Abstract
Much of the regulatory diversity in eukaryotic transcription is provided by coregulators, which are recruited by DNA-binding factors to propagate signaling to basal machinery or chromatin. p160 family members, including the glucocorticoid receptor (GR)-interacting protein 1 (GRIP1), function as coactivators for GR, a ligand-dependent transcription factor of the nuclear receptor superfamily. Unlike other p160s, GRIP1 also potentiates GR-mediated repression of AP1 and NF-κB targets and, surprisingly, transcriptional activation by interferon regulatory factors. What enables GRIP1 activating or repressing properties or discrimination between physiologically antagonistic pathways is unknown. We found that endogenous GRIP1 in mammalian cells undergoes glucocorticoid-induced, GR interaction-dependent phosphorylation and identified one constitutive and six inducible phosphorylation sites and two putative GRIP1 kinases, casein kinase 2 and cyclin-dependent kinase 9. We raised phosphospecific antibodies to the four closely spaced sites in a previously uncharacterized part of GRIP1 which, combined with mutagenesis, revealed the conservation of GRIP1 phosphorylation across several cell types and species and its functional relevance to GR-activated transcription and to response element-specific recruitment of phospho-GRIP1 to native GR targets. We propose that cofactor engagement by GR is neither passive nor stochastic; rather, GR actively imparts modifications that dictate GRIP1 function in a subset of complexes, adding a layer of specificity to GR transcriptional control.
INTRODUCTION
A fundamental question in eukaryotic signal-regulated transcription relates to the molecular determinants of specificity which dictate the composition and function of transcriptional regulatory complexes at their genomic binding sites and ensure the physiologically relevant response of a gene, cell, or tissue to a given environmental stimulus. Glucocorticoid receptor (GR), a ligand-dependent transcription factor of the nuclear receptor (NR) superfamily (14), exemplifies this complexity by conveying the physiological effects of glucocorticoid hormones in virtually every cell in the body and integrating gene expression programs in processes as diverse as metabolism, development, or immune response.
Upon ligand binding, cytoplasmic GR undergoes a dramatic conformational shift, translocates to the nucleus, binds genomic glucocorticoid response elements (GRE), and enucleates the assembly of multiprotein-DNA complexes which alter target gene expression (38). Along with ligand structure and availability, the type of GRE plays an essential role in specifying gene regulation. Usually, GR binding to a palindromic GRE (two conserved hexameric half-sites separated by 3 bp) leads to transcriptional enhancement, whereas GR “tethering” to DNA via interactions with other regulators, e.g., AP1 or NF-κB, represses their activity (25). A recent description of atypical “negative” palindromic GREs with 1- to 2-bp spacers (37) further underscores a key role of binding sites in dictating GR properties.
Like other NRs, to effect transcriptional changes, GR is assisted by numerous coregulators which provide a physical and/or functional link between the liganded GR and basal transcriptional machinery or chromatin. From these, the p160 proteins (steroid receptor coactivator 1 [SRC1]/nuclear receptor coactivator 1 [NCoA1], GR-interacting protein 1 [GRIP1]/transcriptional intermediary factor [TIF2]/NCoA2, and RAC3/AIB1/pCIP/ACTR/NCoA3) act as binding platforms for multiple secondary cofactors with chromatin-modifying activities. For example, the N-terminal bHLH/PAS domain of p160s binds flightless I, CoCoA, GAC63, and the Baf57 subunit of the SWI/SNF chromatin remodeling complex (4, 9, 21, 22), whereas the C-terminal activation domains 1 (AD 1) and 2 recruit histone acetyltransferases (CBP/p300 and pCAF) and arginine methyltransferases (CARM1 and PRMT1), respectively (7, 23, 24, 40). The NR-p160 interaction occurs through activation function 2 (AF2) within the agonist-bound NR ligand-binding domains and one of the three “NR boxes” (LXXLL motifs [15], where “X” is any amino acid) of the centrally located p160 NR interacting domain (10, 15, 18, 29, 40). Despite significant similarities, p160 proteins possess distinct molecular, structural, and functional features (reviewed in reference 43), and remarkably, even within the same organ system and pathway, the three coregulators have nonredundant or even opposing functions (19). With respect to GR actions, GRIP1 has emerged as a particularly divergent member of the p160 family. Indeed, although all three p160s serve as GR coactivators at palindromic GREs, GRIP1 is also recruited to GR-AP1 and GR–NF-κB tethering sites, where it potentiates GR-mediated repression through a unique repression domain (RD) (32, 35). Given that repression of AP1 and NF-κB activities by liganded GR is a key component of the widely exploited immunosuppressive and anti-inflammatory actions of glucocorticoids, these findings suggest that in conjunction with GR, GRIP1 may contribute to controlling inflammation and the immune response.
Surprisingly, recent studies identified GRIP1 as a coactivator for multiple interferon (IFN) regulatory factors (IRFs) at several independent steps of the type I IFN signaling network (5, 11, 31). Furthermore, transcriptome analyses in the mouse liver lacking GRIP1, but not other p160s, revealed a marked downregulation of multiple immune-related genes (19). Combined, these studies place GRIP1 at the intersection of reciprocal, seemingly antagonistic pathways which drive pro- versus anti-inflammatory gene expression programs, yet virtually nothing is known about the molecular cues that trigger GRIP1 activating or repressing properties or enable its selective recruitment to one transcription complex over another. Cofactor availability is viewed as a key variable ensuring sequence-specific transcription complex formation and function. Indeed, we previously reported that in macrophages, which express little GRIP1 protein, GR and IRFs recruit GRIP1 in a mutually exclusive manner and that activation of GR inhibits the IRF-dependent gene expression program and vice versa (11, 31). Although this model may explain cell type-specific patterns in gene expression, in a given cell, the same coregulators are in principle equally available to multiple factors, suggesting either that cofactor recruitment is stochastic or that other mechanisms may contribute to their pathway-specific utilization.
Here, we report that GRIP1 is postranslationally modified in a glucocorticoid-induced, GR interaction-dependent manner. We identify a panel of GRIP1 phosphorylation sites (phosphosites), conserved in multiple cell types and species, and show their functional significance with respect to GR transcriptional activation, suggesting a potential mechanism for regulator-driven coregulator recruitment and an additional layer of specificity in GR transcriptional control.
MATERIALS AND METHODS
Plasmids.
Plasmids XG46TL, MMTV-LTR-Luc, 2×TRE, and pCDNA3-GRIP1 (wild type [WT] and NR box-3 mutant) were previously described (35). pCDNA3.1(+)/myc-His.TIF2 was created by subcloning the TIF2/GRIP1 insert excised from pSG5-TIF2 with BamHI–Acc65I (blunt) into the BglII–SspI sites of pCDNA3.1(+)/myc-His (Invitrogen). GRIP1 phosphorylation site mutants were generated by site-directed mutagenesis using the pSG5-TIF2 backbone. S469 and S487 (individually) and S493 and S499 (together) were mutated to A or D, whereas S565, S699, and S736 were mutated to A only. The pSG5-TIF2 plasmid containing the first four mutations, S469/487/493/499A (4A), or the last three mutations, S565/699/736A, was digested with SbfI–BamHI to swap the two mutation-bearing halves and create the pSG5-TIF2 7A (S469/487/493/499/565/699/736A) mutant. The 4A, 4D, and 7A mutant TIF2 inserts were excised from pSG5 and ligated into pCDNA3.1(+)/myc-His as described above. To generate the 5A (S469/487/493/499/565A) mutant, S565 was mutated to A in the context of pCDNA3.1(+)/myc-His TIF2 4A (S469/487/493/499A).
Mutagenesis primers.
Primers used for mutagenesis were as follows: hTIF2_S469A_F, GCACTCAAAATGAACGCCCCCTCACAAAGCAGC; hTIF2_S469A_R, GCTGCTTTGTGAGGGGGCGTTCATTTTGAGTGC; hTIF2_S487A_F, CCACCTCCATGCTTGCACCAAGGCATCGC; hTIF2_S487A_R, GCGATGCCTTGGTGCAAGCATGGAGGTGG; hTIF2_S493,9A_F, CAAGGCATCGCATGGCCCCTGGAGTGGCTGGCGCCCCTCGAATC; hTIF2_S493,9A_R, GATTCGAGGGGCGCCAGCCACTCCAGGGGCCATGCGATGCCTTG; hTIF2_S565A_F, GCAAAACGCCCCAGTTAATATGAATCCTCCC; hTIF2_S565A_R, GGGAGGATTCATATTAACTGGGGCGTTTTGC; hTIF2_S699A_F, GGACAGCAGTGCCCCTGTGGACTTG; hTIF2_S699A_R, CAAGTCCACAGGGGCACTGCTGTCC; hTIF2_S736A_F, CAAGAGCCGGTGGCCCCCAAGAAGAAAG; hTIF2_S736A_R, CTTTCTTCTTGGGGGCCACCGGCTCTTG; hTIF2_S469D_F, GCACTCAAAATGAACGACCCCTCACAAAGCAGC; hTIF2_S469D_R, GCTGCTTTGTGAGGGGTCGTTCATTTTGAGTGC; hTIF2_S487D_F, CCACCTCCATGCTTGACCCAAGGCATCGC; hTIF2_S487D_R, GCGATGCCTTGGGTCAAGCATGGAGGTGG; hTIF2_S493,9D_F, CAAGGCATCGCATGGACCCTGGAGTGGCTGGCGACCCTCGAATC; and hTIF2_S493,9D_R, GATTCGAGGGTCGCCAGCCACTCCAGGGTCCATGCGATGCCTTG.
Cell culture, treatments, and transfections.
Human U2OS-rGR, U2OS-rGR E773R, U2OS-hGR, U2OS-rGRhTRβ1, SAOS2-rGR (33, 34, 35) osteosarcoma, A549 lung carcinoma, HeLa cervical carcinoma, GrH2 rat hepatoma, and RAW264.7 mouse macrophage-like cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) and 10% fetal bovine serum (HyClone), supplemented with 350 μg/ml G418 (Invitrogen) (for U2OS and SAOS2 stable lines only) and 200 μg/ml hygromycin (Invitrogen) (for U2OS-rGRhTRβ1). Bone marrow-derived macrophages were prepared from 8-week-old C57BL/6 mice as described in reference 11. Dexamethasone (dex), RU486, and triiodothyronine (T3) were from Sigma, and CIP (calf intestinal phosphatase) was purchased from NEB. 5,6-Dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) and 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT) were kindly provided by M. Garabedian (NYU), and flavopiridol was kindly provided by R. Fisher (Mount Sinai). U2OS-rGR, U2OS-rGRhTRβ1, and RAW264.7 cells were transfected using Lipofectamine-Plus reagent (Invitrogen) as described previously (31, 32).
Real-time qPCR.
Total RNA was isolated using the RNeasy Mini Kit (Qiagen). Random-primed cDNA synthesis, quantitative PCR (qPCR) using SYBR green master mix with ROX (Fermentas), and cycle threshold (ΔΔCT) analysis were performed as described elsewhere, with β-actin as the normalization control. Primer pairs for target genes were as described previously (11, 34) except for Per1, for which the primers were hPer1_F (ACTCCCCTATCCGCTTCTGTGCC) and hPer1_R (GGCCCAACACGAAGGCTACCTTG).
Immunoblotting.
Protein extracts were prepared as described previously (31). Immunoblotting with commercial mouse monoclonal or rabbit polyclonal antibodies to GRIP1 (BD Transduction Laboratories or Abcam, respectively), myc tag, and STAT3 (Santa Cruz Biotechnology) was performed using standard protocols. Immunoblotting with phosphospecific antisera was performed at dilutions of 1:500 to 1:5,000, individually optimized to each experiment and cell type. Each blot is a representative of 3 or more independent experiments. For antibody blocking experiments, the relevant phosphopeptide was added to the 1:3,000 dilution of antiserum at 0.25 μg/ml (for phospho-S469 [pS469], pS487, and pS499) or 0.025 μg/ml (for pS493).
Generation of phosphospecific antibodies.
The phosphopeptides C+GSNYALKMN(pS469)PSQSSP-NH2, C+NPGQPTSML(pS487)PRHR-NH2, PRHRM(pS493)PGVAGSPRI+C-NH2, and PGVAG(pS499)PRIPPSQFS+C-NH2 were synthesized and purified by high-pressure liquid chromatography (HPLC) by Anaspec. Peptide conjugation to keyhole limpet hemocyanin (KLH), rabbit immunizations (118-day protocol; 3 rabbits/peptide), prebleed and three production bleeds, and antiserum testing by enzyme-linked immunosorbent assay (ELISA) were performed by Covance. Antisera were further tested by immunoblotting against endogenous GRIP1 in U2OS-rGR cells and relevant phosphopeptide blocking (see “Immunoblotting,” above), and sera derived from one rabbit per group were selected.
Immunoprecipitations and MS.
U2OS-rGR cells were treated as described in the Fig. 3 legend without dex (∼8 × 108 cells/condition), and GRIP1 was immunoprecipitated with 50 μg antibody/condition (Abcam). After separation on SDS-PAGE and Coomassie blue staining, the GRIP1 band was excised and subjected to in-gel reduction, alkylation, and trypsin digestion (16 h at 37°C). Mass spectrometry (MS) analysis was performed at the Rockefeller University Proteomics Resource Center. In brief, generated peptides were extracted with 50% acetonitrile and 0.1% trifluoroacetic acid (TFA) twice and SpeedVac dried. For liquid chromatography-tandem MS (LC–MS-MS) analysis, the peptide mixture was separated by a 60-min gradient elution with the Dionex U3000 capillary–nano-HPLC system (Dionex, Sunnyvale, CA), at a flow rate of 0.250 μl/min, that was directly interfaced with the Thermo-Fisher LTQ-Orbitrap mass spectrometer (Thermo Fisher, San Jose, CA), operated in data-dependent scan mode. The analytical column was a homemade fused-silica capillary column (75-μm inner diameter, 100-mm length; Upchurch, Oak Harbor, WA) packed with C-18 resin (300 A, 5 μm; Varian, Palo Alto, CA). Mobile phase A consisted of 0.1% formic acid, and mobile phase B consisted of 100% acetonitrile and 0.1% formic acid. The 60-min gradient at a 0.250-μl/min flow rate for solvent B went from 0 to 55% in 30 min and then in 10 min to 80%. The experiment consisted of a single full-scan mass spectrum in the Orbitrap (400 to 1,600 m/z, 30,000 resolution) followed by six data-dependent MS-MS scans in the ion trap at 35% normalized collision energy. Data were analyzed with MASCOT software and curated manually.
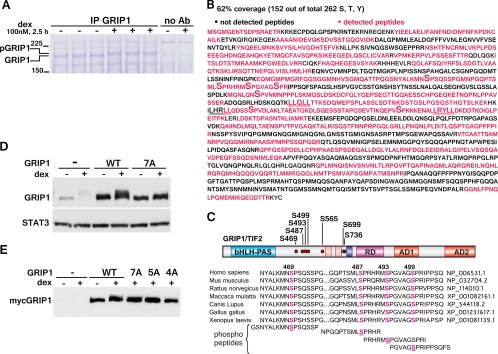
Fig 3.
The identification of GRIP1 phosphorylation sites by MS. (A) U2OS-rGR cells (∼8 × 108 cells/condition) were treated with or without dex, and GRIP1 was immunoprecipitated with 50 μg GRIP1 antibody/condition. After separation on SDS-PAGE gel, the proteins were visualized by Coomassie blue staining. Ab, antibody. (B) MS analysis of GRIP1. Percent coverage was calculated from the results of two independent experiments, and the MS-detected peptides are shown in magenta. LXXLL motifs are underlined; phosphorylated serines are marked by a large “S.” (C) GRIP1 phosphosites identified by MS. The GRIP1 diagram depicts the bHLH-PAS domain, NR boxes, RD, and AD 1 and 2. Six dex-inducible (red dots) and one constitutive (blue dot) phosphorylation site are marked. The alignment of the GRIP1 S469/S487/S493/S499 phosphosite cluster between different species and the peptides used for raising phosphosite-specific antibodies are shown. (D, E) MS-identified phosphosites mediate the GRIP1 mobility shift. myc-tagged WT GRIP1 or the 7A (all identified sites), 5A (S469/487/493/499/565A), or 4A (S469/487/493/499A) mutant was transfected into U2OS-rGR cells (250 ng of GRIP1 plasmid/250,000 cells, overnight), which were then untreated or treated with dex for 2 h and harvested, and GRIP1 electrophoretic mobility was evaluated by immunoblotting with antibodies to GRIP1 (D) and the myc tag (E).
ChIP analysis.
U2OS-rGR cells were seeded into 15-cm dishes and treated the following day as described in the figure legends. Chromatin immunoprecipitations (ChIPs) were performed as described in reference 35 except that formaldehyde cross-linking was done for 10 min at room temperature and sonication was performed using the Bioruptor (Diagenode) according to the manufacturer's instructions. Real-time qPCR was performed with the following primers, using amplification of the 28S rRNA gene as a normalization control: hPER1_−2173_F, ATCCCTGGCTTTGGGACAGGACG; hPER1_−2072_R, CTTGGGAACATCATGTTCTCTTGGC; hPER1_+473_F, GCTGGTTCCTGCTGTTGGCCACA; hPER1_+601_R, CAGAAGACACACAACAGCCAACAGATC; hMT2A_−1197_F, CCTCACTAAACTTTCACTGTGGCAATC; hMT2A_−1077_R, GCCTTAGATCGTCAACCTTGGAAGG; hGILZ_−1869_F, AGTGAATGTTCTTGATGACCCATAAGTATAG; hGILZ_−1744_R, GGACATTCTGTTAACTTTAAGACACAACCTC; hIGFBP1_+1416_F, CCATACCCTGATAGGGTGTTAGAGTG; hIGFBP1_+1561_R, CTGTGGAAATCCATCTCCTTGAGGATC; h28S_rRNA_F, GATCCTTCGATGTCGGCTCTTCCTATC; and h28S_rRNA_R, AGGGTAAAACTAACCTGTCTCACG.
RESULTS
GRIP1 is phosphorylated in response to glucocorticoids.
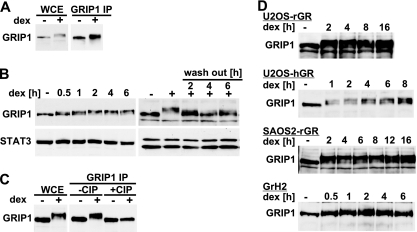
U2OS human osteosarcoma cells with stably integrated wild-type (WT) rat GR (U2OS-rGR) are commonly used for studying GR signaling and transcriptional regulation (8, 33). A 2-h treatment of these cells with a synthetic glucocorticoid, dexamethasone (dex), produced an unexpected dramatic change in the electrophoretic mobility of the transcriptional coregulator GRIP1 toward higher-molecular-weight species (Fig. 1A, left). The identity of the shifted band was further confirmed by immunoprecipitation followed by immunoblotting with GRIP1-specific antibodies (Fig. 1A, right). In a time course experiment, the steady-state total GRIP1 protein levels remained constant, whereas a GRIP1 mobility shift was apparent by 30 min of dex stimulation, plateaued by 1 to 2 h, and persisted for at least 16 h of continuous dex exposure (Fig. 1B and D). The dex-induced GRIP1 mobility shift was substantially reduced 2 h after dex washout (Fig. 1B), suggesting that the process is reversible and tightly regulated.
Fig 1.
GRIP1 is phosphorylated in response to dexamethasone. (A) U2OS-rGR cells were treated with vehicle (−) or 100 nM dex (+) for 2 h. Whole-cell extracts (WCE) or GRIP1 immunoprecipitates (IP) were prepared, and GRIP1 was detected by immunoblotting. (B) GRIP1 modification is dynamic and reversible. U2OS-rGR cells were treated with 100 nM dex for indicated times (left) or treated with dex for 2 h followed by incubation in dex-free medium for indicated times (right). GRIP1 was detected as described for panel A; immunoblotting for STAT3 was used as a loading control. (C) GRIP1 is phosphorylated in a dex-dependent manner. U2OS-rGR cells were treated as shown, and GRIP1 was immunoprecipitated, treated with CIP (50 U at 37°C for 1 h), and detected by immunoblotting. (D) Glucocorticoid-induced GRIP1 phosphorylation occurs in multiple cell types and species. U2OS-rGR, U2OS-hGR, SAOS2-rGR and GrH2 cells were treated with 100 nM dex for indicated times and harvested, and GRIP1 was detected by immunoblotting.
As changes in electrophoretic mobility are often indicative of posttranslational modifications, we used calf intestinal phosphatase (CIP) to remove possible phosphate groups from GRIP1 immunoprecipitates. CIP treatment did not affect the mobility of GRIP1 in untreated cells; however, it fully reversed the dex-induced GRIP1 shift (Fig. 1C), suggesting that GRIP1 is phosphorylated in response to glucocorticoids.
Importantly, a similar sustained shift in GRIP1 electrophoretic mobility was apparent in U2OS-hGR cells ectopically expressing human GR and in SAOS2-rGR osteosarcoma cells (8, 33), as well as in rat hepatoma cells (GrH2), expressing GR endogenously (Fig. 1D), suggesting that glucocorticoid-dependent GRIP1 phosphorylation is not restricted to a specific cell type, ectopically introduced GR, or species.
Glucocorticoid-induced GRIP1 phosphorylation requires the GR-GRIP1 interaction.
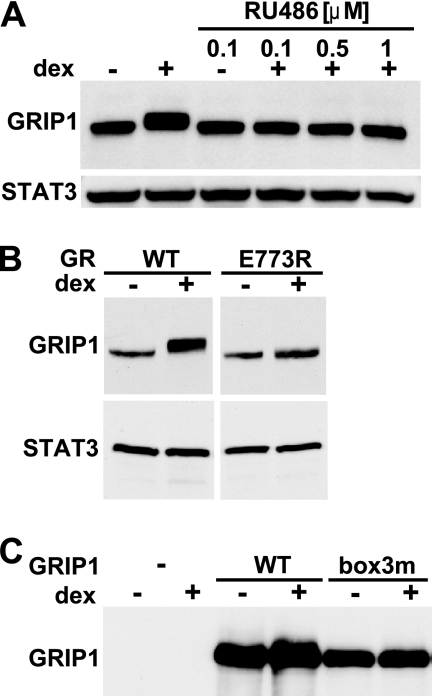
Because GRIP1 was phosphorylated in response to glucocorticoid signaling, we tested whether a physical interaction between GR and GRIP1 was necessary for this modification to occur. We initially used a synthetic GR antagonist, RU486, which occupies the same ligand-binding pocket of GR as dex but promotes a GR conformation that is incompatible with the GRIP1 interaction (6, 10). RU486 treatment not only failed to alter GRIP1 mobility but also reversed dex-induced GRIP1 phosphorylation (Fig. 2A). To rule out possible indirect effects of RU486, we took advantage of U2OS cells stably expressing the GR E773R “charge clamp” AF2 mutant which is competent for ligand binding but fails to bind GRIP1 (10, 34). Figure 2B shows that in U2OS-rGR E773R cells, the GRIP1 dex-dependent mobility shift was abolished. Finally, we disrupted the reciprocal side of the GR-GRIP1 interface and assessed the effect of dex on the GRIP1 NR box-3 mutant that is deficient for GR binding. We found that the transiently overexpressed GRIP1 NR box-3 mutant did not undergo mobility shift in response to dex, although the WT did (Fig. 2C). Of note, the overexpressed WT GRIP1 of either human or mouse origin does not shift as completely as the endogenous protein (Fig. 2C and data not shown), probably because either the kinase or GR becomes limiting. Combined, these results indicate that GRIP1 phosphorylation depends on the glucocorticoid-induced GR-GRIP1 interaction.
Fig 2.
GRIP1 phosphorylation requires a physical interaction with agonist-bound GR. (A) RU486 abolishes GRIP1 phosphorylation. U2OS-rGR cells were treated with 100 nM dex, 100 nM RU486, or dex plus RU486 (0.1 to 1 μM) for 2 h. GRIP1 and STAT3 were detected as described for Fig. 1B. (B) GRIP1 dex-induced phosphorylation requires the GR GRIP1-binding surface. U2OS cells stably expressing WT or E773R rGR were treated with or without dex, and GRIP1 mobility was assessed by immunoblotting. (C) The GRIP1 NR box-3 is required for GRIP1 phosphorylation. U2OS-rGR cells transiently transfected with an empty vector (−) or pcDNA3-GRIP1 expressing the mouse WT or NR box-3 mutant (box3m) GRIP1 were treated with or without dex for 2 h, and GRIP1 mobility was assessed by immunoblotting. At the exposure time selected, only ectopically overexpressed GRIP1 is detectable.
Identification of the GRIP1 sites subject to glucocorticoid-dependent phosphorylation.
To identify the sites of dex-induced phosphorylation in endogenous GRIP1 in its natural mammalian cell context, we performed large-scale immunoprecipitations and mass spectrometry (MS) analysis of GRIP1 from untreated and dex-treated U2OS-rGR cells. Using 8 × 108 cells and 50 μg of anti-GRIP1 antibody per condition, we detected a dex-dependent shift in GRIP1 immunoprecipitates by Coomassie blue staining (Fig. 3A). GRIP1 bands were excised from the gel and subjected to tryptic digestion followed by MS analysis. We recovered ∼62% of the 1,464-amino-acid (aa) human GRIP1 protein in MS-identified peptides (Fig. 3B), which is a considerable degree of coverage given that parts of the GRIP1 polypeptide (e.g., the long peptide at the extreme C terminus) lack trypsin cleavage sites and, thus, are too long to be recovered by MS. Our analysis identified six novel dex-dependent phosphorylation sites (S469, S487, S493, S499, S565, and S736), and one residue (S699) was phosphorylated constitutively (Fig. 3C). The presence of multiple phosphorylation sites (phosphosites) is in agreement with the intermediate GRIP1 mobility shift observed at an early (30 min) time point of dex treatment and the presence of multiple bands probably corresponding to several differentially phosphorylated GRIP1 species in dex-treated cells (Fig. 1B and D). All sites identified are S-P motifs that are conserved in mouse, rat, and human (Fig. 3C), which is consistent with the observation that GRIP1 undergoes the dex-induced mobility shift in all three species (Fig. 1D and data not shown). Interestingly, the site conservation extends to nonmammalian vertebrates, including Gallus gallus and Xenopus laevis (Fig. 3C).
To establish whether phosphorylation of the MS-identified sites was responsible for the GRIP1 electrophoretic mobility shift, we mutated these serine residues individually or in combination to nonphosphorylatable alanine (A) and transfected the myc-tagged GRIP1 derivatives into U2OS-rGR cells. The compound mutation of all seven sites (7A) prevented the dex-induced GRIP1 shift (Fig. 3D); furthermore, a substitution of only five (5A) or a cluster of four (4A) closely spaced sites (S469/487/493/499A) was sufficient to abolish the shift (Fig. 3D and E). Any individual or double mutations within this cluster resulted in a partial dex-induced shift, suggesting that phosphorylation of all four sites collectively accounts for the observed shift in GRIP1's mobility.
A cluster of newly identified GRIP1 phosphorylation sites is modified in various cell types and species.
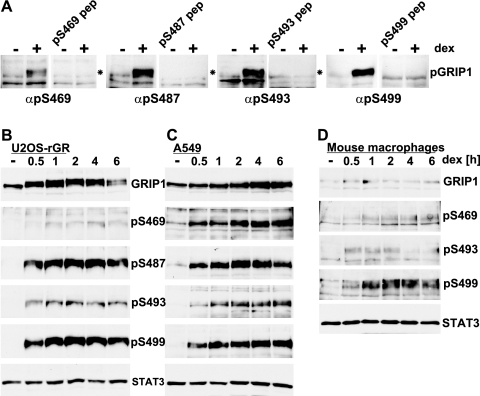
Although mutagenesis studies implicated the MS-identified phosphosites in the GRIP1 mobility shift, we sought to directly assess their phosphorylation in endogenous GRIP1 in vivo. Thus, we generated phosphosite-specific polyclonal antibodies recognizing individually phosphorylated S469, S487, S493, and S499 using 14- to 16-aa synthetic phosphopeptides corresponding to human GRIP1 for immunizations. Of note, while all four sites are conserved in multiple species (Fig. 3C), the peptide containing the S487–P488 motif differs between mouse and human GRIP1 by 4 amino acids, which will preclude the cross-reactivity of our anti-pS487 antibody with mouse GRIP1. Using these phosphospecific antibodies, we observed a striking dex-dependent GRIP1 phosphorylation in U2OS-rGR cells at all four sites, with kinetics coinciding with the mobility shift of total GRIP1 (Fig. 4A and B); in each case, site-specific reactivity against phospho-GRIP1 was blocked by the relevant phosphopeptide (Fig. 4A).
Fig 4.
Detection of MS-identified GRIP1 phosphorylation sites using phosphospecific antibodies. (A) U2OS-rGR cells were untreated or treated with dex for 2 h, and the extracts were subjected to immunoblotting using 1:3,000 dilutions of the indicated antisera in the absence or presence the relevant blocking phosphopeptides. pep, peptide; α, anti. (B to D) U2OS-rGR (B), A549 (C), or primary mouse bone marrow-derived macrophages (D) were treated with dex for indicated periods of time, and whole-cell extracts were prepared. Immunoblotting was performed with antibodies to total GRIP1, antisera to indicated phosphosites, and STAT3 to confirm equal loading.
Interestingly, in A549 human lung epithelial cells, primary mouse macrophages, or HeLa human cervical cells, the GRIP1 mobility appeared relatively unchanged by dex (Fig. 4C and D and data not shown). Although in principle, these findings might reflect cell specificity, an equally plausible interpretation is that a mobility shift reflects the fraction of GRIP1 molecules that have been phosphorylated in response to dex, which in turn is a function of the “signal strength,” determined in part by the relative amounts of GR and GRIP1 in a cell. Consequently, a change in GRIP1 mobility would be visible in cells expressing comparatively high levels of GR, e.g., U2OS-rGR or GrH2 cells, whereas in other cell types, a smaller subpopulation of phosphorylated GRIP1 might be undetectable in a total pool of the protein. Corroborating the latter scenario, immunostaining with GRIP1 phosphosite-specific antibodies revealed that GRIP1 is phosphorylated in response to dex in A549 cells, mouse macrophages, and HeLa cells (Fig. 4C and D and data not shown); notably, in addition to a dramatic increase in band intensity, we observed a slight gradual shift of phosphorylated GRIP1 toward higher-molecular-weight species. The conservation of these four glucocorticoid-inducible phosphorylation sites in multiple cell types and species strongly suggested their potential functional significance.
GRIP1 phosphorylation in vivo is mediated by CK2 and CDK9.
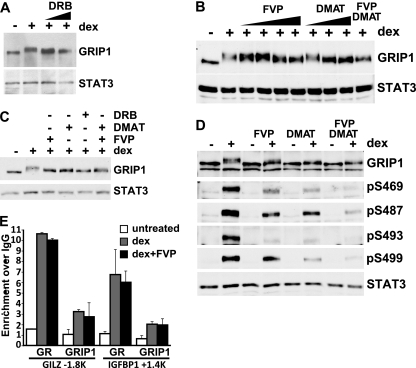
To begin to identify the protein kinases that may phosphorylate GRIP1, we screened a panel of kinase inhibitors for their ability to block the dex-induced mobility shift of GRIP1. As shown in Fig. 5A, cotreatment of U2OS-rGR cells with DRB attenuated the dex-dependent shift but did not block it completely. Since, at the concentrations used, DRB is known to inhibit primarily casein kinase 2 (CK2) and cyclin-dependent kinase 9 (CDK9) (and inhibitors to other CDKs failed to appreciably alter the dex-induced GRIP1 shift [data not shown]), we tested more specific inhibitors for these kinases, i.e., DMAT and flavopiridol (FVP), respectively. Both compounds inhibited the GRIP1 mobility shift in a dose-dependent manner, yet neither inhibitor was able to abrogate the shift when administered simultaneously with dex (Fig. 5B) or after a 2-h pretreatment prior to adding dex (Fig. 5C). Similarly, a simultaneous treatment with DMAT plus FVP failed to block the dex-induced shift completely (Fig. 5B and C), suggesting that additional kinases are probably involved.
Fig 5.
CK2 and CDK9 target a subset of MS-identified GRIP1 phosphorylation sites in vivo. (A) U2OS-rGR cells were untreated or treated for 2 h with 100 nM dex with or without DRB (50 or 100 μM), and GRIP1 and STAT3 were detected by immunoblotting. (B) U2OS-rGR cells were untreated or treated for 2 h with 100 nM dex with or without FVP or DMAT in increasing concentrations (5, 10, 50, and 100 nM and 1, 5, and 10 μM, respectively) or 100 nM FVP plus 10 μM DMAT together, and GRIP1 and STAT3 were detected by immunoblotting. (C) U2OS-rGR cells were untreated or pretreated with 100 nM FVP, 10 μM DMAT, 200 μM DRB, or FVP plus DMAT together, as indicated, for 2 h, followed by the addition of 100 nM dex for an additional 2 h, and GRIP1 and STAT3 were detected by immunoblotting. (D) U2OS-rGR cells were incubated for 2 h with (+) or without (−) 100 nM dex in the absence or presence of FVP, DMAT, or FVP plus DMAT (following pretreatment with inhibitors for 45 min), as indicated, and immunoblotting was performed with antibodies to GRIP1, antisera to indicated GRIP1 phosphosites, and STAT3. (E) U2OS-rGR cells were untreated or pretreated for 30 min with 100 nM FVP, followed by the addition of 100 nM dex for 30 min, and ChIPs were performed with antibodies to GR (35), GRIP1 (Bethyl Laboratories), or normal rabbit IgG as negative control. Genomic regions corresponding to the GILZ and IGFBP-1 GREs at approximately kb −1.8 and +1.4, respectively, were amplified and normalized to corresponding signals at the unrelated 28S rRNA gene, and the results are expressed relative to the signal obtained with normal IgG (set to 1). Error bars show standard errors of the mean.
To examine whether any of the identified GRIP1 phosphosites served as substrates for CK2- and/or CDK9-dependent phosphorylation, we treated U2OS-rGR cells with DMAT or FVP or a combination of the two with and without dex, and assessed phosphorylation using antibodies to pS469, pS487, pS493, and pS499. As shown in Fig. 5D, both inhibitors individually attenuated dex-induced phosphorylation of each of the four phosphosites to various degrees, and DMAT plus FVP together had an additive effect, although residual phosphorylation was still detectable. Importantly, a decrease in GRIP1 phosphorylation did not result from global effects of kinase inhibitors on the ability of GR or GRIP1 to be recruited to target GREs. Indeed, the apparent dex-dependent occupancy of GR and GRIP1 at the established GREs of the GILZ and IGFBP-1 genes (36) was not impaired by either FVP (Fig. 5E) or DRB (not shown). We conclude that both CK2 and CDK9 contribute to phosphorylation of the four serines within the GRIP1 phosphosite cluster in vivo; however, we cannot exclude the involvement of additional kinases.
GRIP1 phosphorylation potentiates GR-mediated activation of transcription.
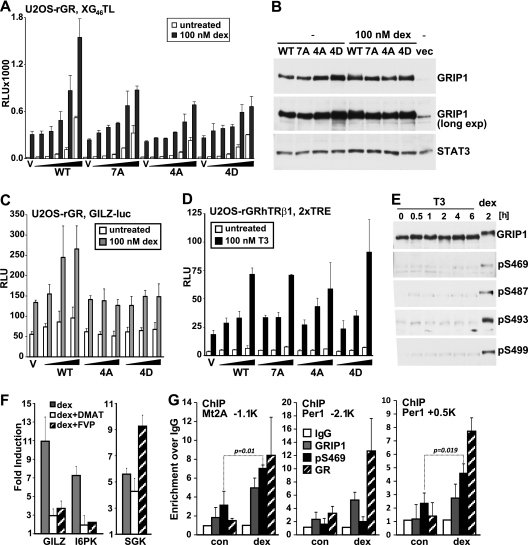
Although GRIP1 serves as a coregulator for a growing number of transcription factors, we reasoned that dex- and GR interaction-dependent modifications may affect the activity of GR transcriptional regulatory complexes. We therefore compared the ability of the overexpressed WT GRIP1, 7A mutant and 4A mutant to potentiate GR-mediated activation. Because U2OS cells express a large amount of endogenous GRIP1, detecting any effect of overexpressed GRIP1 could be challenging. In addition, attributing the effects of overexpressed GRIP1 to a specific interaction with GR in the context of complex genomic sites bound by multiple regulators could be misleading. In contrast, GR-responsive reporters provide an excess of GREs unlikely to be saturated with endogenous GRIP1 or to be bound by non-GR transcription factors, thus enabling a sensitive and specific readout for the effect of overexpressed GRIP1 derivatives on GR transcriptional activation. In U2OS-rGR cells, WT GRIP1 potentiated the activity of the 2×GRE luciferase reporter XG46TL in a dose-dependent manner (Fig. 6A). Interestingly, GRIP1 7A and 4A mutants failed to potentiate GR activity at WT GRIP1 levels, although the expression of all derivatives was equivalent (Fig. 6A and B). Consistent with the conservation of GRIP1 phosphorylation in murine macrophages (Fig. 4D), similar results were obtained in RAW264.7 mouse macrophage-like cells (not shown). Interestingly, overexpression of GRIP1 bearing mutations of the four serine sites to aspartic acid (4D), which in some cases mimics phosphorylation, resulted in a partial loss-of-function phenotype similar to that of the A mutants (Fig. 6A), suggesting that the introduction of a negative charge at phosphosites is not sufficient for GRIP1 coactivator properties.
Fig 6.
GRIP1 phosphorylation facilitates its ability to serve as a GR coactivator. (A, C, and D) U2OS-rGR cells (A, C) and U2OS-rGRhTRβ1 cells (D) were transfected with increasing amounts of WT, 7A, 4A, or 4D GRIP1-expressing plasmid at 0 to 80 ng/25,000 cells along with the XG46TL reporter plasmid (A), 0 to 40 ng/25,000 cells and the GILZ-luc reporter plasmid (C), or 0 to 5 ng/25,000 cells and the 2×TRE reporter plasmid (D) (total amount of expression plasmid is equalized with empty vector, V) and treated with dex (A, C) or T3 (D) overnight as indicated, and luciferase activity was measured, normalized to total protein concentration, and expressed as relative luminescence units (RLU). Each plot is representative of three or more independent experiments; error bars show standard deviations (SD). (B) U2OS-rGR cells transfected with indicated GRIP1 derivatives or empty vector (vec) were treated with or without dex and harvested, and immunoblotting was performed with antibodies to GRIP1 or STAT3. An overexposed blot (long exp) is shown to visualize endogenous GRIP1. (E) U2OS-rGRhTRβ1 cells were treated with 100 nM T3 for indicated times or with dex for 2 h, and extracts analyzed by immunoblotting with total or phosphospecific GRIP1 antibodies as described for Fig. 4. (F) U2OS-rGR cells were untreated or pretreated with 10 μM DMAT or 100 nM FVP, as indicated, for 30 min, followed by the addition of 100 nM dex for 2 h. Total RNA was isolated, reverse transcribed, and subjected to real-time qPCR with primers for indicated genes using amplification of β-actin as an internal control. The expression of GILZ, I6PK, and SGK for each condition is expressed as fold induction over the values with no dex (set to 1). Shown are averages of the results of two independent biological replicates; error bars depict SD. (G) U2OS-rGR cells were untreated (con) or treated with dex for 30 min, and ChIPs were performed with antibodies to GR, GRIP1, or pS469 antiserum or normal rabbit IgG as negative control. Genomic regions corresponding to the Mt2A GRE at kb −1.1 and Per1 GREs at kb −2.1 or +0.5 were amplified, normalized to corresponding signals at the unrelated 28S rRNA gene, and expressed relative to the signals obtained with normal IgG for each locus (set to 1). Shown are averages of the results for three independent biological replicates; error bars depict SD. P values for pS469 (indicated) are calculated using the two-tailed t test.
To assess the role of GRIP1 phosphorylation in GR-dependent activation of an established GR-responsive target, we evaluated the activity of a luciferase reporter driven by the 1-kb fragment derived from the regulatory region of the human GILZ gene, containing four established GREs in their natural albeit nonchromatinized context (41). As shown in Fig. 6C, phosphosite substitution with either A or D similarly attenuated the ability GRIP1 to enhance transcription from the GILZ-derived reporter, suggesting that GRIP1 phosphorylation at the cluster of four sites is required for the optimal GR activation from natural GREs.
To examine whether phosphorylation-deficient GRIP1 mutants retained the ability to potentiate the activity of other NRs, we utilized a derivative of the U2OS-rGR line stably expressing human thyroid hormone receptor β1 (hTRβ1), U2OS-rGRhTRβ1, which provides a nearly identical cellular context for assessing GRIP1 functions and in which overexpressed GRIP1 has been previously shown to serve as an hTRβ1 coactivator (35). We found that all GRIP1 derivatives were equally able to stimulate T3-induced activation of the 2×TRE reporter (Fig. 6D), indicating that phosphorylation-deficient mutants display a specific defect for coactivating GR rather than a generally impaired ability to facilitate transcription. Furthermore, T3 treatment of U2OS-rGRhTRβ1 cells failed to produce a mobility shift of endogenous GRIP1, and no T3-induced phosphorylation of S469, S487, S493, or S499 was detected with phosphospecific antibodies, while dex treatment of the same cells triggered GRIP1 phosphorylation (Fig. 6E), consistent with the functional data.
To evaluate the effect of phosphorylation of endogenous GRIP1 on native GR target genes, we used DMAT and FVP to inhibit CK2 and CDK9 activity, respectively, and analyzed the expression of native GR targets. Each inhibitor attenuated the dex-dependent induction of GILZ (Fig. 6F), consistent with the results of the GILZ-luc reporter assay (Fig. 6C), and of several other GR-responsive genes (30, 34), including I6PK (Fig. 6F), Per1, and IGFBP-1 (not shown). This effect, however, did not reflect the global inability of GR to be recruited to GREs (Fig. 5E) or the loss of its transcriptional regulatory properties, as hormonal induction of the well-established GR target SGK was preserved in the presence of each inhibitor (Fig. 6F), indicating a requirement for GRIP1 phosphorylation at a specific subset of GR-regulated genes.
If GRIP1 phosphorylation affects the regulation of endogenous GR target genes in vivo, GRIP1 phosphoisoforms are expected to be recruited to a subset of natural GREs. Using pS469-specific antiserum, we detected dex-induced enrichment of pS469-GRIP1 at the metallothionein 2A (Mt2A) GRE at approximately kb −1.1 (30) relative to the transcription start site, coinciding with the recruitment of GRIP1 and GR (Fig. 6G, left). Strikingly, while the two established GREs (at approximately kb −2.1 and +0.5) of the Per1 gene were both occupied by GRIP1 and GR in response to a 30-min dex treatment, GRIP1 was S469 phosphorylated at the kb +0.5 GRE only (Fig. 6G, middle and right), suggesting that GRIP1 phosphoisoforms preferentially engage in distinct GR regulatory complexes bound at different GR binding sites even at the same gene.
DISCUSSION
Coregulators are essential components of mammalian transcription complexes, which provide a functional and/or physical link between the activated regulator and basal machinery or chromatin. With an estimated 1,500 transcription factors and up to 350 coregulators encoded by the human genome (28, 39), most coregulators are shared by multiple factors, while the same factor may rely on different coregulators at different target gene-specific and physiological contexts. In the absence of a pairwise factor-cofactor relationship, one of the fundamental questions is how cofactors are “chosen” or “choose” to engage in a particular transcriptional regulatory pathway versus another. Here, we report a panel of novel, glucocorticoid-inducible, GR interaction-dependent posttranslational modifications of GRIP1 which collectively modulate GR-mediated transcription. This finding contrasts with a view whereby coregulator utilization by a given DNA binding factor is “passive” and, in the context of a specific genomic binding site, is dictated primarily by the availability. Instead, we envision a scenario whereby ligand-activated GR, in addition to binding GRIP1, recruits GRIP1 kinases, thus imparting covalent modifications to its own cofactor, perhaps favoring its preferential function at GR transcription complexes over those of other regulators. Furthermore, glucocorticoid-induced phosphorylation at multiple sites and by multiple kinases may combinatorially affect GRIP1 activities at specific genomic sites or protein-protein interactions, thereby tailoring GR actions to specific gene expression programs.
The identity of such glucocorticoid-responsive GRIP1 kinases requires an extensive further investigation. At least two kinases, CK2 and CDK9, appear to target the four GRIP1 phosphosites (S469, S487, S493, and S499) in vivo, but additional kinases are involved, and the contribution of each enzyme to GRIP1 phosphorylation likely differs depending on the specific GR regulatory complex that assembles at a specific GRE. In addition, we cannot as yet exclude the possibility that the involvement of either CK2 or CDK9 is indirect, perhaps by promoting the recruitment or function of another kinase. This possibility remains to be addressed using in vitro and, particularly, ChIP approaches, which will also delineate whether the same phosphosite serves as a substrate for distinct enzymes in a GRE- or cell-specific manner and what triggers their differential engagement.
Of the six identified dex-inducible GRIP1 phosphosites, only S736 was previously reported as a target for mitogen-activated protein kinase (MAPK) family members in response to epidermal growth factor (EGF), which was linked to enhanced activities of estrogen- and androgen-responsive reporters due at least in part to increased GRIP1 protein levels (12, 13, 27). While these findings suggested that modification of S736 is a common regulatory step for several pathways, phosphorylation of this site did not alter GRIP1 level or contribute to dex-induced mobility shift in our system and, consistently, the S736A substitution did not exacerbate the phenotype of the 4A mutant in GR transcription assays. Thus, the functional significance of pS736 with respect to GR-mediated gene activation is unclear and may be gene specific.
What are the mechanistic consequences of GR-dependent GRIP1 phosphorylation? Phosphorylation is often a prerequisite for other posttranslational modifications (e.g., ubiquitination, sumoylation, acetylation, or methylation) which, in principle, could modulate GRIP1 properties. Indeed, a series of elegant studies from the O'Malley lab demonstrated that phosphorylation of NCoA3 (also known as SRC3), another p160 family member, primes it for ubiquitination and turnover (1, 26, 42). Interestingly, GRIP1 was shown to be degraded by the 26S proteasome, in silico analysis of GRIP1 revealed four potential PEST sequences usually required for degradation, and GRIP1 ubiquitin-conjugating enzymes were identified (3, 17, 44). However, glucocorticoid- or phosphorylation-dependent GRIP1 ubiquitination or degradation thus far appear unlikely, as the steady-state GRIP1 protein level was unaffected by dex for up to 16 h, the expression of GRIP1 4A, 4D, 5A, and 7A phosphosite mutants and their ability to enhance hTRβ1-dependent activation were indistinguishable from those of the WT, and our MS analysis detected no evidence of glucocorticoid-dependent ubiquitination. And yet, a compound disruption of GRIP1 phosphosites or inhibition of GRIP1 phosphorylation with kinase inhibitors selectively impaired its ability to serve as a GR coactivator, suggesting that GRIP1 phosphorylation may affect the properties of GR transcriptional activation complexes. Indeed, phosphorylation is a powerful modulator of protein-protein interactions; GR itself is a phosphoprotein which associates with several coregulators in a phosphorylation-dependent manner (16). The cluster of four GRIP1 phosphosites is located between the bHLH-PAS domain and NID, a domain for which no interacting partners have as yet been reported. Conceivably, phosphorylation triggers the formation of a novel functional domain or a binding surface for secondary coregulators. Notably, replacement of the phosphosite cluster with aspartic acid residues (4D) resulted in a coactivation-deficient mutant similar to the 4A mutant, corroborating the idea that a novel GRIP1 interface rather than a negative charge mediates the hormone-dependent assembly or function of GR transcriptional activation complexes.
Thus far, phosphosite mutations affected the ability of GRIP1 to serve as a GR coactivator; whether or not phosphorylation contributes to GRIP1 corepressor function is an intriguing question, as repression by GR is agonist dependent and, therefore, the trigger and interacting partner for GRIP1 are the same in both types of complexes. However, the overall architecture of the GR homodimer bound to a palindromic GRE versus GR-AP1 or GR–NF-κB “tethering” complexes is as divergent as the transcriptional regulatory outcomes of hormonal stimulation. Thus, regardless of the consequences of the phosphosite disruption for repression, determinants in addition to liganded GR, e.g., the genomic sites to which GR complexes are recruited, must dictate whether these residues become substrates for phosphorylation and, if so, which kinases and, ultimately, secondary cofactors associate with GRIP1 in each case. Our data on selective pS469-GRIP1 recruitment to individual GREs of the Per1 gene are consistent with this hypothesis.
Glucocorticoids act in a cell type-specific manner. Indeed, GR represses a number of cell survival genes and triggers apoptosis in U2OS cells, while it induces the expression of CDK inhibitors and cellular senescence in SAOS2 cells, another osteosarcoma cell line (33). This diversity is frequently attributed to the differential expression of GR coactivators and corepressors; however, glucocorticoid-induced GRIP1 phosphorylation may also be cell type specific. Although our phosphospecific antibodies revealed dex-induced phosphorylation at the same sites (S469, S487, S493, and S499) in many cell types, the kinetics of phosphorylation in macrophages appear to differ for individual sites: pS493 peaks by 30 min of dex treatment and decays thereafter, whereas pS469 and pS499 are maximal by 1 to 2 h and are sustained throughout the time course. Whether this dynamics reflects a sequential recruitment of kinases and phosphatases, which varies in different cell types, remains to be determined.
In addition to serving as a cofactor for GR and other NRs, GRIP1 is increasingly recognized as a coregulator for nonreceptor transcription factors. For example, we recently described an unexpected function of GRIP1 as a coactivator for IRF3 and the heterotrimeric ISGF3 complex (11, 31) that are responsible for Toll-like receptor 3 (TLR3)- and type I IFN-inducible gene expression, respectively. Given a unique role of GRIP1 in setting the balance between innate immune and immunosuppressive pathways, these findings underscore a requirement for the molecular cues enabling GRIP1 to discriminate between activating and inhibitory signals or directing it to specific factors or promoters. It is tempting to speculate that in response to IFN or TLR3 ligands, IRF family members recruit GRIP1 kinases with different site specificities, thus promoting distinct GRIP1 phosphopatterns which favor preferential GRIP1 recruitment to IFN target genes.
The identification of GRIP1 as a target for glucocorticoid-dependent phosphorylation points to a previously unrecognized mechanism of specificity in GR-driven gene transcription. Conceivably, certain GRIP1 phosphopatterns may serve as predictive markers of GR actions, such as the ability to suppress inflammation or, conversely, reveal the mechanisms underlying glucocorticoid resistance when GR expression per se is normal. For example, in severe asthma, resistance to glucocorticoid therapy has been correlated with altered GR nuclear localization and GRE binding (for a review, see reference 2). Yet, despite the critical involvement of coregulators in shaping GR transcriptional regulatory networks, the role of cofactors in glucocorticoid resistance remains unexplored. We speculate that GRIP1, a cofactor at the intersection of pro- and anti-inflammatory pathways, is a good candidate for functional alterations whereby a subtle molecular or structural change would shift the balance toward the inflammatory gene expression program, manifesting as resistance. Supporting this idea, in human airway smooth muscle cells, responsible for airway remodeling in asthma (20), a sustained cytokine signaling subverts GRIP1 to facilitate IRF1-dependent proinflammatory rather than GR target gene expression (5). Thus, a dissection of signal-specific GRIP1 modifications and an understanding of their contribution to the regulation of specific groups of genes may facilitate the design of GR ligands that bypass glucocorticoid resistance or display a more dissociated therapeutic profile.
ACKNOWLEDGMENTS
We thank J. Fernandez and H. Deng at the Rockefeller U Proteomics Resource Center for MS data analysis, M. Reily for technical assistance, M. Garabedian (NYU), S. Logan (NYU), L. Ivashkiv (HSS), S. Chen-Kiang (Weill Cornell), and R. Fisher (Mount Sinai) for providing kinase inhibitors, J.-C. Wang (UC Berkeley) for the GILZ-luc reporter construct, H. Samuels (NYU) for providing T3, and K. Zarember and R. Gupte for critical comments on the manuscript.
This work was supported by the grants from the NIH-NIAID and Kirkland Center to I.R. and from the American Heart Association to Y.C.
Footnotes
Published ahead of print 12 December 2011
REFERENCES
- 1. Amazit L, et al. 2007. Regulation of SRC-3 intercompartmental dynamics by estrogen receptor and phosphorylation. Mol. Cell. Biol. 27:6913–6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnes PJ. 2010. Mechanisms and resistance in glucocorticoid control of inflammation. J. Steroid Biochem. Mol. Biol. 120:76–85 [DOI] [PubMed] [Google Scholar]
- 3. Baumann CT, et al. 2001. The glucocorticoid receptor interacting protein 1 (GRIP1) localizes in discrete nuclear foci that associate with ND10 bodies and are enriched in components of the 26S proteasome. Mol. Endocrinol. 15:485–500 [DOI] [PubMed] [Google Scholar]
- 4. Belandia B, Orford RL, Hurst HC, Parker MG. 2002. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 21:4094–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhandare R, et al. 2010. Glucocorticoid receptor interacting protein-1 restores glucocorticoid responsiveness in steroid-resistant airway structural cells. Am. J. Respir. Cell Mol. Biol. 42:9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bledsoe RK, et al. 2002. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110:93–105 [DOI] [PubMed] [Google Scholar]
- 7. Chen D, et al. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174–2177 [DOI] [PubMed] [Google Scholar]
- 8. Chen W, Rogatsky I, Garabedian MJ. 2006. MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol. Endocrinol. 20:560–572 [DOI] [PubMed] [Google Scholar]
- 9. Chen YH, Kim JH, Stallcup MR. 2005. GAC63, a GRIP1-dependent nuclear receptor coactivator. Mol. Cell. Biol. 25:5965–5972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Darimont BD, et al. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Flammer JR, et al. 2010. The type I interferon signaling pathway is a target for glucocorticoid inhibition. Mol. Cell. Biol. 30:4564–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frigo DE, et al. 2006. p38 mitogen-activated protein kinase stimulates estrogen-mediated transcription and proliferation through the phosphorylation and potentiation of the p160 coactivator glucocorticoid receptor-interacting protein 1. Mol. Endocrinol. 20:971–983 [DOI] [PubMed] [Google Scholar]
- 13. Gregory CW, et al. 2004. Epidermal growth factor increases coactivation of the androgen receptor in recurrent prostate cancer. J. Biol. Chem. 279:7119–7130 [DOI] [PubMed] [Google Scholar]
- 14. Gronemeyer H, Gustafsson JA, Laudet V. 2004. Principles for modulation of the nuclear receptor superfamily. Nat. Rev. Drug Discov. 3:950–964 [DOI] [PubMed] [Google Scholar]
- 15. Heery DM, Kalkhoven E, Hoare S, Parker MG. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733–736 [DOI] [PubMed] [Google Scholar]
- 16. Hittelman AB, Burakov D, Iniguez-Lluhi JA, Freedman LP, Garabedian MJ. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18:5380–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoang T, et al. 2004. cAMP-dependent protein kinase regulates ubiquitin-proteasome-mediated degradation and subcellular localization of the nuclear receptor coactivator GRIP1. J. Biol. Chem. 279:49120–49130 [DOI] [PubMed] [Google Scholar]
- 18. Hong H, Kohli K, Garabedian MJ, Stallcup MR. 1997. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol. Cell. Biol. 17:2735–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeong JW, et al. 2006. The genomic analysis of the impact of steroid receptor coactivators ablation on hepatic metabolism. Mol. Endocrinol. 20:1138–1152 [DOI] [PubMed] [Google Scholar]
- 20. Johnson PR, et al. 2001. Airway smooth muscle cell proliferation is increased in asthma. Am. J. Respir. Crit. Care Med. 164:474–477 [DOI] [PubMed] [Google Scholar]
- 21. Kim JH, Li H, Stallcup MR. 2003. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol. Cell 12:1537–1549 [DOI] [PubMed] [Google Scholar]
- 22. Lee YH, Campbell HD, Stallcup MR. 2004. Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity. Mol. Cell. Biol. 24:2103–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. 2005. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl. Acad. Sci. U. S. A. 102:3611–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. 2002. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol. Cell. Biol. 22:3621–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lefstin JA, Yamamoto KR. 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392:885–888 [DOI] [PubMed] [Google Scholar]
- 26. Li C, et al. 2007. Specific amino acid residues in the basic helix-loop-helix domain of SRC-3 are essential for its nuclear localization and proteasome-dependent turnover. Mol. Cell. Biol. 27:1296–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lopez GN, Turck CW, Schaufele F, Stallcup MR, Kushner PJ. 2001. Growth factors signal to steroid receptors through mitogen-activated protein kinase regulation of p160 coactivator activity. J. Biol. Chem. 276:22177–22182 [DOI] [PubMed] [Google Scholar]
- 28. McKenna NJ, et al. 2009. Minireview: evolution of NURSA, the Nuclear Receptor Signaling Atlas. Mol. Endocrinol. 23:740–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nolte RT, et al. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-g. Nature 395:137–143 [DOI] [PubMed] [Google Scholar]
- 30. Reddy TE, et al. 2009. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 19:2163–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reily MM, Pantoja C, Hu X, Chinenov Y, Rogatsky I. 2006. The GRIP1:IRF3 interaction as a target for glucocorticoid receptor-mediated immunosuppression. EMBO J. 25:108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. 2002. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc. Natl. Acad. Sci. U. S. A. 99:16701–16706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rogatsky I, Trowbridge JM, Garabedian MJ. 1997. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol. Cell. Biol. 17:3181–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rogatsky I, et al. 2003. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc. Natl. Acad. Sci. U. S. A. 100:13845–13850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rogatsky I, Zarember KA, Yamamoto KR. 2001. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. EMBO J. 20:6071–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. 2007. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 3:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Surjit M, et al. 2011. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 145:224–241 [DOI] [PubMed] [Google Scholar]
- 38. Tsai MJ, O'Malley BW. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 63:451–486 [DOI] [PubMed] [Google Scholar]
- 39. Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. 2009. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 10:252–263 [DOI] [PubMed] [Google Scholar]
- 40. Voegel JJ, et al. 1998. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17:507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang JC, et al. 2004. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. U. S. A. 101:15603–15608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu RC, Feng Q, Lonard DM, O'Malley BW. 2007. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell 129:1125–1140 [DOI] [PubMed] [Google Scholar]
- 43. Xu J, Wu RC, O'Malley BW. 2009. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat. Rev. Cancer 9:615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yan F, Gao X, Lonard DM, Nawaz Z. 2003. Specific ubiquitin-conjugating enzymes promote degradation of specific nuclear receptor coactivators. Mol. Endocrinol. 17:1315–1331 [DOI] [PubMed] [Google Scholar]