, Tremblay ML, Chabot B. 2011. The DNA damage response pathway regulates the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 286:331–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Singh J, Padgett RA. 2009. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 16:1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smith MJ, Kulkarni S, Pawson T. 2004. FF domains of CA150 bind transcription and splicing factors through multiple weak interactions. Mol. Cell. Biol. 24:9274–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Suñé C, Garcia-Blanco MA. 1999. Transcriptional cofactor CA150 regulates RNA polymerase II elongation in a TATA-box-dependent manner. Mol. Cell. Biol. 19:4719–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Suñé C, et al. 1997. CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 17:6029–6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Svotelis A, Gevry N, Gaudreau L. 2009. Chromatin immunoprecipitation in mammalian cells. Methods Mol. Biol. 543:243–251 [DOI] [PubMed] [Google Scholar]
- 65. Tous C, et al. 2011. A novel assay identifies transcript elongation roles for the Nup84 complex and RNA processing factors. EMBO J. 30:1953–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang ET, et al. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang GS, Cooper TA. 2007. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 8:749–761 [DOI] [PubMed] [Google Scholar]
- 68. Wang KC, Cheng AL, Chuang SE, Hsu HC, Su IJ. 2000. Retinoic acid-induced apoptotic pathway in T-cell lymphoma: Identification of four groups of genes with differential biological functions. Exp. Hematol. 28:1441–1450 [DOI] [PubMed] [Google Scholar]
- 69. Wilhelm BT, et al. 2011. Differential patterns of intronic and exonic DNA regions with respect to RNA polymerase II occupancy, nucleosome density and H3K36me3 marking in fission yeast. Genome Biol. 12:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang X, et al. 1994. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl. Acad. Sci. U. S. A. 91:6924–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yuryev A, et al. 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. U. S. A. 93:6975–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.