Abstract
HIV-1 Vpr-binding protein (VprBP) has been implicated in the regulation of both DNA replication and cell cycle progression, but its precise role remains unclear. Here we report that VprBP regulates the p53-induced transcription and apoptotic pathway. VprBP is recruited to p53-responsive promoters and suppresses p53 transactivation in the absence of stress stimuli. To maintain target promoters in an inactive state, VprBP stably binds to nucleosomes by recognizing unacetylated H3 tails. Promoter-localized deacetylation of H3 tails is a prerequisite for VprBP to tether and act as a bona fide inhibitor at p53 target genes. VprBP knockdown leads to activation of p53 target genes and causes an increase in DNA damage-induced apoptosis. Moreover, phosphorylation of VprBP at serine 895 impairs the ability of VprBP to bind H3 tails and to repress p53 transactivation. Our results thus reveal a new role for VprBP in regulation of the p53 signaling pathway, as well as molecular mechanisms of cancer development related to VprBP misregulation.
INTRODUCTION
VprBP was first identified as a protein that can interact with HIV-1 viral protein R by coimmunoprecipitation assays (37). VprBP is a 1,507-amino-acid protein that contains conserved domains, including YXXY repeats, the Lis homology motif, and WD40 repeats. Despite the lack of molecular characterization of VprBP, recent studies suggest that VprBP can specifically associate with DDB1 to act as a substrate recognition subunit of the CUL4-DDB1 ubiquitin E3 ligase complex (12, 20, 26, 33, 36, 38). Through binding to Vpr, VprBP allows Vpr to modulate the intrinsic catalytic activity of the CUL4-DDB1 complex, which in turn leads to the induction of G2 phase cell cycle arrest in the virus-infected cells. The direct interaction of tumor suppressor Merlin with VprBP is shown to be an integral part of the mechanism by which Merlin inhibits CUL4-DDB1 ubiquitin E3 ligase to suppress tumorigenesis (22). Furthermore, the observation that VprBP-depleted cells activate DNA damage checkpoints and increase the cellular level of CDK inhibitor p21 suggests that VprBP is involved in the control of cell cycle arrest and apoptosis (11).
p53 is an important tumor suppressor which induces either cell cycle arrest or apoptosis in response to DNA damage (27, 30, 34). p53 regulates these processes mainly by acting as a sequence-specific DNA binding factor that regulates transcription of a number of target genes. p53 regulates the transcription reaction, to a large extent, at the level of chromatin, which establishes a physical barrier for the binding of transcription factors to the promoter region of a target gene. The most dynamic parts of chromatin are amino-terminal domains (called histone “tails”) of core histones, which protrude from the DNA. The major contributions of individual histone tails in gene transcription are made through their posttranslational modifications (3, 18, 21, 29, 35). Among various modifications, histone acetylation has been implicated as a critical mark for activation of p53 target genes (1, 5, 7, 10, 13). While acetylation of all four histone tails has been linked to active transcription, there is an emerging body of evidence to support that acetylation of H3 and H4 tails is particularly important for transcriptional activation of p53 target genes (1, 5, 7, 10, 13, 23). When cells are exposed to stress conditions, p53 recruits histone acetyltransferases (HATs) to establish distinct histone acetylation at its target gene promoters, which will in turn allow the transcriptional machinery to initiate the high level of transcription. Because histone acetylation is actively regulated by a competitive action of HAT and histone deacetylase (HDAC) (15, 25, 31, 32), the deregulation of this chromatin-remodeling process can lead to aberrant repression of p53 target genes. Given this reversible nature of histone acetylation, cells need to employ additional factors that can recognize and lock in a distinct (de)acetylation status of promoter nucleosomes. In relation to the present study, the cellular depletion of VprBP leads to the increased expression of the p53 target gene p21 (11). These results raise questions about whether VprBP is able to downregulate p53-mediated transcription and, if so, how this would affect cellular responses to DNA damage.
In this study, we demonstrate that VprBP is recruited to promoters by p53 and attenuates p53-dependent transcription. This occurs through VprBP interaction with histone H3 tails and inhibition of their acetylation at promoter regions. HDAC1-mediated deacetylation of H3 tails contributes to the stable localization of VprBP at p53 target promoters. VprBP is overexpressed in three types of cancer cell lines, and RNA interference (RNAi) against VprBP augments DNA damage-induced apoptotic cell death. Furthermore, VprBP phosphorylation by DNA-activated protein kinase (DNA-PK) inhibits its interaction with promoter nucleosomes and reactivates p53 target genes. Together, these results reveal a hitherto-unknown role of VprBP in repressing p53-dependent transcription and a distinct regulatory mechanism governing VprBP function under stress conditions.
MATERIALS AND METHODS
Cell culture and constructs.
U2OS, 293T, LD611, and MCF7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). MCF10-2A cells were grown in a 1:1 mixture of DMEM and DMEM-F12 supplemented with 20 ng/ml epidermal growth factor, 100 ng/ml cholera toxin, 0.01 mg/ml insulin, 500 ng/ml hydrocortisone, and 5% horse serum. Urotsa cells were grown in DMEM (low glucose) containing 10% FBS. MLC cells were grown in T medium containing 10% FBS. LNCaP cells were grown in RPMI-1640 with 10% FBS. Wild-type and VprBPflox/− MEF cells were propagated in DMEM supplemented with 10% FBS as previously described (26).
The p53ML601-14 plasmid was constructed as recently described (17). To generate the glutathione S-transferase (GST)-p53 and GST-HDAC1 constructs, the corresponding cDNAs were amplified by PCR and inserted into the BamHI and EcoRI sites of pGEX-4T1. Bacterial expression constructs of VprBP deletion mutants were generated by PCR tagging of the corresponding cDNA and subcloning them into the pET-15b or pET-11d vector in frame with 5′ hexa-His or Flag sequences, respectively. For mammalian expression of HDAC1 and p53, the corresponding cDNAs were amplified by PCR and ligated into the correct reading frame of pIRES containing the 5′ Flag coding sequence or pCDNA3.1/His. Further details of plasmid construction are available upon request. Expression vectors for core histones, Flag-p53, and GST-histone tails were as described previously (1, 2, 24).
Antibodies.
Rabbit polyclonal antibody against S895-phosphorylated VprBP was generated by using the VTAAApSPVSLPRKKC peptide as an immunogen. Bleeds were passed three times through a nonphospho peptide column to remove antibody that recognizes the peptide without phosphorylation. The final flowthrough was loaded on a phosphopeptide column, which was washed three times with 0.1 M Tris buffer (pH 8.0). Bound antibody was eluted with 0.1 M glycine (pH 2.5) and neutralized with 1 M Tris buffer. For the dot blot assay, serial dilutions of unmodified or phosphorylated peptides were spotted on a polyvinylidene difluoride (PVDF) membrane and blotted with the phospho-S895 antibody. For peptide competition assays, the antibody was preincubated with unmodified or phosphorylated peptides at room temperature for 60 min before immunoblotting. Other antibodies used in this study are as follows: anti-H3, anti-acetyl H3, anti-H4, anti-acetyl H4, and anti-Myc antibodies (Abcam); anti-VprBP antibody (Proteintech Group); anti-HDAC1 antibody (Active Motif); anti-His antibody (Novagen); anti-p53 antibody (Santa Cruz Biotech); and anti-Flag antibody (Sigma).
Preparation of recombinant proteins and H3 tail peptides.
Recombinant histones were expressed in Escherichia coli Rosetta 2(DE3)pLysS cells (Novagen) and purified as described previously (1, 9). GST-fused proteins were expressed in E. coli Rosetta 2(DE3)pLysS cells and purified on glutathione-Sepharose 4B beads as described previously (16). Bacterially expressed His-tagged and Flag-tagged VprBP fragments were initially purified with Ni-nitrilotriacetic acid (NTA) and M2 agarose, respectively, and further purified with Q Sepharose and SP-HP columns according to standard procedures. VprBP and p300 proteins were expressed as His-tagged proteins in insect (Sf9) cells using a baculovirus vector and purified by Ni-NTA affinity chromatography. The peptides corresponding to the N-terminal tail of H3 (amino acids 1 to 28) were synthesized by Genemed Synthesis Inc. (South San Francisco, CA) by solid-phase Fmoc/tBu chemistry using an automated peptide synthesizer. The synthesized peptides were purified by reverse-phase high-performance liquid chromatography (RP-HPLC) on a Zorbax SB300 C8 column (9.4 mm by 25 cm) using a water (0.1% trifluoroacetic acid [TFA])-to-acetonitrile (0.1% TFA) gradient, and peptide purity was confirmed using electrospray ionization mass spectrometry (ES-MS) and amino acid analysis.
Reconstitution of nucleosome arrays and mononucleosomes.
For nucleosome array reconstitution, the p53ML601-14 plasmid was digested with EcoRI and HindIII, and the 3.4-kb p53ML-601 array DNA fragment was gel purified. For mononucleosome reconstitution, the 207-bp p53RE DNA fragment containing p53 response elements was PCR amplified from the p53ML601-14 plasmid using a pair of 5′ biotinylated primers. The nucleosome array and mononucleosome were reconstituted by salt gradient dialysis and purified by sedimentation in a 5 to 30% (vol/vol) glycerol gradient (14). Micrococcal nuclease (Sigma) digestion of nucleosome arrays (2 μg) was performed as described previously (1). Reconstituted nucleosome arrays and mononucleosomes were analyzed on a 1% agarose nucleoprotein gel stained with ethidium bromide.
In vitro transcription and modification assays.
In vitro transcription assays were as described previously (16) except that 100 ng of p53ML-601 nucleosome arrays or an equimolar amount of DNA templates was used for each reaction. VprBP (25 or 50 ng) was added before, after, or together with p300 (20 ng) and acetyl-coenzyme A (Ac-CoA) (10 μM). Transcription assays using DNA-PK were as described above, but VprBP was preincubated with DNA-PK (50 ng) and ATP (10 mM) for 60 min. For the chromatin HAT assay, p53ML-601 nucleosome arrays (200 ng) were preincubated with p53 (15 ng) for 20 min and then with p300 (20 ng), Ac-CoA (10 μM), and/or VprBP (25 or 50 ng) for another 60 min. HAT reactions were analyzed by Western blotting with H3, acetyl-H3, H4, and acetyl-H4 antibodies. To identify posttranslational modifications of VprBP, in vitro modification assays were performed using a panel of kinases (DNA-PK, ATM, and ATR), acetyltransferases (p300, Tip60, and PCAF), and methyltransferases (SET7, G9a, and PRMT1), supplemented with [γ-32P]ATP, [3H]Ac-CoA, and S-[3H]adenosylmethionine ([3H]SAM). The reactions were subjected to SDS-PAGE and autoradiography. To determine VprBP phosphorylation sites, DNA-PK phosphorylation reactions were resolved by 15% SDS-PAGE and visualized by Coomassie blue staining. The band corresponding to the VprBP 751–909 fragment was excised from the gel and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
H3 tail- and nucleosome-binding assays.
To analyze the VprBP-tail interaction in vitro, GST-histone tails (2 μg) were immobilized on glutathione-Sepharose beads and incubated with full-length or truncated VprBP (2 μg) for 16 h at 4°C in 750 μl of binding buffer (25 mM Tris-HCl, pH 7.8, 0.2 mM EDTA, 20% glycerol, 200 mM KCl, 0.1% Nonidet P-40). The beads were washed three times with binding buffer, and bound proteins were subjected to SDS-PAGE followed by immunoblotting. For H3 tail peptide binding experiments, His-VprBP (2 μg) was coupled with Ni-NTA agarose beads and incubated with unmodified or acetylated H3 peptides (2 μg) for 16 h at 4°C in the binding buffer. After washing the beads three times, tail peptides bound to the VprBP protein were resolved by 4 to 20% SDS-PAGE and analyzed by silver staining. To study the effect of phosphorylation on VprBP-H3 tail interaction, the wild-type or S895-mutated version of Flag-VprBP 751–1507 was phosphorylated by DNA-PK and incubated with immobilized GST-H3 tails. After extensive washing with washing buffer (25 mM Tris-HCl, pH 7.8, 0.2 mM EDTA, 20% glycerol, 300 mM KCl, 0.1% Nonidet P-40), the bound VprBP protein was subjected to SDS-PAGE and detected by Western blotting using anti-Flag antibody. For nucleosome binding assays, 2 μg DNA equivalents of mononucleosomes were preacetylated by p53 (150 ng) and p300 (200 ng) supplemented with Ac-CoA (10 μM) and immobilized on streptavidin-agarose (Novagen). After the beads were washed to remove p53, p300, and Ac-CoA, His-tagged VprBP was added to mononucleosomes and incubated in 500 μl of binding buffer at 4°C for 16 h. The beads were washed three times with binding buffer, and nucleosome-bound VprBP was detected by Western blotting using the anti-His antibody.
Protein-protein interaction.
For GST pulldown assays, His-tagged VprBP (2 μg) was incubated with GST-p53 (2 μg) or GST-HDAC1 (2 μg) immobilized on glutathione-Sepharose beads in 750 μl of binding buffer for 16 h at 4°C with gentle rotation. After beads were washed three times with binding buffer, bound VprBP was resolved by 8% SDS-PAGE and detected by Western blot analysis using anti-His antibody. Similar pulldown assays were carried out using Flag-p53 (2 μg) and GST-HDAC1 (2 μg), and p53 binding was determined by Western blotting with anti-Flag antibody. For in vivo interaction studies, 293T cells were transiently transfected with expression vectors encoding Myc-VprBP, Flag-p53, Flag-HDAC1, and His-HDAC1. Two days after transfection, cells were solubilized, and the cleared lysates were subjected to immunoprecipitation with anti-Flag antibody. The bound proteins were eluted by boiling in SDS sample buffer and analyzed by Western blot analysis. For coimmunoprecipitation of endogenous proteins, 293T cell lysates (1 mg) were immunoprecipitated using anti-p53 or anti-HDAC1 antibody and immunoblotted with anti-VprBP, anti-HDAC1, or anti-p53 antibody.
qRT-PCR, immunofluorescence, and cell viability and apoptosis assays.
For qRT-PCR analysis of p53 target gene expression, total RNA was isolated using the RNeasy minikit (Qiagen) according to the manufacturer's instructions. Quantitative reverse transcription-PCR (qRT-PCR)was performed using the IQ SYBR green Supermix (Bio-Rad) and the IQ5 real-time cycler (Bio-Rad). Assay results were normalized to β-actin mRNA levels. All reactions were run in triplicate, and data presented are the averages of data from three individual experiments. Sequences of the primers used for qRT-PCR are available upon request. Immunofluorescence was essentially as described previously (16).
The cytotoxicity of etoposide treatment was estimated by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay according to the standard protocol. Briefly, wild-type and VprBPflox/− mouse embryonic fibroblast (MEF) cells were first transfected with Cre recombinase peptides to delete the VprBP gene (28) and treated with or without etoposide (100 μM). After cells were washed with phosphate-buffered saline (PBS), 1 ml of MTT (0.5 mg/ml) was added to cells for 3 h at 37°C. The MTT formazan precipitate was dissolved with 1 ml of MTT solvent, and cellular proliferation was determined from the conversion of MTT to formazan using a microplate reader, model 680 (Bio-Rad) at a wavelength of 570 nm with background subtraction at 650 nm. For the apoptosis assay, LD611, LNCaP, and MCF7 cells were cultured in 100-mm plates and treated with or without etoposide (100 μM) for 24 h. After the treatment, the cells were harvested with trypsin-EDTA, resuspended in the binding buffer, and incubated with fluorescein isothiocyanate (FITC)-conjugated annexin V and propidium iodide (FITC annexin V apoptosis detection kit I; BD Pharmingen), according to the manufacturer's protocol. The numbers of apoptotic cells were monitored with flow cytometry.
ChIP, ChIP and re-ChIP, and RNA interference.
Chromatin immunoprecipitation (ChIP) assays with U2OS cells, either treated or not treated with etoposide, were performed using the ChIP assay kit from Upstate/Millipore according to the manufacturer's protocol and as recently described (16). Antibodies specific to VprBP, H3, acetyl-H3, p53, and HDAC1 were used for immunoprecipitation. ChIP assays with U2OS cells expressing Myc-tagged VprBP were performed 2 days after transfection with or without 8 h of etoposide treatment using anti-Myc antibody. Sequences of the primers used for quantitative real-time PCR (qPCR) are available upon request. All samples were run in triplicate, and results were averaged. The positions of the PCR primers within the promoter regions are shown in Fig. 5. For re-ChIP assays, complexes from the initial ChIP using anti-p53 antibody were eluted with 100 μl of 10 mM DTT at 37°C for 30 min and diluted 1:20 in immunoprecipitation (IP) dilution buffer. The eluates were reimmnunoprecipitated with control IgG or anti-VprBP antibody.
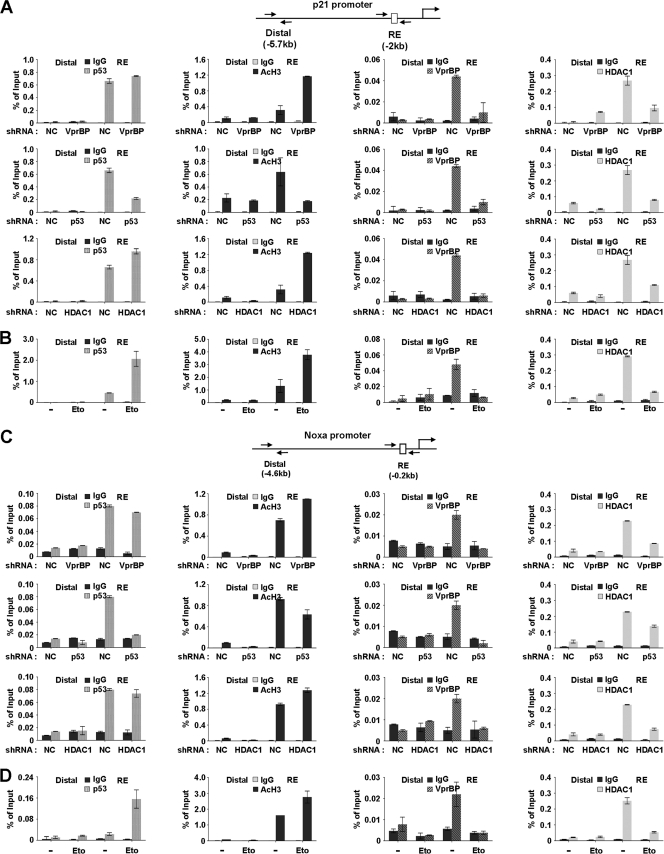
Fig 5.
Dynamics of promoter occupancy of VprBP, p53, and HDAC1. (A) VprBP depletion-promoted H3 acetylation at the p21 promoter. VprBP, p53, and HDAC1 were depleted as for Fig. 4A, and ChIP assays of promoter and distal regions were performed using antibodies specifically recognizing VprBP, p53, HDAC1, and acetyl H3. Input DNA and immunoprecipitated DNA were quantified by qPCR analyses using distal and proximal primer sets. The results are shown as percentages of input, and the error bar indicates the means ± SE. (B) DNA damage-induced dissociation of VprBP. U2OS cells were treated with or without etoposide (100 μM) for 8 h and then analyzed by ChIP analysis of p21 promoter as described for panel A. (C) VprBP depletion-promoted H3 acetylation at the Noxa promoter. ChIP analyses were essentially as described for panel A but over the Noxa gene promoter. (D) DNA damage-induced dissociation of VprBP. U2OS cells were either DMSO treated or etoposide (100 μM) treated for 8 h and then analyzed by ChIP analysis of the Noxa promoter as described for panel B.
For short hairpin RNA (shRNA)-based knockdown experiments with U2OS cells, DNA oligonucleotides encoding shRNAs specific for VprBP mRNA (5′-AATCACAGAGTATCTTAGA-3′), p53 mRNA (5′-GACTCCAGTGGTAATCTAC-3′), and HDAC1 mRNA (5′-GCAGATGCAGAGATTCAAC-3′) were subcloned into the H1 promoter-driven vector pSUPER.puro (OligoEngine). U2OS cells were transfected with the indicated shRNA expression constructs and then subjected to ChIP analysis. For gene knockdown experiments with LD611, LNCaP, and MCF7 cells, 21 nucleotide small interfering RNA (siRNA) duplexes with 3′ dTdT overhangs corresponding to VprBP mRNA (5′-UCACAGAGUAUCUUAGAGA-3′) were synthesized by the DNA Core Facility at the University of Southern California. Cells were transfected with siRNA oligonucleotides (60 μM) or negative-control siRNA (Ambion). At 48 h posttransfection, cells were treated with dimethyl sulfoxide (DMSO) or etoposide (100 μM) and subjected to qRT-PCR analysis.
RESULTS
VprBP regulates cell viability and p53 target gene expression.
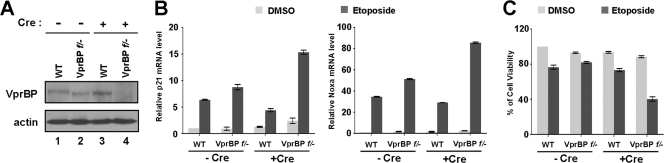
To examine the functional involvement of VprBP in regulating p53 responses more precisely, we employed conditional VprBPflox/− MEF cells that allow switching VprBP expression on and off under the control of Cre recombinase. As determined by Western blot analysis, the transfection of wild-type MEF cells with cell-permeating TAT-Cre recombinase did not affect VprBP expression (Fig. 1A, lanes 1 and 3), but the same treatment of VprBPflox/− MEF cells with the TAT-Cre recombinase resulted in ablation of VprBP expression (lanes 2 and 4). We first measured the level of transcription of two p53 target genes, the p21 and Noxa genes, by quantitative reverse transcription-PCR (qRT-PCR). As summarized in Fig. 1B, a distinct increase in DNA-damage-induced transcription of the p21 and Noxa genes was detectable after Cre-mediated deletion of the VprBP gene in VprBPflox/− MEF cells but not in wild-type MEF cells.
Fig 1.
Effects of VprBP knockout on cell viability. (A) Conditional knockout of VprBP expression. Wild type and VprBPflox/− MEF cells were treated with Cre recombinase peptides for 72 h, and VprBP deletion was confirmed by Western blot analysis using anti-VprBP antibody. (B) Transcriptional activation of the p21 and Noxa genes by VprBP knockout. Wild-type and VprBPflox/− MEF cells were treated with etoposide (100 μM) for 24 h, and total RNA was isolated and analyzed by qRT-PCR analysis. Averages and standard deviations of data from three independent experiments are shown. (C) Decreased cell viability by VprBP knockout. Wild-type and VprBPflox/− MEF cells were treated with etoposide as for panel B. Cell viability was measured by analysis of MTT conversion. All reactions were carried out in triplicate.
To address whether the observed contribution of VprBP to p53 target genes is accompanied by an alteration in cell proliferation, we next investigated its effect on cell viability. MTT assays of wild-type and VprBPflox/− MEF cells showed no significant difference in cell viability upon treatment with the DNA-damaging agent etoposide without Tat-Cre recombinase transfection (Fig. 1C, −Cre). In contrast, similar assays revealed that cell viability upon etoposide treatment was markedly reduced by Cre recombinase-mediated deletion of the VprBP gene in VprBPflox/− MEFs (Fig. 1C, +Cre). These observations imply that VprBP can contribute to the regulation of p53 transactivation and cell survival.
VprBP represses p53-mediated transcription from chromatin.
To further define the functional role of VprBP, we prepared recombinant VprBP (Fig. 2A) and examined its effects on p53-mediated transcription using a cell-free assay system. For reconstitution of linear nucleosome arrays, we employed a p53ML-601 array template containing five copies of the p53 response element and the adenovirus major late core promoter and, on both sides, seven direct repeats of the 601 nucleosome positioning sequence (17). Unmodified nucleosomal arrays were assembled from recombinant histone octamers and p53ML-601 array DNA by salt gradient dialysis. After confirming the successful reconstitution by partial micrococcal nuclease digestion (data not shown), transcription assays were carried out with p53 and p300 HAT as summarized in Fig. 2B. Transcription from p53ML-601 nucleosome arrays was completely dependent upon p53, p300 HAT, and Ac-CoA, whereas transcription from the p53ML-601 DNA template showed a dependence only on the activator p53 (Fig. 2C, lanes 1 to 3). When the effect of VprBP was examined, we found that addition of VprBP prior to p300 significantly decreased the level of transcription from nucleosome array templates (Nuc array, lanes 4 and 5). However, no detectable change in transcription was observed upon simultaneous addition of VprBP and p300 or sequential addition of VprBP after p300 (Nuc array, lanes 10, 11, 16, and 17). A similar transcription analysis with histone-free DNA templates also failed to show any effect of VprBP (Fig. 2C, DNA). Addition of VprBP to transcription reactions carried out with two Gal4-based activators (i.e., Gal4-VP16 and Gal4-CTF) showed no obvious changes in transcription (data not shown). These results confirm that the repressive effect of VprBP on chromatin transcription is highly dependent on p53.
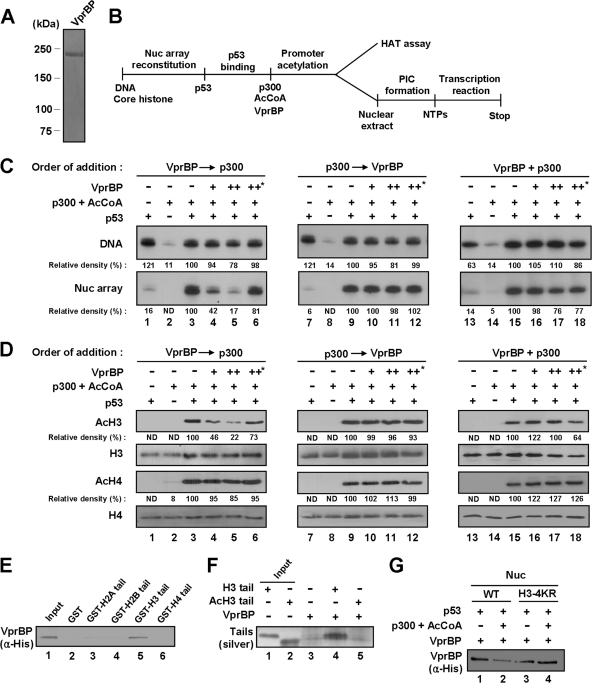
Fig 2.
VprBP-mediated repression of chromatin transcription and H3 acetylation. (A) Analysis of purified VprBP by 8% SDS-PAGE and Coomassie blue staining. (B) Outline of chromatin HAT and transcription assays. Abbreviations: AcCoA, acetyl-CoA; PIC, preinitiation complex; NTPs, nucleotide triphosphates. (C) Repressive effect of VprBP on chromatin transcription. p53ML-601 nucleosome array or histone-free p53ML-601 DNA was transcribed in the presence of p53, p300, Ac-CoA, and/or VprBP as summarized for panel B. Prior to transcription, p300 and VprBP were added together or sequentially as indicated. Heat-inactivated VprBP was used in control transcription reactions, marked by an asterisk (lanes 6, 12, and 18). Data were quantitated by using a phosphorimager, and the results shown are representative of three independent experiments. (D) Repressive effect of VprBP on H3 acetylation. The p53ML-601 nucleosome array was incubated with p53, p300, Ac-CoA, and/or VprBP as summarized in panel B. Acetylation of nucleosome arrays was detected by Western blotting with anti-acetyl H3 and H4 antibodies (AcH3 and AcH4). Western blot analyses of H3 and H4 confirmed equal loading of histones (H3 and H4). (E) Selective interaction of VprBP with H3 N-terminal tail. His-tagged VprBP was tested for binding to GST (lane 2) or GST-histone tail fusion (lanes 3 to 6) proteins. VprBP binding to histone tails was determined by Western blot analysis using anti-His antibody. Lane 1 represents 10% of VprBP used in the binding reactions. (F) Selective interaction of VprBP with unmodified H3 tail. Unmodified (H3) and acetylated (AcH3) H3 peptides (amino acids 1 to 28) were synthesized and incubated with His-tagged VprBP immobilized on Ni-NTA agarose beads. After extensive washing, bound H3 peptides were resolved in a 4 to 20% SDS-PAGE gel and silver stained. Input lanes 1 and 2 represent 10% of tail peptides used in the binding reactions. (G) Preferential binding of VprBP to unmodified nucleosome. Nucleosomes containing wild-type or mutant H3 were reconstituted on biotinylated 207-bp p53 RE and incubated with p300, p53, and/or Ac-CoA. After reconstituted nucleosomes and free DNA were immobilized on streptavidin-agarose beads, the interaction assays were performed with VprBP. The presence of VprBP in the beads was analyzed by Western blotting with anti-His antibody.
Because the primary role of acetylated H3 and H4 tails has been well illustrated in p53-mediated transcription (1, 6, 10, 13), a possible interpretation of these results is that VprBP could repress p300-mediated acetylation of H3 and H4 at the promoter region. This possibility was investigated by checking whether VprBP is also repressive for acetylation of H3 and H4 within reconstituted nucleosome arrays. Western blots of HAT reactions confirmed that p300-mediated histone acetylation in the context of nucleosome arrays is completely dependent on p53, which is known to recruit p300 for promoter-targeted acetylation (Fig. 2D, lanes 1 to 3). In parallel experiments with VprBP, we observed a significant inhibition of H3 acetylation but not H4 acetylation by addition of VprBP prior to p300 (lanes 4 and 5). As expected from transcription assays, there was no apparent repression of H3 acetylation, when VprBP was added after or simultaneously with p300 (lanes 10, 11, 16, and 17). When the same concentration of the heat-inactivated VprBP was tested for transcription and HAT assays, no inhibitory effect was detected (Fig. 2C and D, lanes 6, 12, and 18), further supporting specificity of VprBP-induced repression of H3 acetylation and chromatin transcription.
VprBP binds to unmodified H3 tail in the context of the nucleosome.
A potential mechanism underlying the inhibitory action of VprBP in transcription is that VprBP interacts with histone tails, especially H3 tails, to repress their acetylation. To test this possibility, we examined the interaction of His-VprBP with GST-histone tail fusion proteins prebound to glutathione-Sepharose beads. After extensive washing of the beads, VprBP binding was determined by Western blotting with anti-His antibody. As shown in Fig. 2E, we found that VprBP binding was highly selective for H3 tail. Because VprBP lost its repressive activity when nucleosomal arrays were preacetylated by p300 (Fig. 2C), we further analyzed the effect of tail acetylation on H3 tail-VprBP interaction using synthetic tail peptides. When incubated with immobilized His-VprBP, unmodified H3 tail peptides showed a distinct interaction with VprBP, as confirmed by silver staining after SDS-PAGE of the reactions (Fig. 2F, lane 4). In contrast, a parallel experiment with H3 tail peptides bearing lysine acetylations at K9, K14, K18, and K23 showed no detectable interaction of VprBP (lane 5). Additionally, similar binding experiments with mononucleosomes reconstituted on a 207-bp p53RE DNA fragment showed VprBP binding both to nucleosomes containing wild-type H3 and to nucleosomes containing lysine-mutated H3 (Fig. 2G, lanes 1 and 3). Notably, however, the observed VprBP-nucleosome interaction was inhibited by p300-mediated H3 acetylation, as reflected by an apparent decrease in binding of VprBP to wild-type H3 nucleosome but not to H3-mutated nucleosome (Fig. 2G, lane 2 versus lane 4).
The LisH motif of VprBP is required for its repressive action.
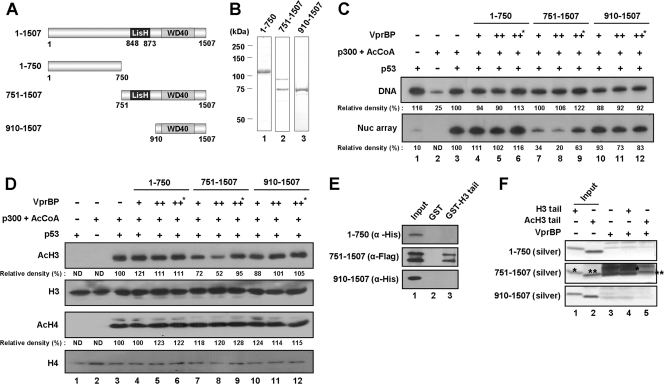
VprBP contains an evolutionally conserved Lis homology (LisH) motif in the central region and several WD40 repeats in the carboxyl-terminal region (Fig. 3A). Since these motifs are often critical for known functions of full-length proteins, we were interested in determining which part of VprBP is required for its repressive action. To this end, three different VprBP fragments were prepared from bacteria (Fig. 3B) and tested in transcription and HAT assays. The N-terminal fragment (residues 1 to 750) and C-terminal fragment containing WD40 repeats (residues 910 to 1507) showed no detectable change in p53-dependent, p300-mediated transcription of nucleosome arrays (Fig. 3C, Nuc array, lanes 4, 5, 10, and 11). In striking contrast, the fragment carrying both the C-terminal region and the central region containing the LisH motif (residues 751 to 1507) repressed the nucleosome array transcription to a level comparable to that observed with full-length VprBP (Nuc array, lanes 7 and 8). As expected, transcription from histone-free DNA was not altered by these VprBP fragments in all cases (Fig. 3C, DNA). In HAT assays with nucleosome arrays, we also observed a significant inhibition of p300-mediated acetylation of H3 by residues 751 to 1507 containing the LisH motif but not by residues 1 to 750 or 910 to 1507 (Fig. 3D, AcH3).
Fig 3.
Requirement of the LisH motif for VprBP action. (A) Schematic illustration of VprBP deletion mutants. Numbers indicate amino acid residues. The Lis homology motif (LisH) and WD40 repeat motif (WD40) are indicated. (B) Preparation of VprBP deletion mutants. His-tagged or Flag-tagged VprBP proteins were prepared as described in Materials and Methods and analyzed on an 8% SDS-PAGE gel stained with Coomassie blue. Lane 1, amino acids 1 to 750; lane 2, amino acids 751 to 1507; lane 3, amino acids 910 to 1507. (C) Differential effects of VprBP deletion mutants on chromatin transcription. Transcription reactions were carried out essentially as described for Fig. 2C, but mixtures contained VprBP deletion mutants which were added prior to p300. The asterisk indicates the usage of heat-inactivated VprBP. (D) Differential effects of VprBP deletion mutants on H3 acetylation. HAT assays were carried out as described for Fig. 2D but with the indicated VprBP deletion mutants. The heat-inactivated VprBP (marked with an asterisk) was also included. (E) Preferential binding of VprBP LisH motif to H3 tail. GST pulldown assays were performed with GST-H3 tail and VprBP deletion mutants as indicated. Input (lane 1) represents 10% of mutant proteins used in the binding experiments. (F) Preferential binding of VprBP LisH motif to unmodified H3 tail. Binding assays were performed using synthetic H3 tail peptides as for Fig. 2F but with the indicated VprBP deletion mutants. The positions of unmodified and acetylated H3 tail peptides are indicated by single and double asterisks, respectively. Lanes 1 and 2 show 10% of tail peptides used in the binding reactions.
Because VprBP binds to unmodified H3 tails to act as a transcription repressor (Fig. 2E and F), the above results argue that the LisH motif in the central region might be important for the interaction between VprBP and H3 tails. In fact, GST pulldown assays with the deletion mutants of VprBP revealed that only residues 751 to 1507 can interact with H3 tails (Fig. 3E, lane 3), indicating that the LisH motif is responsible for the interaction of VprBP with H3 tails. Similar binding experiments with unmodified or acetylated H3 tail peptides also showed that, in contrast to N-terminal (residues 1 to 750) and C-terminal (residues 910 to 1507) fragments, the fragment (residues 751 to 1507) carrying both central and C-terminal regions can bind to these peptides (Fig. 3F, lanes 4 and 5). Importantly, however, the fragment showed a marked preference for unmodified H3 peptides over acetylated H3 peptides (compare the asterisk-marked bands).
VprBP downregulates p53 target genes by modulating histone acetylation.
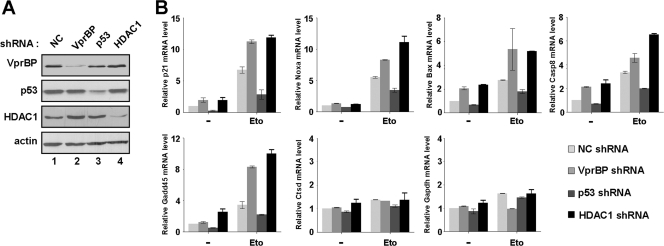
To assess the in vivo relevance of our in vitro results, we determined whether VprBP can antagonize H3 acetylation at p53 target promoters by RNAi-complemented ChIP assays. Based on recent indications that HDAC1 acts as a key repressor of p53 transactivation by blocking histone acetylation (8, 19), we also examined the promoter localization of HDAC1 and its relationship with VprBP occupancy at the promoter. As first confirmed by Western blotting, the transfection of U2OS cells with shRNAs against VprBP, p53, and HDAC1 efficiently depleted their respective target proteins (Fig. 4A). Interestingly, in contrast with a recent report showing that the expression level of p53 is relatively high in VprBP-depleted cells (11), our data demonstrated no detectable alteration in the p53 protein level after VprBP knockdown. In checking the transcription of p53-responsive genes, such as p21, Gadd45, Bax, Casp8, and Noxa, by qRT-PCR, we found that RNAi depletion of p53 led to a severe reduction in transcription after DNA damage, indicating that p53 is the major regulator of these genes (Fig. 4B, Eto). In contrast, upon depletion of VprBP or HDAC1, there was a significant enhancement in DNA-damage-induced transcription of the target genes to a comparable extent. Similar experiments without DNA damage showed, although to a much lower extent, a detectable increase in transcription after depletion of VprBP or HDAC1 (Fig. 4B, −). The fact that the individual depletion of VprBP and HDAC1 did not cause any changes in non-p53 target genes, the Ctsd and Gapdh genes, further supports that VprBP and HDAC1 are required to maintain the repressed state of p53 target genes.
Fig 4.
Stimulatory effects of VprBP knockdown on p53 transcriptional targets. (A) Depletion of VprBP, p53, and HDAC1. U2OS cells were transfected with VprBP shRNA (lane 2), p53 shRNA (lane 3), HDAC1 shRNA (lane 4), or control shRNA (lane 1), and Western blot analyses were performed using the indicated antibodies. Actin was used as an internal control. (B) Relative effect of p53, VprBP, or HDAC1 knockdown on p53 target gene transcription. Cells depleted of VprBP, p53, or HDAC1 were treated with 100 μM etoposide for 12 h, and mRNA levels were analyzed by qRT-PCR. Averages and standard deviations of data from three independent experiments are shown.
When ChIP experiments were performed with VprBP-depleted U2OS cells under normal conditions, we detected a dramatic decrease in HDAC1 occupancy and a concomitant accumulation of acetylated H3, but a minimal change in the level of p53, at the p21 promoter region (Fig. 5A, top panel). In ChIP experiments using p53-depleted cells, a significant reduction in H3 acetylation and a near-complete loss of VprBP were evident (middle panel). This observation strongly suggests that the initial recruitment of VprBP and HDAC1 to the p21 promoter is mediated by p53. One anomaly in this result is that the dissociation of HDAC1 from the p21 promoter coincides with the reduction of H3 acetylation (middle panel, AcH3 and HDAC1). This may be due to the failure of p53-mediated recruitment of a HAT, which would initially acetylate H3 at the promoter. Also consistent with our results indicating a stable association of VprBP with unmodified H3 tail (Fig. 2F and G), our ChIP assays after HDAC1 knockdown showed a significant decrease in promoter localization of VprBP but only little change in p53 occupancy (bottom panel). To explore the possible effect of VprBP on other p53 target genes, we also carried out the same analysis on the Noxa gene. Consistent with the results from the p21 gene, shRNA-mediated depletion of VprBP at the Noxa promoter region was found to coincide with dissociation of HDAC1 and an apparent increase in H3 acetylation (Fig. 5C). Depletion of p53 or HDAC1 also caused a reduction in immunoprecipitation of the promoter region using VprBP antibody (middle and bottom panel), strongly supporting that VprBP plays an inhibitory role in other p53 target genes as well.
To further investigate the repressive role of VprBP, we performed ChIP analyses with U2OS cells that were either untreated or treated with etoposide. Predictably, etoposide-induced DNA damage resulted in an accumulation of p53 and a large increase in H3 acetylation at the p21 promoter (Fig. 5B, p53 and AcH3, RE). In correlation with the increased level of H3 acetylation, the release of VprBP from the promoter region was evident (VprBP, RE). ChIP assays also showed that the dissociation of VprBP from the promoter coincides with the decrease in HDAC1 occupancy at the promoter region (HDAC1, RE), suggesting that HDAC1 binding and H3 deacetylation at the p21 promoter are dependent on VprBP. In contrast, a minimal alteration in the levels of p53, VprBP, H3 acetylation, and HDAC1 at the distal region was observed upon DNA damage in all cases (Fig. 5B, Distal). To demonstrate the coexistence of p53 and VprBP at the p21 promoter, we also performed a ChIP–re-ChIP experiment. Expectedly, the VprBP re-ChIP assay with anti-p53 immunoprecipitates resulted in the precipitation of the p21 promoter DNA fragment (Fig. 6A, p21 promoter), indicating that p53 and VprBP were bound together on the same p21 promoter. We repeated these experiments with the Noxa gene; similar results were obtained from these parallel experiments (Fig. 5C and D and Fig. 6A, Noxa promoter). These results support the idea that VprBP exerts its inhibitory effects on transcription of other p53 target genes.
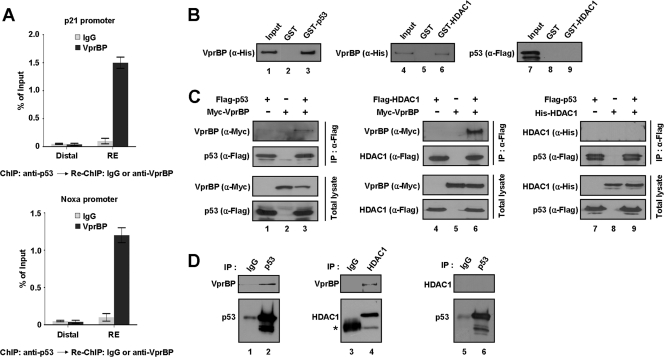
Fig 6.
Direct interaction of VprBP with p53 and HDAC1. (A) ChIP and re-ChIP assays were performed for the p21 or Noxa promoter in U2OS cells. Chromatin fragments immunoprecipitated with anti-p53 antibody in the first ChIP were used as an input and were subjected to Re-ChIP analysis using control IgG or anti-VprBP antibody. (B) VprBP interaction with p53 and HDAC1 in vitro. His-tagged VprBP or Flag-tagged p53 was incubated with GST or GST-fused p53 or HDAC1, and bound proteins were analyzed by Western blotting using anti-His and anti-Flag antibodies as indicated. “Input” corresponds to 10% of the material used in the binding reactions. (C) VprBP interaction with p53 and HDAC1 in vivo. 293T cells were transfected for 48 h with epitope-tagged forms of VprBP, p53, and/or HDAC1. Whole-cell lysates were prepared and subjected to immunoprecipitation with anti-Flag antibody followed by Western blotting with indicated antibodies (IP: α-Flag). Cell extracts were also analyzed by Western blotting to confirm that equivalent amounts of proteins from each lysate were used for immunoprecipitation (Total lysate). (D) Interaction of endogenous VprBP with p53 and HDAC1. Whole-cell extracts were prepared from 293T cells and immunoprecipitated with anti-p53 (α-DO1) or anti-HDAC1 (α-HDAC1) antibody. The precipitates were analyzed by Western blotting with antibodies against VprBP, HDAC1, and p53 as indicated. The asterisk in lane 3 indicates a nonspecific band containing IgG heavy chain.
VprBP physically interacts with p53 and HDAC1.
To determine if the recruitment and repressive action of VprBP at p53 target genes reflect its direct interaction with p53 and HDAC1, we performed a series of binding studies. In initial binding experiments, GST-p53 and GST-HDAC1 immobilized on Sepharose beads were incubated with His-VprBP and Flag-p53, and the bound proteins were detected by Western blotting after extensive washing. As shown in Fig. 6B, His-VprBP was specifically precipitated from the reaction by both GST-p53 and GST-HDAC1 (lanes 3 and 6). In contrast, pulldown experiments in which GST-HDAC1 was used under the same conditions failed to show any detectable binding to Flag-p53 (lane 9). To validate these in vitro results in vivo, extracts from 293T cells transiently expressing Myc-VprBP, Flag-p53, and/or Flag- or His-HDAC1 were immunoprecipitated using anti-Flag antibody. Consistent with in vitro binding data, our Western blot analysis showed a strong interaction of VprBP with p53 and HDAC1 (Fig. 6C, lanes 3 and 6) but no detectable interaction between p53 and HDAC1 (lane 9). The interaction of VprBP with p53 and HDAC1 under physiological conditions was further tested by Western blotting of anti-p53 and anti-HDAC1 immunoprecipitates from extracts of 293T cells. Again, both p53 and HDAC1 were coimmunoprecipitated with VprBP, but HDAC1 failed to show coimmunoprecipitation of p53 (Fig. 6D).
Proapoptotic effect of VprBP knockdown in cancer cells.
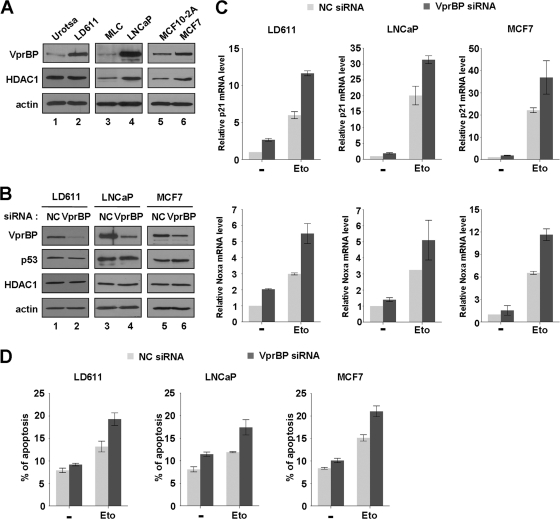
Given demonstrated action of VprBP as a negative regulator of p53, we next assessed the expression level of VprBP in three human cancer cell lines expressing wild-type p53: a bladder cancer cell line (LD611), a prostate cancer cell line (LNCaP), and a breast cancer cell line (MCF7). Based on our Western blotting of these transformed cancer cells, a much higher level of VprBP expression was evident than with nontransformed cells (Urotsa, MLC, and MCF10-2A) (Fig. 7A, VprBP). Prostate (LNCaP) and breast (MCF7) cancer cells also showed a higher level of HDAC1 than the normal cells (HDAC1, lanes 4 and 6 versus lanes 3 and 5), arguing for the functional connection between VprBP and HDAC1 as observed at the p53 target genes.
Fig 7.
Stimulation of p53 transcription and apoptosis by VprBP knockdown. (A) Western blot analysis of normal and cancer cells. Exponentially growing normal (Urotsa, MLC, and MCF10-2A) and cancer (LD611, LNCaP, and MCF7) cells were subjected to Western blot analysis using anti-VprBP, anti-p53, and anti-HDAC1 antibodies. Actin served as a control for equal protein loading. (B) RNAi-mediated depletion of VprBP. LD611, LNCaP, and MCF7 cells were transfected with a control siRNA (lanes 1, 3, and 5) or a siRNA directed against VprBP (lanes 2, 4, and 6) for 72 h and analyzed by Western blotting using the indicated antibodies. All analyses were performed in parallel for each of the three cell lines. (C) Activation of the p21 and Noxa genes by VprBP depletion. Cells were first transfected with siRNA targeting VprBP or an irrelevant control (NC) as in panel B and treated with or without etoposide (100 μM) for 24 h. Transcription levels of the p21 and Noxa genes were analyzed by qRT-PCR. (D) Upregulation of DNA damage-induced apoptosis by VprBP depletion. After transfected with either a control siRNA or a VprBP siRNA, cells were DMSO treated or etoposide treated as for panel C. The apoptotic status of cells was analyzed by assessing annexin V-FITC fluorescent intensity. Averages and standard deviations are shown for data from three independent experiments.
In line with the repressive action of VprBP on p53 transcription activity, a possible effect of VprBP depletion on expression of the p21 and Noxa genes was examined in these cancer cell lines. Because the shRNA-mediated knockdown of VprBP was not efficient in these cell lines (data not shown), we employed synthetic siRNA to achieve high-efficiency knockdown, as confirmed by Western blot analysis (Fig. 7B, VprBP). When these VprBP-depleted cells were treated with etoposide, the expression of the p21 and Noxa genes was significantly stimulated, supporting VprBP acting as a transcriptional repressor of p53 (Fig. 7C, Eto). Although the p53 target genes exhibited very weak transcription without etoposide treatment, a moderate enhancement in the target gene expression was also observed by VprBP depletion under this uninduced condition (Fig. 7C, −). Of note, we did not see any changes in the cellular level of p53 after VprBP depletion (Fig. 7B, p53). Thus, it does not seem likely that the observed action of VprBP on p53 target genes is related to VprBP-triggered degradation of p53.
Because p53-mediated transcription of the p21 and Noxa genes represents the early stage of apoptosis, DNA damage-triggered apoptosis that is compromised by VprBP overexpression in cancer cells may be reactivated by VprBP depletion. To test this, we depleted VprBP in the cancer cell lines and then subjected them to apoptosis analysis. As summarized in Fig. 7D, VprBP depletion resulted in an apparent increase in the proportion of apoptotic cells, suggesting that endogenous VprBP plays an important role in attenuating DNA damage-induced apoptosis.
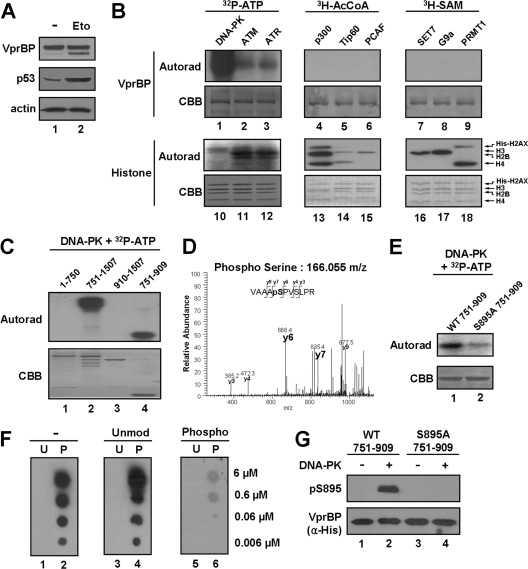
VprBP is phosphorylated at serine 895 by DNA-PK.
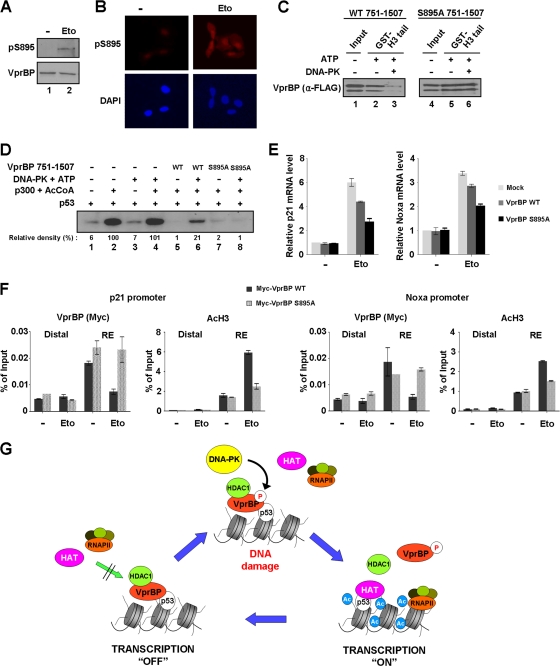
Knowing that VprBP-H3 tail interaction is a key event in the inhibition of p53-mediated transcription, we next sought to explore whether the inhibitory effect of VprBP is reversible. Interestingly, although the cellular level of p53 increased during the course of damage response, the level of VprBP remained fairly constant (Fig. 8A). These results led us to speculate that VprBP may be postsynthetically modified upon DNA damage such that its interaction with the H3 tail is hindered. To this end, we performed radioactive in vitro modification assays with a panel of kinases (DNA-PK, ATM, and ATR), acetyltransferases (p300, Tip60, and PCAF), and methyltransferases (SET7, G9a, and PRMT1), all of which are known to participate in the DNA damage response. SDS-PAGE and autoradiography of the radioactively labeled modification reactions revealed that VprBP can be modified by DNA-PK but not by other factors (Fig. 8B). In order to delineate the location of the VprBP phosphorylation sites, in vitro kinase assays were repeated with different subregions of VprBP with DNA-PK. Our assays revealed that the main phosphorylation site for DNA-PK is localized within amino acids 751 to 909 of VprBP (Fig. 8C). As a more direct approach toward localizing phosphorylation, mass spectrometric sequencing was performed on the VprBP 751–909 fragment after the phosphorylation reaction. Our analysis identified only a singly phosphorylated peptide that could have arisen from phosphorylation of serine 895 (S895), which is in close proximity to the LisH motif (Fig. 8D). Consistent with the mass spectrometric results, S895-mutated VprBP is phosphorylated by DNA-PK at a much lower level than the wild-type protein (Fig. 8E). To further investigate the role of VprBP phosphorylation, we raised and purified a rabbit polyclonal antibody that reacts with S895-phosphorylated VprBP. The specificity of the purified antibody was verified by dot blot and peptide competition assays using the unmodified and phosphorylated peptides (Fig. 8F). In addition, the affinity-purified antibody reacted strongly with phosphorylated wild-type VprBP but not with unmodified wild-type VprBP and phosphorylated S895-mutated VprBP (Fig. 8G).
Fig 8.
Phosphorylation of VprBP by DNA-PK. (A) Western blot analysis of VprBP and p53 after DNA damage. U2OS cells were DMSO treated or etoposide treated for 12 h, and cell lysates were subjected to Western blotting using the indicated antibodies. (B) Posttranslational modification of VprBP. In vitro modification assays were performed with a panel of kinases, acetyltransferases, and methyltransferases using VprBP (lanes 1 to 9) or H2AX histone octamer (lanes 10 to 18) as a substrate. The reactions were analyzed by SDS-PAGE and autoradiography as described in Materials and Methods. (C) Determination of the phosphorylation domain of VprBP. The indicated domains of VprBP were generated in E. coli and subjected to in vitro kinase assays using DNA-PK and [γ-32P]ATP. (D) Mapping VprBP phosphorylation sites by mass spectrometry. A representative MS/MS spectrum of the major phosphopeptide sequence, which identified phosphorylation at S895, is shown. The theoretical fragment mass of phosphoserine is highlighted. (E) Mutational analysis of the identified VprBP phosphorylation site. An in vitro kinase assay was performed using DNA-PK and [γ-32P]ATP as for panel C but with VprBP 751–909 mutated at S895. (F) Generation of a phospho-S895-specific antibody. Dot blotting of unmodified (U) and phosphorylated (P) peptides was performed in the presence of either the unmodified or the phosphorylated peptides during the antibody binding step. (G) Specificity of phospho-S895 antibody. Wild-type and mutant VprBP 751–909 proteins were phosphorylated by DNA-PK and cold ATP and analyzed by Western blot analysis using the phospho-S895 antibody.
Phosphorylation inhibits transrepression activity of VprBP.
We next assessed the contribution of S895 phosphorylation to the H3 tail binding and transrepressive activities of VprBP. As determined by Western blotting, etoposide-induced DNA damage resulted in a rapid phosphorylation of VprBP at S895, but no change in the cellular concentration of VprBP was observed (Fig. 9A). Immunofluorescence analysis also indicated a considerable increase in VprBP phosphorylation in cells that had been treated with etoposide (Fig. 9B). To check the effect of VprBP phosphorylation on VprBP-H3 tail interaction, the glutathione-Sepharose-bound GST-H3 tail was incubated with unphosphorylated or phosphorylated Flag-VprBP, and the bead-bound VprBP fraction was analyzed by Western blotting with anti-Flag antibody (Fig. 9C). The interaction between the H3 tail and wild-type VprBP was reduced by ∼80% following the phosphorylation of VprBP (lanes 2 and 3), but there was no disruption of H3 tail-mutant VprBP interaction after the phosphorylation reaction (lanes 5 and 6).
Fig 9.
Alleviation of VprBP-induced repression by phosphorylation. (A) Western blotting of S895 phosphorylation after DNA damage. Cell extracts were prepared 8 h after etoposide treatment of U2OS cells and analyzed by Western blot analysis using VprBP and phospho-S895 VprBP antibodies. (B) Immunostaining of S895 phosphorylation after DNA damage. U2OS cells were treated with etoposide as for panel A and stained with phospho-S895 VprBP antibody. Increased staining for S895 phosphorylation is evident after etoposide-induced DNA damage. (C) Impaired interaction of VprBP with H3 tail upon phosphorylation. In vitro binding experiments using immobilized H3 tails were performed with phosphorylated or unphosphorylated wild type (WT) (lanes 1 to 3) and S895-mutated (lanes 4 to 6) VprBP. VprBP binding to H3 tails was determined by Western blotting using anti-Flag antibody. (D) Stimulatory effect of VprBP phosphorylation on transcription. Transcription reactions were identical to those in Fig. 3C, but wild-type and mutant VprBP 751–1507 fragments were premodified by DNA-PK. (E) Differential effects of wild-type and S895-mutated VprBP on p21 and Noxa transcription. U2OS cells were transfected with Myc-tagged versions of wild-type and S895-mutated VprBP for 48 h and treated with etoposide for additional 12 h. p21 and Noxa mRNA levels were analyzed by using qRT-PCR. (F) Phosphorylation-dependent dissociation of VprBP from the promoter. U2OS cells were transfected with wild-type and S895-mutated VprBP as for panel E. ChIP assays after etoposide treatment were essentially as described for Fig. 5B. (G) Model for VprBP inhibition of p53-mediated transcription. Under unstressed conditions, VprBP is brought to p53-responsive promoters by p53 and acts as a molecular rheostat to block transcription initiation by binding to unacetylated H3 tails. For the most efficient repression, VprBP cooperates with HDAC1 to remove and block H3 acetylation. Upon DNA damage, the majority of VprBP is phosphorylated by DNA-PK and dissociated from the promoter nucleosomes. HAT then acetylates H3 tails and establishes an active promoter environment for p53 to achieve the most efficient transcription of proapoptotic genes. In this way, cells would achieve more accurate and efficient regulation of the p53 transcriptional network. See Discussion for further details.
To analyze the functional effect of VprBP phosphorylation, transcription assays were performed as in Fig. 3C, except that DNA-PK was added to phosphorylate VprBP in the reaction. As shown in Fig. 9D, DNA-PK antagonized the repressive action of VprBP and produced a distinct enhancement of p53-dependent, p300-mediated transcription from p53ML-601 nucleosome arrays (lanes 5 and 6). On the contrary, the ability of DNA-PK to enhance the nucleosome array transcription was significantly compromised when wild type VprBP was replaced by S895-mutated VprBP (lanes 7 and 8). These results, together with the results from the binding assays (Fig. 9C), strongly suggest that the phosphorylation of S895 is the major cause of impaired repressive action of VprBP. Additional support for the regulatory role of S895 phosphorylation in VprBP activity came from transfection experiments in which wild-type or mutant VprBP was expressed in U2OS cells and the impact of DNA damage on p53 transactivation was measured by qRT-PCR. The data shown in Fig. 9E indicate that blocking DNA damage-induced S895 phosphorylation by mutation has a positive influence on VprBP-induced repression of p53 transactivation.
In view of the antagonizing effect of phosphorylation on VprBP-H3 tail interaction and VprBP-induced repression, ChIP analysis was performed with U2OS cells ectopically expressing wild-type and S895-mutated forms of VprBP. As expected, we observed a specific localization of wild-type VprBP at the p21 and Noxa promoters in the absence of DNA damage and its dissociation following etoposide-induced DNA damage (Fig. 9F, VprBP, Myc-VprBP WT). In correlation with the release of VprBP, the rapid accumulation of H3 acetylation at the promoter regions was detected after DNA damage (AcH3, Myc-VprBP WT). Remarkably, however, parallel ChIP analyses with mutant VprBP showed no apparent change in VprBP occupancy (VprBP, Myc-VprBP S895A) and a slight increase in H3 acetylation around the promoter regions in response to DNA damage (AcH3, Myc-VprBP S895A). We therefore conclude that VprBP phosphorylation at S895 occurs upon DNA damage and that this phosphorylation is critical for DNA damage-induced activation of p53 response genes.
DISCUSSION
Although VprBP was originally implicated in HIV-1 replication and pathogenesis based on its interaction with the HIV-1 protein Vpr, a recent study revealed that VprBP knockdown increases the level of the endogenous p21 protein under normal growth conditions (11). However, it was unknown whether transcription is the primary target for VprBP and if so how VprBP modulates transcription of p53 target genes, such as the p21 gene. In this study, we explored the hypothesis that VprBP could generate a repressive environment at p53 target genes and antagonize DNA damage-induced apoptosis. Taking advantage of our well-defined in vitro transcription system, we demonstrated that VprBP can act as a negative regulator of p53-mediated chromatin transcription when added prior to p300 HAT. It is worth noting that the use of recombinant histones for chromatin assembly allowed us to exclude the effect of any prior modifications that could influence the repressive action of VprBP. In striking contrast, the repressive properties of VprBP cannot be recapitulated by the addition of p300 HAT prior to VprBP or the simultaneous addition of VprBP and p300 HAT, pointing to a mechanism that is dependent upon the unacetylated state of nucleosomes. Consistent with this notion, we found that VprBP specifically interacts with unacetylated H3 tails protruding from inactive nucleosomes. Thus, apart from our demonstration of VprBP as a new repressor in regulating p53 transactivation potential, these results point to a direct link between the unacetylated state of H3 tails and the repressive property of VprBP.
Our studies also show that the LisH motif of VprBP plays a dominant role in repressing p53-mediated chromatin transcription. These results can be explained by an intrinsic ability of the LisH motif to recognize unmodified H3 tails. In fact, our interaction studies confirmed the requirement of the LisH motif for the interaction between VprBP and unmodified H3 tails. Notably, our results contrast with those of a recent study showing that the LisH motif in transducin beta-like protein 1 and its receptor (TBL1 and TBLR1) binds to the hypoacetylated H4 tail for chromatin targeting by the nuclear receptor corepressor complex (4). This may reflect a low degree of sequence homology (only 30%) in their LisH motifs, which could discriminate between related but distinct regions of H3 and H4 tails. Thus, whether LisH motifs in different proteins recognize different histone tails is an intriguing question that needs to be addressed in future studies.
In accord with our in vitro studies, RNAi-complemented ChIP analyses demonstrate that VprBP is necessary for the maintenance of repressed states of p53 target genes under normal unstressed conditions. Importantly, the ability of VprBP in establishing this repressed environment relies on its initial recruitment by p53 as well as its ability to recognize unmodified H3 tails. One important aspect revealed in this regard is that HDAC1 acts as a gatekeeper to prevent H3 acetylation at the p21 and Noxa promoters, supporting its role in stimulating stable action of VprBP at the promoter regions. Because VprBP is required for initial recruitment of HDAC1 to the promoters, functional significance of the VprBP-HDAC1 interaction should also be emphasized. In further support of the interplay between VprBP and HDAC1 in constraining p53 transcription, DNA damage-induced activation of p53 target genes was accompanied by dissociations of VprBP and HDAC1 and concomitant accumulation of H3 acetylation. The critical role of VprBP in the p53 signaling pathway is further underscored by the finding that a high level of VprBP is detected in three human cancer cell lines, which reinforces our hypothesis that misregulation of VprBP expression maintains the repressive state of p53 target genes and interferes with the apoptotic capability of cells. In agreement with this hypothesis, VprBP knockdown in these cancer cells results in an apparent stimulation of p53 transcription and apoptotic cell death in response to etoposide treatment. These data constitute a powerful argument that overexpression of VprBP in cancer cells antagonizes transcription of p53 target genes encoding proapoptotic factors.
Another intriguing observation from our study is that VprBP is phosphorylated by DNA-PK at S895, and mutation of this serine residue greatly diminishes the ability of VprBP to repress p53-dependent chromatin transcription. The finding that DNA-PK-mediated phosphorylation of VprBP leads to the dissociation of VprBP from promoter nucleosomes and the reactivation of p53 target genes also points to the functional importance of this modification in limiting the repressive action of VprBP upon DNA damage (Fig. 9F). Because S895 is located immediately C-terminal to the LisH motif of VprBP, phosphorylation may affect the structural integrity of the LisH motif and therefore alter its affinity for H3 tails. Mechanistically, it is also reasonable to speculate that VprBP phosphorylation might antagonize the initial localization of VprBP at the promoter. In this respect, it will be interesting to distinguish the effects of S895 phosphorylation on VprBP dissociation versus VprBP association at the promoter nucleosomes. There are also other possible aspects of the regulation of VprBP activity by S895 phosphorylation. For instance, we do not yet know the consequence of VprBP phosphorylation for the p53-VprBP interaction, which is critical for the promoter-targeted recruitment of VprBP. Such early regulation of VprBP would benefit the fine-tuning of p53 transcriptional responses. Therefore, further characterization of physiological and oncogenic activities of VprBP has broad implications for the repertoire and complexity of the regulatory mechanisms underlying p53 signaling pathways.
ACKNOWLEDGMENTS
We acknowledge Peter Jones for the Urotsa and LD611 cell lines, Michael Lieber for DNA-PK, Igor Pogribny for the MCF10-2A cell line, Daniela Rhodes for p601-7, Pradip Roy-Burman for the MLC and LNCaP cell lines, Jacek Skowronski for pCG-Myc-VprBP, Michael Kastan for pcDNA3.1-Flag-ATM, and Karlene Cimprich for pBJ5.1-Flag-ATR. We also thank Deborah Johnson and members of the An laboratory for their instructive discussions and valuable suggestions.
This work was supported by NIH grant R01GM84209, ACS Research Scholar grant DMC-1005001, and Department of Defense grant W81XWH0910330, awarded to W.A.
Footnotes
Published ahead of print 19 December 2011
REFERENCES
- 1. An W, Kim J, Roeder RG. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735–748 [DOI] [PubMed] [Google Scholar]
- 2. An W, Roeder RG. 2003. Direct association of p300 with unmodified H3 and H4 N termini modulates p300-dependent acetylation and transcription of nucleosomal templates. J. Biol. Chem. 278:1504–1510 [DOI] [PubMed] [Google Scholar]
- 3. Berger SL. 2007. The complex language of chromatin regulation during transcription. Nature 447:407–412 [DOI] [PubMed] [Google Scholar]
- 4. Choi HK, et al. 2008. Function of multiple Lis-homology domain/WD-40 repeat-containing proteins in feed-forward transcriptional repression by silencing mediator for retinoic and thyroid receptor/nuclear receptor corepressor complexes. Mol. Endocrinol. 22:1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Donner AJ, Szostek S, Hoover JM, Espinosa JM. 2007. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell 27:121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doyen CM, et al. 2006. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol. Cell. Biol. 26:1156–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doyon Y, Selleck W, Lane WS, Tan S, Cote J. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24:1884–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. 2005. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 25:10338–10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dyer PN, et al. 2004. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375:23–44 [DOI] [PubMed] [Google Scholar]
- 10. Espinosa JM, Emerson BM. 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell 8:57–69 [DOI] [PubMed] [Google Scholar]
- 11. Hrecka K, et al. 2007. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. U. S. A. 104:11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang J, Chen J. 2008. VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene 27:4056–4064 [DOI] [PubMed] [Google Scholar]
- 13. Ivanov GS, et al. 2007. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol. Cell. Biol. 27:6756–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jaskelioff M, Gavin IM, Peterson CL, Logie C. 2000. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol. 20:3058–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Katan-Khaykovich Y, Struhl K. 2002. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 16:743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim K, et al. 2008. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J. Biol. Chem. 283:9113–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim K, et al. 19 December 2011, posting date Functional interplay between p53 acetylation and H1.2 phosphorylation in p53-regulated transcription. Oncogene. doi:10.1038/onc.2011.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- 19. Lagger G, et al. 2003. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol. Cell. Biol. 23:2669–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Rouzic E, et al. 2007. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6:182–188 [DOI] [PubMed] [Google Scholar]
- 21. Li B, Carey M, Workman JL. 2007. The role of chromatin during transcription. Cell 128:707–719 [DOI] [PubMed] [Google Scholar]
- 22. Li W, et al. 2010. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell 140:477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu G, Xia T, Chen X. 2003. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J. Biol. Chem. 278:17557–17565 [DOI] [PubMed] [Google Scholar]
- 24. Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260 [DOI] [PubMed] [Google Scholar]
- 25. MacDonald VE, Howe LJ. 2009. Histone acetylation: where to go and how to get there. Epigenetics 4:139–143 [DOI] [PubMed] [Google Scholar]
- 26. McCall CM, Miliani de Marval PL, Chastain PD, II, et al. 2008. Human immunodeficiency virus type 1 Vpr-binding protein VprBP, a WD40 protein associated with the DDB1-CUL4 E3 ubiquitin ligase, is essential for DNA replication and embryonic development. Mol. Cell. Biol. 28:5621–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Menendez D, Inga A, Resnick MA. 2009. The expanding universe of p53 targets. Nat. Rev. Cancer 9:724–737 [DOI] [PubMed] [Google Scholar]
- 28. Peitz M, Pfannkuche K, Rajewsky K, Edenhofer F. 2002. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl. Acad. Sci. U. S. A. 99:4489–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peterson CL, Laniel MA. 2004. Histones and histone modifications. Curr. Biol. 14:R546–R551 [DOI] [PubMed] [Google Scholar]
- 30. Riley T, Sontag E, Chen P, Levine A. 2008. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9:402–412 [DOI] [PubMed] [Google Scholar]
- 31. Roth SY, Denu JM, Allis CD. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81–120 [DOI] [PubMed] [Google Scholar]
- 32. Shahbazian MD, Grunstein M. 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76:75–100 [DOI] [PubMed] [Google Scholar]
- 33. Tan L, Ehrlich E, Yu XF. 2007. DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J. Virol. 81:10822–10830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vousden KH, Prives C. 2009. Blinded by the light: The growing complexity of p53. Cell 137:413–431 [DOI] [PubMed] [Google Scholar]
- 35. Wang GG, Allis CD, Chi P. 2007. Chromatin remodeling and cancer, part I: covalent histone modifications. Trends Mol. Med. 13:363–372 [DOI] [PubMed] [Google Scholar]
- 36. Wen X, Duus KM, Friedrich TD, de Noronha CM. 2007. The HIV1 protein Vpr acts to promote G2 cell cycle arrest by engaging a DDB1 and Cullin4A-containing ubiquitin ligase complex using VprBP/DCAF1 as an adaptor. J. Biol. Chem. 282:27046–27057 [DOI] [PubMed] [Google Scholar]
- 37. Zhang S, Feng Y, Narayan O, Zhao LJ. 2001. Cytoplasmic retention of HIV-1 regulatory protein Vpr by protein-protein interaction with a novel human cytoplasmic protein VprBP. Gene 263:131–140 [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y, et al. 2008. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20:1437–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]