Abstract
Cyclin-dependent kinase 16 (CDK16, PCTK1) is a poorly characterized protein kinase, highly expressed in the testis and the brain. Here, we report that CDK16 is activated by membrane-associated cyclin Y (CCNY). Treatment of transfected human cells with the protein kinase A (PKA) activator forskolin blocked, while kinase inhibition promoted, CCNY-dependent targeting of CDK16-green fluorescent protein (GFP) to the cell membrane. CCNY binding to CDK16 required a region upstream of the kinase domain and was found to be inhibited by phosphorylation of serine 153, a potential PKA phosphorylation site. Thus, in contrast to other CDKs, CDK16 is regulated by phosphorylation-controlled cyclin binding. CDK16 isolated from murine testis was unphosphorylated, interacted with CCNY, and exhibited kinase activity. To investigate the function of CDK16 in vivo, we established a conditional knockout allele. Mice lacking CDK16 developed normally, but male mice were infertile. Spermatozoa isolated from their epididymis displayed thinning and elongation of the annulus region, adopted a bent shape, and showed impaired motility. Moreover, CDK16-deficient spermatozoa had malformed heads and excess residual cytoplasm, suggesting a role of CDK16 in spermiation. Thus, CDK16 is a membrane-targeted CDK essential for spermatogenesis.
INTRODUCTION
The PCTAIRE kinases 1 to 3 belong to the family of cyclin-dependent kinases (CDKs) (29). About half of the 26 human CDK family members, mostly those involved in cell cycle regulation or transcription, are well understood in terms of their regulation and function, but little is known about the others. Among those poorly characterized CDK family members are the PCTAIRE (PCTK) and the closely related PFTAIRE (PFTK) kinases. The names of these kinases are derived from the primary sequences of an alpha-helix known to be involved in cyclin binding of other CDKs. Although the cyclin partners of PFTK1 and -2 and PCTK1 to -3 have not been fully revealed, they have recently been classified as cyclin-dependent kinases 14 to 18 (27).
These kinases are found only in the animal kingdom and are characterized by a highly conserved catalytic domain as well as N- and C-terminal extensions that are more divergent from or absent in other CDKs. These additional domains play important roles as they mediate protein-protein interactions (8) and were shown to regulate kinase activity (12). PCTAIRE and PFTAIRE sequence-containing kinases can also be found in some unicellular eukaryotes but are absent in yeasts and plants. Whether these related genes are true homologs is uncertain, because they lack either the typical N-terminal extensions (e.g., in kinetoplasts) or conserved domains within these extensions (e.g., in choanoflagellates). Insects appear to have only one member of these kinases, e.g., the Drosophila melanogaster CDK14 homolog L63, which is essential for development (42).
Vertebrate PCTKs, i.e., CDK16 to -18, are very similar and differ only within their N- and C-terminal domains. Human PCTK1/CDK16 was cloned more than 20 years ago (30), but its function and regulation have remained elusive. CDK16 mRNA and protein can be detected in many tissues and cell lines (7, 30). When immunoprecipitated from HeLa cells, CDK16 showed kinase activity toward myelin basic protein (MBP) that oscillated during the cell cycle with a peak in S and G2 phases (7). Several experiments suggested that PCTKs might be activated by a regulatory subunit (12, 16), but despite the identification of many interacting proteins (8), the identity of this postulated cyclin remained elusive. In contrast, Caenorhabditis elegans PCT-1 and the Drosophila PFTK1/CDK14 homolog, L63, have both been shown to interact with, and become activated by, the cyclin Y orthologues CYY-1 and CG14939, respectively (10, 34).
In human cell lines, CDK16 is phosphorylated at several residues, including Ser119 and Ser153, which have been shown to be substrates for protein kinase A (PKA) in vitro, and other residues in the N- and C-terminal extensions (9, 11, 12, 32, 36). Phosphorylation of S119 and S153 creates 14-3-3 consensus binding motifs, whose binding activity has been experimentally confirmed (12), but the function of this modification and 14-3-3 binding has not been resolved yet.
Although CDK16 activity has been shown to be cell cycle regulated, it remained uncertain whether CDK16 itself plays a role in cell cycle progression (7). The expression of PCTKs in neurons and differentiating spermatids (4, 21) points to functions unrelated to proliferation. Such a non-cell-cycle-related function has recently been discovered in neurons of PCT-1 mutant nematodes, which displayed a misbalance of antero- and retrograde axonal vesicle transport (34). Vertebrate PCTKs have also been implicated in the regulation of intracellular vesicles (25, 35) and in the translocation of glucose transporters (44) and neurite outgrowth (12).
In the mouse, CDK16 is expressed in various tissues with highest levels in brain and testis (5). In the brain, high levels of CDK16 are found in the cytoplasm of cerebellar Purkinje cells, as well as in cells of the hippocampus and the neocortex (5, 21). In testis, CDK16 expression increases during differentiation but is absent in mature spermatozoa, as CDK16 is retained within the residual body, a cytoplasmic compartment that is shed at the end of spermatogenesis (5). Here we describe a genetic approach to dissect the function of CDK16 in vivo and define its mode of activation and regulation.
MATERIALS AND METHODS
Reagents.
Chemicals, enzymes, and oligonucleotides were purchased from Sigma (Vienna, Austria), Promega (Mannheim, Germany), MWG Biotech (Ebersberg, Germany), and Microsynth AG (Balgach, Switzerland), unless stated otherwise. Full-length cDNA clones were obtained from Imagenes (Berlin, Germany), and small interfering RNA (siRNA) pools were from Dharmacon (Lafayette, CO). The following antibodies (Abs) were used: for CDK16, mouse monoclonal antibody (MAb) G6 (kindly provided by J. Gannon, CRUK, South Mimms, United Kingdom [12]) and rabbit polyclonal antibody (rbAb) JG46 (12) recognize the N-terminal residues of CDK16, while rbAb C-16 (SC-174; Santa Cruz Biotechnology Inc., Heidelberg, Germany) is specific for the C terminus; for CCNY, rbAb A302-376A-1 (Bethyl Laboratories Inc., Montgomery, TX); for pan14-3-3, rbAb K-19 (SC-629; Santa Cruz); for yellow fluorescent protein (YFP), rbAb SG4-1 (affinity purified, raised against full-length recombinant green fluorescent protein [GFP]); for FLAG, MAb M2 (F3165, Sigma); for CREB, mouse MAb 86B10 (Cell Signaling, Millipore), and for pS133CREB, rb MAb 87G3 (Cell Signaling); as mouse IgG control for immunoprecipitation (IP) experiments, purified mouse IgG2b κ (BioLegend Europe BV, Uithoorn, The Netherlands); and secondary anti-mouse and anti-rabbit horseradish peroxidase (HRP)-conjugated antibodies (Dako, Glostrup, Denmark). The CDK16 phosphoserine 153-specific antibodies were generated by immunizing rabbits with the peptide SRRLRRV S(PO3H2) LSEIG and purified by sequential purification using nonphosphorylated and phosphorylated peptide affinity chromatography (Eurogentech SA, Seraing, Belgium).
Cell lines and RNAi.
HeLa, U2OS, and HEK293 cells were grown as described previously (48). For RNA interference (RNAi) experiments, HeLa and 293A cells were transfected with 50 nM CCNY-specific Dharmacon ON-Target Plus siRNA pools for 24 h as described previously (3).
Plasmids, mutagenesis, and yeast two-hybrid screening.
For yeast two-hybrid screening, the open reading frame of CDK16 was amplified by PCR and subcloned into SpeI/PacI-digested plasmids pB27 and pB29 (Hybrigenics, Paris, France) for N- and C-terminal LexA DNA binding domain fusion proteins, respectively. Epitope tag and fluorescent protein expression vectors were generated using Gateway technology (Invitrogen). The open reading frames of human CDK16, CCNI, CCNK, CCNY, CCNYL1, and RINGO-A with and without stop codons were PCR amplified from pEF-YFP-PCTK1 (kindly provided by R. Gräser, Tumor Biology Center, Freiburg, Germany [12]), IRAUp969B1119D, IRATp970A0911D, DKFZp547F237Q, and IRATp970E1099D, respectively, and cloned into pDONR207 (Invitrogen). After sequence confirmation, the open reading frames were transferred into Gateway-adapted vectors for the expression of FLAG-tagged proteins (ΔT-FLAG-DEST) and fluorescent protein fusion proteins (pEGFP-N-DEST, pEGFP-C-DEST, pCherry-N-DEST, pECFP-N-DEST, and pECFP-C-DEST) in human cells. Mutagenesis was carried out using standard protocols (15, 23). All coding sequences were confirmed by DNA sequencing. Plasmid pB27-PCTK1 was used to carry out a yeast two-hybrid interaction screen (Hybrigenics), in which more than 14 million interactions were screened in an automated arrayed mating assay using yeasts transformed with a human testis cDNA GAL4-activation domain fusion protein library.
Immunoblotting and IP.
Immunoblotting was essentially performed as described previously (48). For overexpressed protein immunoprecipitation (IP), subconfluent HEK293A cells were transfected in 6-well plates with 1 μg YFP-CCNY (wild type [wt] or mutant) together with 3 μg FLAG-PCTK1 (wild-type or mutant) plasmid using Lipofectamine 2000 (Invitrogen). After 48 to 72 h, cells were harvested and lysed in IP buffer (150 mM NaCl, 20 mM NaF, 50 mM Tris, pH 7.4, 0.1% NP-40, 1 mM EGTA, and Roche complete protease inhibitors) for 1 h and spun down at 14,000 × g for 30 min, and supernatants (SN) were taken. Two hundred micrograms of total proteins was used for immunoprecipitation with 4.9 μg anti-FLAG M2 or 1.5 μg anti-YFP SG4-1 Ab coupled to Affiprep beads (Bio-Rad Laboratories, Vienna, Austria) for 1 to 2 h at 4°C. The beads were washed with IP buffer, and proteins were eluted with SDS sample buffer, boiled, resolved by SDS-PAGE, and subjected to immunoblotting. For endogenous CDK16 IP, 2 to 3 mg of testis and brain (CDK16+/0 and CDK16−/0) lysates (see below) was used for IP with 30 μg G6 Ab or with the same amount of IgG isotype control (coupled to Affiprep beads) for 2 h at 4°C.
Recombinant proteins.
CCNY was shuttled from pENTR-CCNY into a pMAL-DEST, a Gateway-compatible maltose-binding protein fusion protein expression vector (unpublished data), using LR clonase (Invitrogen). A CDK16 variant lacking the amino-terminal 106 residues (ND106) and harboring the S153A mutation was cloned into pET21d and expressed as hexahistidine-tagged protein (Novagen, EMD4 Biosciences). Proteins were transformed into Escherichia coli strain Arctic Express (Stratagene, Agilent Technologies, Santa Clara, CA) and induced with 100 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 18°C for 12 h. Recombinant proteins were purified from cleared lysates using amylose beads (NEB GmbH, Frankfurt am Main, Germany) and Ni-NTA beads (Qiagen, Hilden, Germany), respectively. Purified proteins were mixed and assayed for activity using MBP as a model kinase substrate.
Kinase assays.
For kinase assays, HEK293A cells were Lipofectamine 2000 transfected in 6-well plates with 1 μg YFP-CCNY together with 3 μg FLAG-CDK16 and CDK16 immunoprecipitated using FLAG M2 antibody-bound beads. After washing in IP buffer, half of the beads were boiled in SDS sample buffer to check for IP efficiency by immunoblotting, while the second half were washed once more with kinase buffer (15 mM EGTA, 25 mM NaF, 250 mM Na-β-glycerophosphate, and 5 mM dithiothreitol [DTT]); used for in vitro kinase assays, carried out at 30°C for 10 min after addition of 1 μg/ml MBP as substrate and [γ-32P]ATP and magnesium acetate; and analyzed by SDS-PAGE and autoradiography.
To detect endogenous CDK16 kinase activity, 200 μg of testis lysate (Cdk16−/0 or Cdk16+/0) was used to immunoprecipitate CDK16 using JG46 (or preimmune serum as negative control). The samples were incubated for 1 h before addition of 10 μl of protein A Sepharose (Pharmacia LKB Biotechnology, Uppsala, Sweden) and further incubation for 30 min. Samples were washed at 4°C three times in IP buffer and once in kinase buffer. The kinase assay was performed by adding 13 μg of desalted and dephosphorylated testis lysate as substrate, MnCl2, [γ-33P]ATP, and 20 μM cold ATP (adapted from reference 20) and incubated for 20 min at 30°C.
Knockout (KO) construct and gene targeting.
For targeting vector construction, a 13-kb Sv129 mouse genomic DNA fragment was cloned from a λ-KO2 (52) library by targeted insertion of a loxP-site-flanked tetracycline resistance cassette (TcR) into Cdk16 intron 6. After release of the plasmid from the phage clone and Cre-recombinase-mediated removal of TcR, a second loxP site along with an FLP recombination target (FRT) site-flanked neomycin resistance (NeoR) marker was inserted into intron 14 by recombineering in EL250 cells (52). After sequencing of exons 7 to 14, the conditional gene targeting plasmid pKO2-PCTK1-KO-II was SalI linearized and electroporated into R1 embryonic stem (ES) cells (31). Two hundred forty G418 (1 mg/ml)-resistant clones were screened by PCR for the presence of a wild-type or the targeted allele, and positive clones were verified by Southern blot analysis using an external 5′ probe on NdeI-digested and a 3′ probe on DraIII-digested genomic DNA, respectively.
Microinjection, mouse husbandry, and genotyping.
Clone 177 was used for microinjection into C57BL/6 blastocysts, and chimeric male mice were bred to C57BL/6 females. After verification of germ line transmission by PCR, mice were backcrossed to C57BL/6 and to Flpe-deleter animals (ACTB-FLPe [37]) to remove the NeoR cassette and then to Cre-deleter mice (PGK-Cre transgenic [39]) to delete exons 7 to 14. All mouse breeding was performed according to the requirements of the Animal Protection Act. Genotyping was performed on DNA samples prepared from 1-mm tail clips of 3- to 4-week-old animals (46) using primers ON952 (GGGTACCTATGCCACTGTCT) and ON953 (GGCCTTGTTCTAAACCTAAG) to detect wild-type (wt) (590-bp) and floxed (530-bp) alleles and primer pair ON1059 (AGGACTGTGGTAAAGATGGGGG) and ON1060 (CCACAAGCCAGGACTACCTAACAC) to detect the knockout allele (600 bp). The Cre and Flp transgenes were detected as 600-bp and 400-bp fragments using primer pairs ON1965 (GCCTGCATTACCGGTCGATGCA) and ON1966 (GTGGCAGATGGCGCGGCAACAC) as well as ON1971 (CTTCAGGGGAAAAAATAGCATCATG) and ON1972 (CCAGATGCTTTCACCCTCACTTAG), respectively. Fertility was assessed by keeping knockout males in the presence of 2 wild-type females over a period of 6 months. The same females were subsequently mated with wild-type males and gave birth to offspring.
Light and electron microscopy.
Phase-contrast microscopy was performed on a Zeiss Axiovert 200 M microscope (Carl Zeiss, Vienna, Austria) using Plan NeoFluar 40×, numerical aperture (NA) 0.75, and Plan Apochromat 63×, NA 1.4, objectives. Epifluorescence microscopy was performed as described previously (48). Confocal images were obtained on a Leica TCS-SP5 DMI6000 microscope (Leica, Mannheim, Germany) using an HCX PL APO lambda blue 63×, NA 1.4, oil UV objective and 488-nm laser excitation at a pixel resolution of 96 nm.
Flat embedding of sperm for longitudinal sectioning for transmission electron microscopy (TEM) was performed by modifying previously described methods (13). Briefly, a droplet of fresh sperm from the epididymis was sandwiched between 2 sapphire coverslips (ϕ = 3 mm; 50 μm thick) with an electron microscopy copper slot grid as spacer and subjected to high-pressure freezing, freeze substitution, and plastic embedding as described previously (14). For scanning electron microscopy (SEM), sperm from the epididymis was suspended in room-temperature (RT)-warm M2 medium (Sigma-Aldrich) according to reference 53, followed by several rinses and mild centrifugation in M2 and finally fixation with 2.5% (vol/vol) glutaraldehyde in 0.1 M phosphate buffer (pH 7.4). The fixed suspension was gently centrifuged with a cytocentrifuge (usually used for blood cells) onto poly-l-lysine-coated coverslips and processed for SEM according to standard protocols (14).
Sperm isolation and handling.
Two-month-old male mice (Cdk16−/0 and Cdk16+/0) were sacrificed, and the epididymis was isolated, cleaned of blood vessels and adipose tissue, washed in phosphate-buffered saline (PBS), placed in a glass-bottomed dish, cut gently into pieces in 1 ml of modified CZB-H buffer (81.6 mM NaCl, 4.8 mM KCl, 1.2 mM MgSO4, 1.7 mM CaCl2, 1.18 mM KH2PO4, 0.1 mM EDTA, 31 mM Na-dl-lactate, 5.6 mM glucose, 25 mM NaHCO3, 0.3 mM Na-pyruvate, 10 μg/ml phenol red, 5 mg/ml bovine serum albumin [BSA], 1 mM l-glutamine, 1 mM Pen/Strep), and incubated for half an hour at 37°C to allow the sperm to swim out. After this swim-out period, movies were made to determine the motility and morphology of the spermatozoa. Spermatozoa were placed on poly-l-lysine-coated coverslips and dried for 60 min on ice, washed with PBS, and fixed with 3% formaldehyde (FA) freshly prepared from paraformaldehyde for 20 min, followed by permeabilization with 0.1% Triton X-100 (TX100) for 5 min and routine immunostaining (47) using Sept7-specific antibodies.
RESULTS
CDK16 interacts with cyclins.
We screened a human testis cDNA library for proteins able to interact with human CDK16 expressed in yeast. Among 14.7 million interactions tested, we obtained 71 clones, representing 21 potential candidates, including cyclins I, K, Y, and Y-like 1, and the CDK activator SPDY-A/RINGO (Table 1). When expressed as YFP-tagged proteins, none of these cyclins and cyclin-like proteins localized like CDK16-GFP, which, like the endogenous protein, was cytoplasmatic (Fig. 1A). CCNY-YFP and CCNY-like1-YFP localized to the plasma membrane (18) (Fig. 1A), while the other proteins either localized exclusively to the nucleus (SPD-A) or were found in the cytosol and in the nucleus (CCNI and K). Coimmunoprecipitation (co-IP) experiments, however, revealed that FLAG-tagged CDK16 specifically interacted with CCNY (Fig. 1B).
Table 1.
CDK16-interacting proteinsa
| Category | Gene | RefSeq | GID | No. of clones (no. of independent clones) | Length of longest in-frame ORF (nt) | Domain present in prey clone | Known function |
|---|---|---|---|---|---|---|---|
| Cyclins | CCNI | NM_006835.2 | 17738314 | 9 (1) | 768 | Cyclin fold | Interacts with CDK5 |
| CCNK | NM_001099402.1 | 150417988 | 6 (3) | 779 | Cyclin fold | Interacts with CDK9 | |
| CCNY | NM_145012.4 | 190341111 | 2 (1) | 1,043 | Cyclin fold | Interacts with CDK14 | |
| CCNYL1 | NM_152523.1 | 22749084 | 1 | 1,334 | Cyclin fold | None | |
| SPDYA | NM_182756.2 | 60498968 | 1 | 802 | None | Interacts with CDK1/2 | |
| FBXO8 | NM_012180.2 | 48928043 | 2 (2) | 1,514 | Cyclin fold like F-box; Sec7 domain | Targets Arf6 | |
| Others | ADO23/MIF4GD | NM_020679.2 | 23510351 | 1 | 788 | MIF4G domain | mRNA metabolism |
| C3orf67 | NM_198463.2 | 142359507 | 5 (1) | 759 | None | None | |
| PTS | NM_000317.2 | 164663900 | 2 (1) | 556 | Catalytic | BH4 synthesis | |
| TMEM99 | NM_145274.2 | 116268112 | 1 | 360 | None | None | |
| UAP1 | NM_003115.4 | 156627574 | 5 (1) | 1,001 | Catalytic | Glycosylation | |
| Promiscuous domains | TZFP | NM_014383.1 | 7657664 | 1 | 666 | Zn-finger | Transcription |
| ZBTB16 | NM_001018011.1 | 66932931 | 1 | 434 | Zn-finger | Transcription | |
| Unclear | MGC26597 | BC028580.2 | 34190093 | 1 | 882 | PIP5K | Processed pseudogene |
| BC041636 | BC041636.1 | 27370971 | 2 (2) | 883 | None | None (noncoding?) | |
| Genomic clones | NA | NA | 9 (9) | NA | NA | NA | |
| Known false positive | SNRP70 | NM_003089.4 | 57634537 | 16 (9) | 577 | RRM | Splicing |
Abbreviations: NA, not available; GID, gene identifier; ORF, open reading frame; nt, nucleotide.
Fig 1.
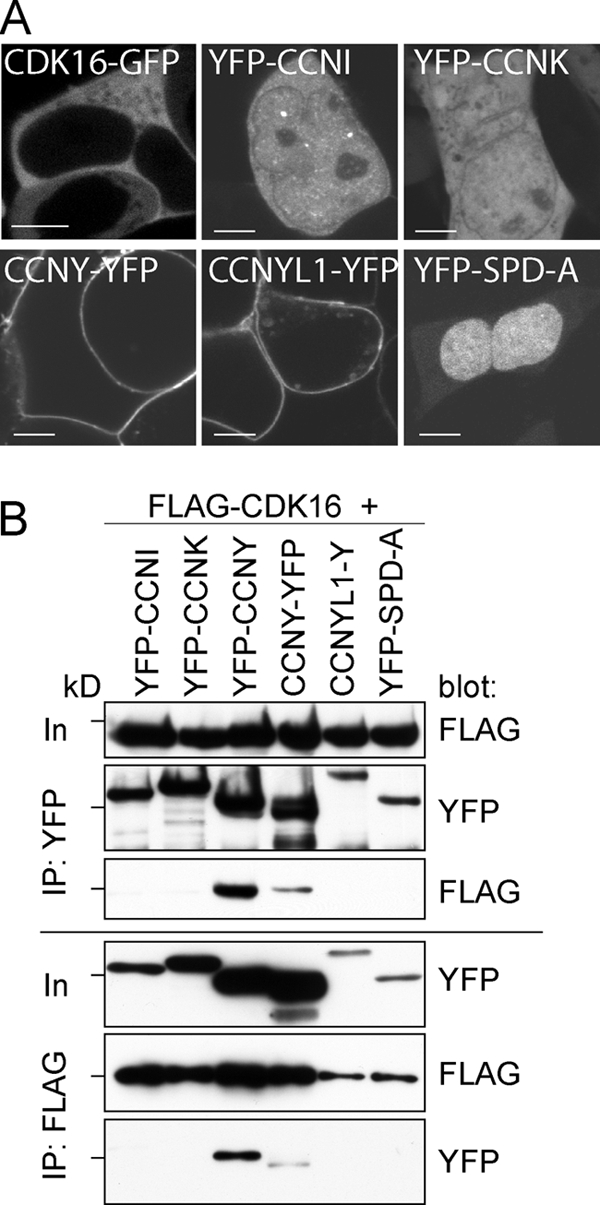
CDK16 interacts with CCNY. (A) 293A cells were transfected for 48 h with expression plasmids for the indicated proteins and imaged on a Leica SP5 confocal microscope using a 63× objective. Bars = 5 μm. (B) 293A cells were transfected for 60 h, and FLAG-tagged CDK16 and YFP-tagged cyclins were immunoprecipitated using anti-FLAG and anti-YFP antibodies and analyzed by immunoblotting. Input (In) is 5% of the protein amount used for IP. The 72-kDa marker is indicated.
CDK16 interacts with CCNY via a novel mechanism.
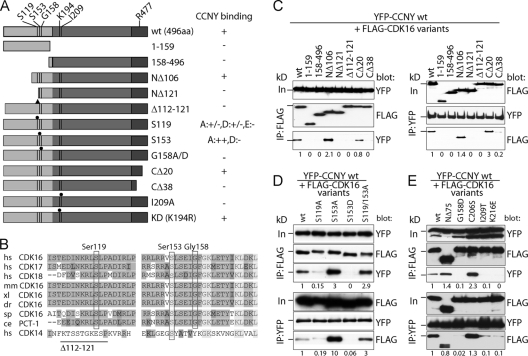
To further characterize this interaction, we generated several point, truncation, and internal deletion mutants of FLAG-tagged CDK16 and YFP-tagged CCNY (Fig. 2A and B and Fig. 3) and tested their interaction in co-IP assays. Deletion of the first 157 amino acid residues of CDK16 prevented binding to coexpressed CCNY (Fig. 2C), showing that, in contrast to other known CDK-cyclin pairs, such as cyclin A-CDK2, the CDK domain of CDK16 is not sufficient for cyclin binding. The N-terminal domain (1 to 159) by itself, however, was also unable to bind CCNY, indicating that CDK16 contains more than one domain required for CCNY binding. Mapping of the N-terminal binding domain revealed that the first 105 residues of CDK16 were not essential, while a further deletion of 16 residues (NΔ121) abrogated CCNY binding. Sequence comparison of CDK16s of different species revealed conserved domains in the N-terminal extension, including residues 112 to 121 (Fig. 2B). Deletion of these conserved residues blocked the ability of CDK16 to interact with CCNY (Fig. 2C). Interestingly, mutation of S119 into an alanine strongly reduced the amounts of coimmunoprecipitated CCNY (>75% reduction), suggesting that phosphorylation of this residue might be required for CCNY binding (Fig. 2D). Phosphorylation-dependent CCNY binding could, however, not be mimicked by changing S119 into phosphomimetic residues (data not shown). In contrast, when S153, which, like S119, was described as a phosphorylated residue, was changed to alanine, increased cyclin binding was observed, while binding was lost in an S153D phosphomimetic mutant (Fig. 2D). CCNY binding was also lost when G158 was changed to aspartate (Fig. 2E; see also Fig. 7A) or alanine (data not shown), suggesting that the N-terminal extension might adopt different phosphorylation-state-dependent conformations.
Fig 2.
Mapping of CDK16 residues required for CCNY binding. (A) Scheme of N-terminally FLAG-tagged CDK16 variants analyzed for interaction with YFP-tagged CCNY. (B) Amino acid sequences of ∼60 residues upstream of the conserved kinase domain were aligned with each other. Identical residues are shown in medium gray and similar ones in dark gray, whereas completely conserved residues are shown in light gray. The positions of human Ser119, Ser153, and Gly158 as well as the deletion of residues 110 to 121 are indicated. hs, human; mm, mouse; xl, frog; dr, zebrafish; sp, sea urchin; ce, nematode. (C) 293A cells were transfected with expression plasmids for YFP-CCNY together with FLAG-tagged CDK16 variants for 60 h. IPs were performed as in Fig. 1, and proteins were immunoblotted for the presence of the interaction partner. (D and E) Cells were transfected as in panel C to express phosphosite mutants of FLAG-CDK16 and tested for YFP-CCNY binding. Input (In) is 10% of the protein amount used for IP. The position of the 72-kDa protein marker band is indicated. Proteins were quantified by scanning densitometry using ImageJ. Relative levels normalized to the values of the immunoprecipitated partner proteins are shown.
Fig 3.
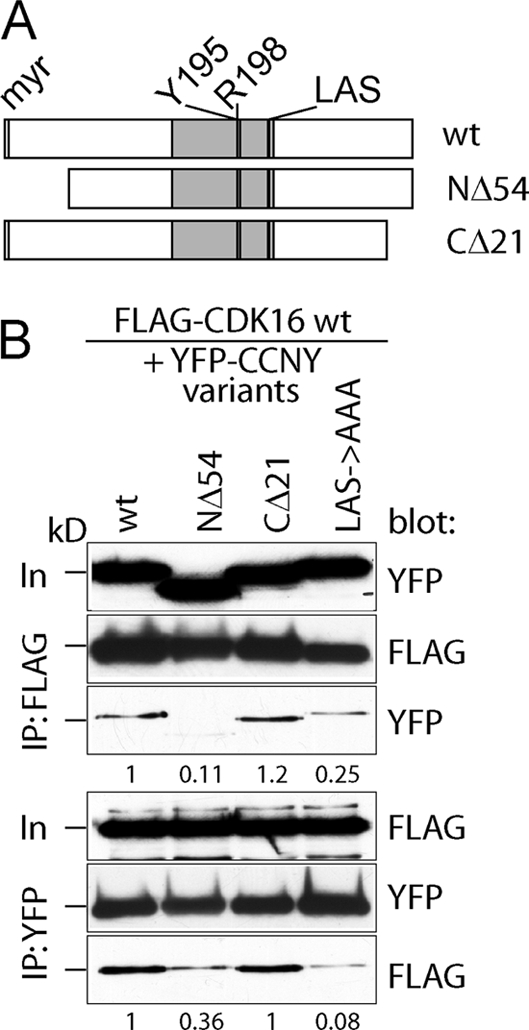
Mapping of CCNY residues required for CDK16 binding. (A) Scheme of N-terminally YFP-tagged CCNY variants analyzed for interaction with FLAG-tagged CDK16. (B) Similarly to Fig. 2C, CCNY variants were analyzed for their ability to interact with CDK16 by transient-transfection and immunoprecipitation experiments followed by immunoblotting.
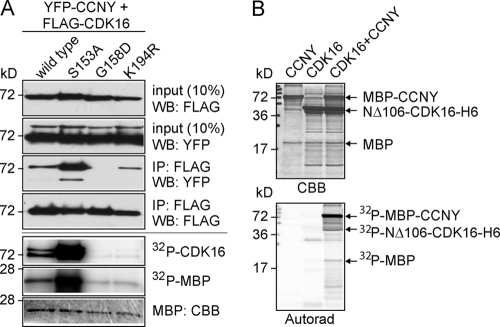
Fig 7.
CCNY activates CDK16. (A) FLAG-tagged wild-type, S153A, G158D, and K194R human CDK16 were expressed with YFP-tagged human CCNY in 293A cells for 48 h before IP-kinase assays using myelin basic protein (MBP) and CDK16 autophosphorylation as readout. (B) Recombinant hexahistidine-tagged CDK16 lacking the N-terminal 106 amino acid residues and harboring the S153A mutation as well as bacterially expressed and purified maltose binding protein-CCNY (MBP-CCNY) was either used alone or in combination for an in vitro kinase assay using myelin basic protein as a model substrate. Input proteins are visualized by Coomassie brilliant blue (CBB) staining (top panel), phosphorylated proteins, and autoradiography (bottom panel).
To define the role of the CDK domain in CCNY binding, we changed C206 within the PCTAIRE helix to a Ser to generate a PSTAIRE motif that is known to mediate cyclin binding in other CDKs, but we found that this mutant still interacted with CCNY. In contrast, mutations I209T and K216E abrogated the ability of CDK16 to bind CCNY (Fig. 2E), suggesting that the PCTAIRE helix and its surroundings are required for cyclin binding. Sequence alignment of the C-terminal extensions of PCTKs revealed a conserved domain (461 to 473 in human CDK16) followed by a less well conserved region (data not shown). Deletion of the C-terminal 20 residues (477 to 496) had no effect on CCNY binding, while it was lost when the C-terminal 38 residues (461 to 496) were missing (Fig. 2C). Whether this indicates a role for the C terminus in cyclin binding or points to a structural function of this domain will require further experiments.
Next, we searched for CCNY domains important for CDK16 binding. We first expressed a previously described splice variant of CCNY that lacks the first 54 residues and was reported to be located in the nucleus (24). As shown in Fig. 3, this truncated version of CCNY failed to interact with CDK16. Deletion of the last 21 amino acid residues had no impact on CDK16 binding, while larger truncations severely affected the stability of CCNY, suggesting that they are required for proper folding of the molecule. The alignment of the cyclin box of CCNY with those of other cyclins revealed only a few conserved residues, including Y195, R198, L222, and S224, which correspond to residues in cyclin A that flank the CDK-interaction domain (Fig. 3A). Substitution of any of these residues with alanines prevented CCNY binding to CDK16 (Fig. 3 and data not shown). Thus, the binding of CCNY to CDK16 requires an intact cyclin box that interacts with the kinase domain. In addition, however, CCNY binding requires the N-terminal domain of CDK16, including two residues that might be subject to phosphorylation.
CCNY recruits PCTK1 to the plasma membrane in a phosphorylation-dependent manner.
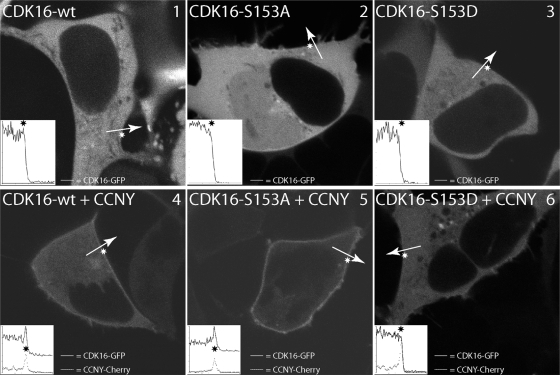
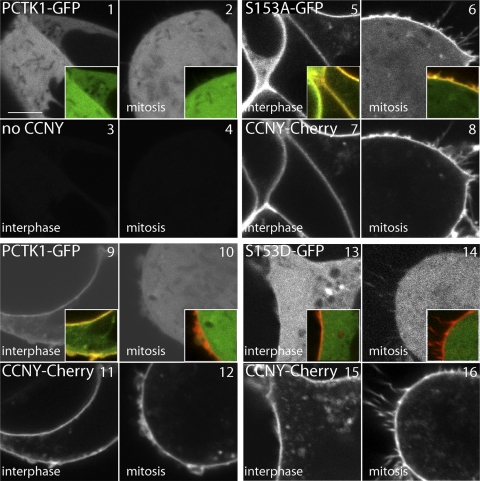
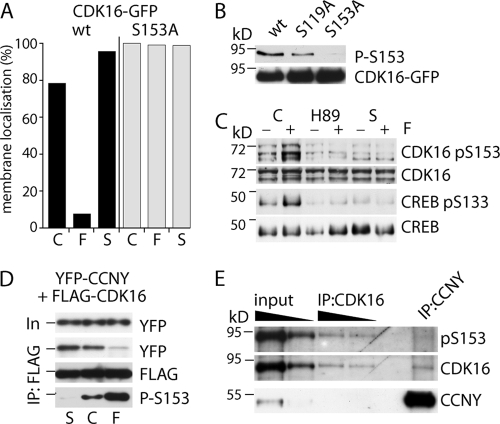
Because CCNY is targeted to the plasma membrane by N-terminal myristoylation, we next examined whether CDK16 can be detected at the plasma membrane as well. As can be seen in Fig. 4 (panel 1), CDK16-GFP expressed alone in 293A cells was found exclusively in the cytoplasm (top left panel). However, when expressed together with CCNY-Cherry (Fig. 4, panel 4), a fraction of CDK16-GFP was recruited to the plasma membrane (insets show signal intensities along the indicated arrow with the asterisk marking the position of the cell membrane). Interestingly, membrane targeting was lost in mitosis, suggesting a cell-cycle-dependent regulation of CDK16 (Fig. 5, panel 10). Consistent with the coimmunoprecipitation experiments, cells expressing CDK16-S153A (Fig. 4, panel 5) displayed stronger membrane localization in a CCNY-dependent manner, while the S153D mutant failed to localize (Fig. 4, panel 6), as did the CDK16-G158A mutant. Interestingly, CDK16-S153A membrane localization was also preserved in mitosis (Fig. 5, panel 6), while CDK16-S153D was cytoplasmatic in interphase as well as in mitosis (Fig. 5, panels 13 and 14). Thus, the interaction between CCNY and CDK16 appears to be dynamic and regulated by S153 phosphorylation.
Fig 4.
CDK16 interacts with CCNY in an S153-dependent manner. 293A cells were transfected with expression vectors for CDK16-GFP and CCNY-Cherry either alone (top) or in combination (bottom) as indicated and analyzed by live cell confocal imaging 24 h later. Insets show line scans (solid line, CDK16; dashed line, CCNY) across the cell membrane (asterisk).
Fig 5.
Cell-cycle-dependent interaction of CCNY with CDK16. 293A cells were transfected with fluorescent fusion protein expression vectors as indicated and analyzed by live cell confocal imaging 24 h later. Each quartet of images shows an interphase and mitotic cell in the GFP and Cherry fluorescence channels, respectively. Size bar = 5 μm.
Because S153 has been shown to be a PKA substrate in vitro, we tested whether forskolin treatment, which elevates cyclic AMP (cAMP) levels and activates PKA, would affect CDK16-GFP localization in cells coexpressing CCNY-Cherry. As can be seen in Fig. 6A, 5 μM forskolin (F) prevented membrane targeting of CDK16, which could be reversed by addition of 0.4 μM staurosporine (S), a broad-spectrum kinase inhibitor. This effect required S153 since no changes of CDK16 localization could be detected when using the S153A mutant.
Fig 6.
CCNY binding to CDK16 is regulated by S153 phosphorylation. (A) Membrane localization of CDK16wt-GFP and CDK16-S153A-GFP in the presence of CCNY-Cherry was determined as in Fig. 4 before (=control, C) and 2 to 5 min after addition of 5 μM forskolin (F) and again 15 to 30 min after further addition of 0.4 μM staurosporine (S). (B) 293A cells expressing GFP fusions of CDK16wt, S119A, and S153A were probed for S153 phosphorylation. (C) 293 cells were treated for 1 h with vehicle (control, C), 30 μM H89, or 0.4 μM staurosporine (S) before addition of 5 μM forskolin (+) or were left untreated (−) for an additional 15 min, after which cells were lysed for immunoblot analysis to detect CDK16, CDK-pS153, CREB, and p-S133 CREB, as indicated. (D) Cellular extracts from 293A cells treated as in panel B were analyzed by co-IP experiments and probed using anti-GFP and FLAG antibodies and pS153-specific antibodies. (E) 293A cells coexpressing CDK16-GFP and FLAG-CCNY were subjected to co-IP experiments and probed using pS153.
To further define the role of S153 phosphorylation, we generated a phosphopeptide-specific polyclonal antibody, which reacted with wild-type CDK16-GFP but not with CDK16-S153A (Fig. 6B). To investigate whether PKA was the upstream inhibitory CDK16 kinase, we treated 293A cells for 1 h with the PKA inhibitor H89 (30 μM) followed by a 15-min treatment with 5 μM forskolin. As can be seen in Fig. 6C, forskolin increased the phosphorylation of CDK16 at S153 and of the PKA target CREB at S133 (Fig. 6C, lanes 1 and 2), which was prevented by H89 (lanes 3 and 4) as well as by staurosporine (lanes 5 and 6).
To assess the impact of S153 phosphorylation on the ability of CDK16 to interact with CCNY, 293 cells were treated with forskolin, which strongly increased S153 phosphorylation and reduced the binding to CCNY (Fig. 6D). Staurosporine, in contrast, strongly reduced S153 phosphorylation and promoted the interaction with CCNY (Fig. 6D). To investigate whether S153 phosphorylation is indeed incompatible with CCNY binding, we coexpressed CDK16-GFP with FLAG-CCNY and analyzed the phosphorylation status of CDK16 bound to CCNY. As can be seen in Fig. 6E, no phospho-S153 could be detected in CCNY-bound CDK16. To rule out the possibility that the phosphopeptide antibody simply failed to detect the levels of phosphorylated protein present in the coimmunoprecipitates, we compared it to similar levels from CDK16-GFP immunoprecipitates, in which phospho-S153 could readily be detected (Fig. 6E, lanes 3 and 4). Thus, S153 phosphorylation of CDK16 blocks CCNY binding.
CCNY and PCTK1 are coexpressed and form an active kinase complex in vivo.
To investigate whether CCNY not only bound CDK16 but also activated it, we coexpressed YFP-CCNY and FLAG-tagged CDK16 in 293A cells to perform in vitro kinase assays. Recombinant CDK16 was immunoprecipitated using anti-FLAG antibodies and tested for autophosphorylation as well as activity against MBP. As shown in Fig. 7A, wild-type CDK16, but not the catalytic inactive mutant K194R, was activated by CCNY. Consistent with the above-described binding assays, CDK16-S153A bound to CCNY more strongly and was hyperactivated, while the CCNY-nonbinding G158D mutant was not. To rule out the possibility that the observed phosphorylation of MBP and of CDK16 was due to a copurifying contaminating kinase, CCNY was expressed as a maltose-binding protein fusion protein in bacteria, purified, and mixed with purified hexahistidine-tagged CDK16 lacking the N-terminal 106 amino acid residues and harboring the S153A mutation. As can be seen in Fig. 7B, CDK16 was active only in the presence of recombinant CCNY and phosphorylated MBP, itself, and most prominently maltose binding protein-CCNY. Mass spectrometric analysis of in vitro-phosphorylated maltose binding protein-CCNY revealed that CDK16 phosphorylated CCNY at residue S336 (data not shown).
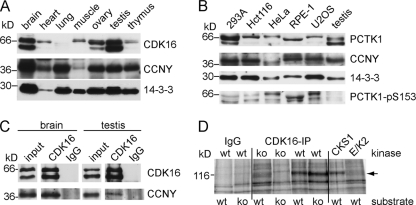
To further investigate the function and regulation of CDK16 by CCNY, we determined the expression of these two proteins in mouse tissues. Murine CDK16 could be detected in at least three different isoforms in many tissues with highest levels in brain and testis (Fig. 8A). The natures of these different isoforms are currently not known but might correspond to different splice variants or protein modifications. Multiple CDK16 isoforms were also detected in human cell lines, in which CDK16 was also found to be phosphorylated at S153 (Fig. 8B). For detection of murine CCNY, we used an RNAi-verified antibody (data not shown) and found that CCNY was ubiquitously expressed as an ∼38-kDa protein (Fig. 8A). In contrast to human cell lines, CDK16 was little phosphorylated at S153 in murine testis (Fig. 8B). When CDK16 immunocomplexes from murine brain and testis lysates were analyzed by immunoblotting, they were found to contain CCNY, demonstrating that the CDK16-CCNY complex exists in vivo (Fig. 8C). No such interaction could be detected when endogenous CDK16 was immunoprecipitated from 239 cells, suggesting that CDK16 is inactive in these cell lines (data not shown).
Fig 8.
Endogenous CDK16 interacts with CCNY and forms an active kinase in the testis. (A) Mouse tissue lysates were analyzed by immunoblotting for expression of CDK16, CCNY, and 14-3-3 proteins (as loading control). (B) Total cell extracts were prepared from the indicated human cell lines and murine testis and probed for CDK16, CCNY, 14-3-3 proteins, and CDK16-pS153 using specific antibodies. (C) CDK16-immunoprecipitates from 2 mg of tissue were analyzed by immunoblotting using CCNY-specific antibodies. Control reactions were performed using preimmune isotype control antibodies. Input lanes show 1% of the amount used for IP. (D) Endogenous CDK16 was immunoprecipitated from testis lysates of Cdk16 wild-type (wt) or Cdk16-knockout (ko) mice and incubated with tissue extracts obtained from wt or ko mice. As controls, kinases binding to GST-CKS1 and recombinant active cyclin E/Cdk2 (E/K2) were used to phosphorylate proteins in tissue extracts using [γ-32P]ATP. The phosphorylated band at 116 kDa is indicated by an arrow.
To determine whether the endogenous CCNY/CDK16 complex found in the testis is also active, we immunopurified CDK16 from testis lysates of wild-type and CDK16-knockout mice (see below) and incubated it with desalted and dephosphorylated testis lysates from wild-type and knockout animals (20). As can be seen in Fig. 8D, similar to glutathione S-transferase (GST)–CKS1 pulldown experiments (to pull down CDKs), CDK16 immunoprecipitates phosphorylated a 116-kDa protein band. This band was, however, not phosphorylated using control IP antibodies or IPs using lysates from CDK16-knockout mice and not by active recombinant cyclin E/CDK2. We tried to identify the candidate protein by excising proteins with a molecular mass range of 90 to 130 kDa from polyacrylamide gels and subjecting them to tryptic digestion and peptide mass fingerprinting. Among the proteins identified were several known phosphoproteins, e.g., the 115-kDa protein USO1, a conserved regulator of vesicle transport (1, 38, 40, 41). Whether or not p115 is a target of CDK16 is currently being investigated.
In summary, CCNY interacts with and activates CDK16 in vitro and in vivo and active CDK16 can be detected in lysates from mouse testis. Because no bona fide substrate of CDK16 had been identified yet that could give a clue about the function of CDK16 in vertebrates, we decided to generate a CDK16-knockout mouse model to learn whether CDK16 has a nonredundant essential function, which could help to further define the physiological and molecular function of CDK16.
Generation and analysis of CDK16-knockout mice.
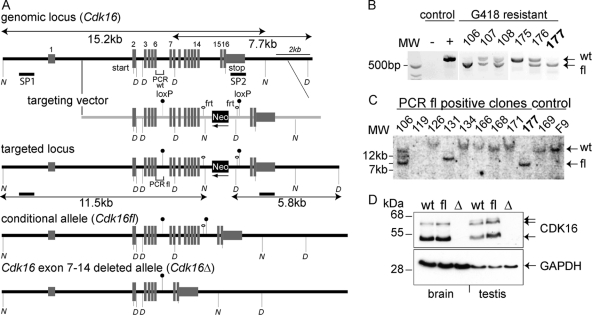
Because previous attempts at generating CDK16-knockout mice using conventional gene replacement vectors have failed (12), we generated a mouse strain harboring a conditional knockout allele of Cdk16. CDK16 is located in a gene-dense locus between Ube1x and Usp11 on the X chromosome. To generate a conditional allele, we introduced loxP sites in introns 6 and 14 of a genomic Cdk16 fragment (Fig. 9A) and electroporated male R1 embryonic stem cells to obtain 2 correctly targeted clones (Fig. 9B and C), which were used to generate germ line-transmitting chimeric mice. Conditional floxed Cdk16 strain (Cdk16fl) male mice were crossed to Cre-recombinase-deleter females (Cdk16+/+[Cre+]) to obtain heterozygous female Cdk16+/Δ mice, which were viable, fertile, and without apparent phenotype. Cdk16Δ (having Cdk16 exons 7 to 14 deleted) male offspring, generated by crossing Cdk16+/Δ mice to wild-type (Cdk16+/0) males, were born at the expected frequency (11 out of 21 males). These mice lacked CDK16 expression (Fig. 9D) but did not display any obvious phenotype and developed indistinguishably from control littermates.
Fig 9.
Generation of conditional Cdk16-knockout mice. (A) A 129/Sv genomic DNA clone encompassing Cdk16 exons 2 to 16 was cloned from a λ-KO2 phage library (52) and modified to harbor an FRT site-flanked NeoR gene along with the second loxP site in intron 14. Shown are schematic representations of the wt and the targeted as well as the conditional (floxed) and the exon-7-to-14-knockout allele. (B and C) G418-resistant R1 embryonic stem cells were prescreened by PCR (B) (positions of primers are indicated in panel A) and verified (C) by Southern blotting using NdeI (N)-digested genomic DNA and an external probe (SP1, shown in panel A). (D) Detection of Pctk1 expression in brain and testis lysates obtained from wt, floxed, and Cdk16-knockout mice. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) detection was used as a loading control.
CDK16 is required for spermatogenesis.
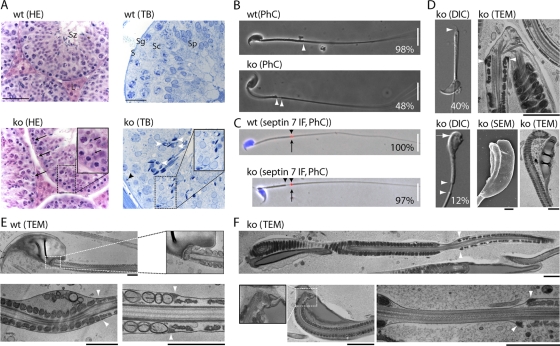
All tested adult Cdk16Δ knockout males (n = 8), however, were found to be infertile. To assess whether CDK16 was also required in females, we generated Cdk16Δ/Δ female mice which were healthy and fertile (n = 3), suggesting that CDK16 has a unique role in spermatogenesis. Examination of the testis by histological comparison of hematoxylin-eosin (HE)-stained sections of 45-, 125-, and 152-day-old wt and Cdk16Δ males revealed little difference. Seminiferous tubules of all spermatogenesis stages were present and contained the defining composition of Sertoli cells, spermatogonia, and spermatids as well as spermatozoa in both genotypes. In contrast to the wt, however, Cdk16Δ testis showed a mild degree of focal dysplasia, and upon closer examination of stage IX tubuli (Fig. 10A, 152d Cdk16Δ, ko), we noticed the presence of condensed sperm nuclei (arrows) close to the basal membrane, which indicates phagocytosis of abnormal spermatids by Sertoli cells. Abnormal, degenerated cells (white arrowheads) as well as displaced sperm heads (black arrowhead) could also be detected in toluidine blue (TB)-stained semithin testis sections from Cdk16Δ but not wt animals. These findings suggested a defect in the terminal steps of spermatogenesis.
Fig 10.
CDK16 is required for spermatogenesis. (A) Testis were obtained from 152-day-old mice, fixed in Bouin's solution or glutaraldehyde, and embedded in paraffin or epoxy resin, respectively. Sections of 6 or 1 μm were stained with hematoxylin-eosin (HE) or toluidine blue (TB), respectively. L, Leydig cells; S, Sertoli cells; Sg, spermatogonia; Sc, spermatocytes; Sp, spermatids; Sz, spermatozoa. Size bars = 100 μm and 50 μm. (B) Sperm were isolated from the testis of wt and Cdk16-knockout 2-month-old mice and evaluated by phase-contrast (PhC) microscopy (wild type, n = 95; knockout, n = 91). (C) Spermatozoa obtained as in panel B were stained for Sept7 by immunofluorescence (IF; red, arrow) and DNA (Hoechst 33342 [blue]; wild type, n = 70; knockout, n = 75). Size bar in panels B and C = 20 μm. Arrowheads in panels B and C indicate the annulus region. (D) Spermatozoa obtained from the epididymis of wt and Cdk16-knockout mice were analyzed by differential interference contrast (DIC) or by transmission electron microscopy (TEM) as well as scanning electron microscopy (SEM). The relative frequencies of the observed defects are shown. (E and F) Transmission electron microscopy of spermatozoa isolated from wt and CDK16-deficient mice. Size bar = 1 μm. In panels D to F, arrowheads indicate the annulus region.
To investigate sperm morphology and motility, we isolated sperm from the testis as well as from the epididymis. All spermatozoa isolated from the testis of Cdk16Δ mice (but none of those isolated from wt mice) displayed a thinning at the annulus, which normally connects the midpiece with the principal piece of the flagellum (Fig. 10B, arrowheads). In addition, 40% (n = 91) were found to be bent (Fig. 10D), while 12% displayed maloriented heads (Fig. 10D, lower panels). Because the annulus is formed by septins, which assemble in ring-like structures important for establishing and demarcating membrane domains (6), we stained spermatozoa for septin 7. As shown in Fig. 10C, septin 7 was expressed and confined to a small ring-like structure in both genotypes, but this domain was dislocated in 97% of Cdk16-deleted sperm (n = 75, arrows in Fig. 10C). Spermatozoa isolated from the caudal epididymis of 2-month-old wt male mice displayed normal motility, while spermatozoa from knockout mice were hypomotile due to structural abnormalities, such as bent tails. Interestingly, while approximately 50% of spermatozoa isolated from the testis were straight, the vast majority of the spermatozoa isolated from the cauda epididymis were found to be bent.
Transmission electron microscopy revealed microtubule breakage in the bent annulus area (Fig. 10D, top) and the presence of cytoplasmic organelles within the cytoplasmic droplet surrounding the sperm head (Fig. 10D, bottom). Furthermore, the thinning of the annulus region, as observed by light microscopy, was caused by the displacement of the circular fibrous sheath together with the annulus, i.e., the septin ring, without disruption of the plasma membrane (Fig. 10F, arrowheads). Finally, the neck region, which connects the head with the midpiece, was irregular in some mutant sperm with maloriented heads (Fig. 10F). In summary, Cdk16-knockout male mice displayed asthenozoospermia, characterized by dyskinesia and multiple morphological alterations, including malformed sperm heads, excess of cytoplasm, and structural defects of the annulus region.
DISCUSSION
Our data show that CDK16 is activated by membrane-bound CCNY, which might explain previous findings that linked CDK16 activity to membrane-associated processes such as vesicle formation (35), fusion (25), and transport (34). Activation of CDK16 by CCNY features several new mechanisms, including novel interaction domains and an important role of CDK phosphorylation that has not been found in other cyclin-CDK pairs. In contrast to conventional CDKs, CCNY binding to CDK16 required the kinase domain as well as part of the N-terminal extension. This additional region also contains one of the two serines (S119 and S153) that were previously identified as kinase targets in vitro as well as in vivo (9, 11, 12, 32). A CDK16 mutant harboring the nonphosphorylatable S153A mutant showed enhanced binding, while the phosphomimetic S153D mutant almost completely failed to bind CCNY. By using a phosphoepitope-specific antibody, we could confirm phosphorylation of S153 in vivo and the absence of this phosphorylation when CDK16 was complexed with CCNY. Thus, the N-terminal domain is required for CCNY binding and this interaction is controlled by phosphorylation of CDK16 on S153.
Phosphorylation of these serines creates binding sites for 14-3-3 proteins (12), suggesting that CDK16, depending on its phosphorylation status, might bind either to CCNY at the cell membrane or, when phosphorylated, to a 14-3-3 protein in the cytoplasm. Consistent with this hypothesis is the kinetics of phosphorylation versus dephosphorylation of CDK16 at residue 153. Stimulation of phosphorylation of S153 by forskolin led to an immediate (within minutes) delocalization from the plasma membrane and loss of CCNY binding, while the action of the kinase inhibitor was much more delayed. One possible explanation for this difference is that 14-3-3-bound CDK16 might be protected from the effects of phosphatases, which is under investigation. The kinase responsible for Ser153 phosphorylation in vivo is currently unknown, but the sensitivity of S153 phosphorylation to forskolin and the kinase inhibitor H89 strongly implies PKA, consistent with previous findings (12), but further experiments are required to establish PKA as a CDK16-inhibitory kinase. The identification of CDK16 regulatory kinase will also help to understand where and under which conditions CDK16 is phosphorylated.
Analysis of knockout mice revealed that CDK16 is not essential on the cellular level, since Cdk16-deficient mice are viable. Gene targeting experiments to define the function of cyclins and CDKs in animal model organisms in vivo continue to yield surprising results (26). For example, CDK2, which was thought to be essential for cellular proliferation, was found to be dispensable for development and similar results were obtained for various other CDKs and cyclins (28). Although not required for cellular proliferation in knockout mice, CDK2 was found to be essential for meiosis and, thus, gametogenesis (33). Similarly, CDK4, cyclin A1, cyclin D2, cyclin E, and others lack an essential function in somatic cells but are essential for the development of sperm and oocytes (49). In contrast to CDK16, however, targeting of the “conventional” CDKs and/or their associated cyclins led to arrest in spermatogonium proliferation or disruption of meiosis. These defects result in a depletion of more mature spermatids from the tubules of the testis and usually result in azoospermia. CDK16 deficiency, however, does not affect meiosis but is required for the terminal differentiation steps in spermatogenesis. Consistent with this function, the resulting phenotype is asthenozoospermia. Our finding that CDK16, which is widely expressed in many different cell types, has no essential function in somatic cells (at least no function whose corruption would translate into an immediate phenotype) suggests that other CDK members are likely to step in to take over the function of CDK16. The most likely candidates for such redundant functions are CDK17/18 as well as CDK14/15, which can also associate with, and become activated by, CCNY.
In Drosophila, CCNY (CG14939) and CDK14/PFTK (L63) are essential genes required for coactivation of the Wnt coreceptor LRP6 (10). In contrast, vertebrates have several CCNY and CCNY-like cyclins and 5 CCNY-dependent CDKs (CDK14 to -18), suggesting a high degree of functional redundancy (10). However, with respect to mitosis, we find that CDK16, in contrast to CDK14/PFTK, does not interact with CCNY due to phosphorylation of Ser153. Thus, CDK16 and CDK14, although activated by the same cyclin, might be active at different times during the cell division cycle to carry out different functions. Interestingly, the domains surrounding Ser153 and Ser119 are unique to PCTKs, i.e., CDK16 to -18. Although the N-terminal extension of CDK14 is also required for CCNY binding (18), no evidence for a role of phosphorylation in the regulation of this cyclin-CDK interaction has been obtained so far.
Another critical missing link in our understanding of PCTK kinases is the lack of a bona fide substrate. We have identified a potential 116-kDa substrate that is expressed in murine testis, and we are currently trying to identify this candidate CDK16 substrate. The identification of testis substrates will also be essential for understanding the spermatogenesis failure phenotype in CDK16-deficient mice. Alternatively, CDK16 might function as a scaffolding protein rather than a protein kinase. However, the paucity of proteins indentified in our yeast two-hybrid interaction screens does not strongly support this hypothesis. The phenotype of these mutant mice resembled at least three previously published knockout phenotypes. Like mice lacking Spem1 (53), CDK16-deficient sperm displayed bent sperm heads that were enclosed in residual cytoplasm and were frequently disconnected from the flagellum. This defect can be explained by a failure in the terminal step of spermatogenesis, i.e., spermiation, during which the cytoplasm is removed and mature spermatozoa are released from their intimate relationship with Sertoli cells. Defects in this process can cause phagocytosis of spermatozoa by Sertoli cells (50), which we could also observe in our knockout animals. Unfortunately, the molecular function of SPEM1 is currently unclear, and in our preliminary experiments, recombinant SPEM1 was not phosphorylated by CDK16. Thus, whether CDK16 and SPEM1 function in the same pathway or not remains to be established.
Lack of CDK16 activity additionally caused an annulus defect similar to that seen in Sept4- and Tat1-knockout mice (17, 19, 45). In contrast to the depletion of septin 4, but similar to the depletion of TAT1, CDK16 malfunction did not interfere with the formation of the septin ring but prevented its proper localization. Septins are known CDK substrates (2), and the PCTK-related protein CDK14 has been shown to interact with septin 8 (51), suggesting that the septins might be CDK16 substrates, too, and we are currently testing this possibility.
The weakening of the annulus as well as the malformation of the sperm head and neck resulted in two points of weakened mechanical stability. When isolated from the caudal epididymis, the flagella were frequently found in a hairpin-like bend and displayed maloriented or disconnected sperm heads. Because these defects were less prominent in sperm isolated from more proximal parts, these changes are likely due to mechanical stress during the transit through the epididymis and/or the onset of motility. Thus, the infertility of CDK16-deficient mice can be explained by structural abnormalities as well as secondary hypomotility.
Teratoasthenozoospermia, i.e., infertility due to malformed and hypomotile spermatozoa, is a frequently observed but poorly understood cause of human male infertility, and patients with abnormal annulus structures have been described (22, 43), suggesting that CDK16 might be a human fertility gene, a possibility we are currently addressing by sequencing the CDK16 genes of infertile men. In summary, we have established the regulation of CDK16 by CCNY and described CDK16 as an essential gene for spermatogenesis. It is likely that additional CDK16 functions in other tissues are redundantly buffered by homologs and further research in the functions of these kinases is required to fully understand the physiological as well as pathophysiological roles of theses cyclin-dependent kinases.
ACKNOWLEDGMENTS
We are grateful to S. Maurer, K. Gutleben, and A. Flörl for technical support; C. Wandke for advice; H. Ebner for electron microscopy; A. Villunger and R. Fässler for support in mouse genetics; K. Rossi for animal care; R. Gräser, J. Gannon, and E. Stefan for materials; S. Dirnhofer for histology; and A. Helmberg, R. Kofler, and W. Sachsenmaier for support.
This work was funded by Austrian Science Foundation (FWF) grants P20860-B12 (S.G.), SFB021 “Cell proliferation and cell death in tumors” (S.G.), and P19486-B12 (M.W.H.) and Austrian National Bank Jubiläumsstiftung (ÖNB) grants 12787 (S.G.) and 11050 (M.W.H.) as well as by support from the Tiroler Zukunftsstiftung (SG) and from the assets of Eva von Lachmüller, Brixen, Italy.
P. Mikolcevic performed the biochemical characterization of CDK16, mouse husbandry and genotyping, mouse mutant phenotype analysis, and manuscript preparation; R. Sigl generated gene targeting vector and targeted embryonic stem cells and performed mouse breeding, genotyping, and mouse mutant phenotype analysis; V. Rauch performed genotyping and immunoprecipitation experiments; M. W. Hess performed transmission electron microscopy; K. Pfaller performed scanning electron microscopy; M. Barisic performed immunofluorescence microscopy; L. J. Pelliniemi performed flat embedding of sperm and phenotype analysis; M. Boesl performed generation of chimeric mice; S. Geley designed and performed experiments and wrote the manuscript.
We declare no conflicts of interest.
Footnotes
Published ahead of print 19 December 2011
REFERENCES
- 1. Allan BB, Moyer BD, Balch WE. 2000. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science 289:444–448 [DOI] [PubMed] [Google Scholar]
- 2. Amin ND, et al. 2008. Cyclin-dependent kinase 5 phosphorylation of human septin SEPT5 (hCDCrel-1) modulates exocytosis. J. Neurosci. 28:3631–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barisic M, et al. 2010. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol. Biol. Cell 210:1968–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Besset V, Rhee K, Wolgemuth DJ. 1998. The identification and characterization of expression of Pftaire-1, a novel Cdk family member, suggest its function in the mouse testis and nervous system. Mol. Reprod. Dev. 50:18–29 [DOI] [PubMed] [Google Scholar]
- 5. Besset V, Rhee K, Wolgemuth DJ. 1999. The cellular distribution and kinase activity of the Cdk family member Pctaire1 in the adult mouse brain and testis suggest functions in differentiation. Cell Growth Differ. 10:173–181 [PubMed] [Google Scholar]
- 6. Caudron F, Barral Y. 2009. Septins and the lateral compartmentalization of eukaryotic membranes. Dev. Cell 16:493–506 [DOI] [PubMed] [Google Scholar]
- 7. Charrasse S, Carena I, Hagmann J, Woods-Cook K, Ferrari S. 1999. PCTAIRE-1: characterization, subcellular distribution, and cell cycle-dependent kinase activity. Cell Growth Differ. 10:611–620 [PubMed] [Google Scholar]
- 8. Cole AR. 2009. PCTK proteins: the forgotten brain kinases? Neurosignals 17:288–297 [DOI] [PubMed] [Google Scholar]
- 9. Daub H, et al. 2008. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31:438–448 [DOI] [PubMed] [Google Scholar]
- 10. Davidson G, et al. 2009. Cell cycle control of wnt receptor activation. Dev. Cell 17:788–799 [DOI] [PubMed] [Google Scholar]
- 11. Dephoure N, et al. 2008. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 105:10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graeser R, et al. 2002. Regulation of the CDK-related protein kinase PCTAIRE-1 and its possible role in neurite outgrowth in Neuro-2A cells. J. Cell Sci. 115:3479–3490 [DOI] [PubMed] [Google Scholar]
- 13. Hawes P, Netherton CL, Mueller M, Wileman T, Monaghan P. 2007. Rapid freeze-substitution preserves membranes in high-pressure frozen tissue culture cells. J. Microsc. 226:182–189 [DOI] [PubMed] [Google Scholar]
- 14. Hess MW, et al. 2010. 3D versus 2D cell culture: implications for electron microscopy. Methods Cell Biol. 96:649–670 [DOI] [PubMed] [Google Scholar]
- 15. Higuchi R, Krummel B, Saiki RK. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hirose T, Tamaru T, Okumura N, Nagai K, Okada M. 1997. PCTAIRE 2, a Cdc2-related serine/threonine kinase, is predominantly expressed in terminally differentiated neurons. Eur. J. Biochem. 249:481–488 [DOI] [PubMed] [Google Scholar]
- 17. Ihara M, et al. 2005. Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev. Cell 8:343–352 [DOI] [PubMed] [Google Scholar]
- 18. Jiang M, Gao Y, Yang T, Zhu X, Chen J. 2009. Cyclin Y, a novel membrane-associated cyclin, interacts with PFTK1. FEBS Lett. 583:2171–2178 [DOI] [PubMed] [Google Scholar]
- 19. Kissel H, et al. 2005. The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev. Cell 8:353–364 [DOI] [PubMed] [Google Scholar]
- 20. Knebel A, Morrice N, Cohen P. 2001. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38delta. EMBO J. 20:4360–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le Bouffant F, Le Minter P, Traiffort E, Ruat M, Sladeczek F. 2000. Multiple subcellular localizations of PCTAIRE-1 in brain. Mol. Cell. Neurosci. 16:388–395 [DOI] [PubMed] [Google Scholar]
- 22. Lhuillier P, et al. 2009. Absence of annulus in human asthenozoospermia: case report. Hum. Reprod. 24:1296–1303 [DOI] [PubMed] [Google Scholar]
- 23. Li S, Wilkinson MF. 1997. Site-directed mutagenesis: a two-step method using PCR and DpnI. Biotechniques 23:588–590 [DOI] [PubMed] [Google Scholar]
- 24. Li X, Wang X, Liu G, Li R, Yu L. 2009. Identification and characterization of cyclin X which activates transcriptional activities of c-Myc. Mol. Biol. Rep. 36:97–103 [DOI] [PubMed] [Google Scholar]
- 25. Liu Y, Cheng K, Gong K, Fu AK, Ip NY. 2006. Pctaire1 phosphorylates N-ethylmaleimide-sensitive fusion protein: implications in the regulation of its hexamerization and exocytosis. J. Biol. Chem. 281:9852–9858 [DOI] [PubMed] [Google Scholar]
- 26. Malumbres M, Barbacid M. 2005. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 30:630–641 [DOI] [PubMed] [Google Scholar]
- 27. Malumbres M, et al. 2009. Cyclin-dependent kinases: a family portrait. Nat. Cell Biol. 11:1275–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malumbres M, Ortega S, Barbacid M. 2000. Genetic analysis of mammalian cyclin-dependent kinases and their inhibitors. Biol. Chem. 381:827–838 [DOI] [PubMed] [Google Scholar]
- 29. Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934 [DOI] [PubMed] [Google Scholar]
- 30. Meyerson M, et al. 1992. A family of human cdc2-related protein kinases. EMBO J. 11:2909–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 90:8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Olsen JV, et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3:ra3. [DOI] [PubMed] [Google Scholar]
- 33. Ortega S, et al. 2003. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 35:25–31 [DOI] [PubMed] [Google Scholar]
- 34. Ou CY, et al. 2010. Two cyclin-dependent kinase pathways are essential for polarized trafficking of presynaptic components. Cell 141:846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palmer KJ, Konkel JE, Stephens DJ. 2005. PCTAIRE protein kinases interact directly with the COPII complex and modulate secretory cargo transport. J. Cell Sci. 118:3839–3847 [DOI] [PubMed] [Google Scholar]
- 36. Rikova K, et al. 2007. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131:1190–1203 [DOI] [PubMed] [Google Scholar]
- 37. Rodriguez CI. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140 [DOI] [PubMed] [Google Scholar]
- 38. Sapperstein SK, Walter DM, Grosvenor AR, Heuser JE, Waters MG. 1995. p115 is a general vesicular transport factor related to the yeast endoplasmic reticulum to Golgi transport factor Uso1p. Proc. Natl. Acad. Sci. U. S. A. 92:522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schwenk F, Baron U, Rajewsky K. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Short B, Haas A, Barr FA. 2005. Golgins and GTPases, giving identity and structure to the Golgi apparatus. Biochim. Biophys. Acta 1744:383–395 [DOI] [PubMed] [Google Scholar]
- 41. Sohda M, Misumi Y, Yano A, Takami N, Ikehara Y. 1998. Phosphorylation of the vesicle docking protein p115 regulates its association with the Golgi membrane. J. Biol. Chem. 273:5385–5388 [DOI] [PubMed] [Google Scholar]
- 42. Stowers RS, Garza D, Rascle A, Hogness DS. 2000. The L63 gene is necessary for the ecdysone-induced 63E late puff and encodes CDK proteins required for Drosophila development. Dev. Biol. 221:23–40 [DOI] [PubMed] [Google Scholar]
- 43. Sugino Y, et al. 2008. Septins as diagnostic markers for a subset of human asthenozoospermia. J. Urol. 180:2706–2709 [DOI] [PubMed] [Google Scholar]
- 44. Tang X, et al. 2006. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proc. Natl. Acad. Sci. U. S. A. 103:2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Toure A, et al. 2007. The testis anion transporter 1 (Slc26a8) is required for sperm terminal differentiation and male fertility in the mouse. Hum. Mol. Genet. 16:1783–1793 [DOI] [PubMed] [Google Scholar]
- 46. Truett GE, et al. 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29:52–54 [DOI] [PubMed] [Google Scholar]
- 47. Wandke C, Geley S. 2006. Generation and characterization of an hKid specific monoclonal antibody. Hybridoma 25:41–43 [DOI] [PubMed] [Google Scholar]
- 48. Wolf F, Wandke C, Isenberg N, Geley S. 2006. Dose-dependent effects of stable cyclin B1 on progression through mitosis in human cells. EMBO J. 25:2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wolgemuth DJ. 2003. Insights into regulation of the mammalian cell cycle from studies on spermatogenesis using genetic approaches in animal models. Cytogenet. Genome Res. 103:256–266 [DOI] [PubMed] [Google Scholar]
- 50. Yan W. 2009. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Mol. Cell. Endocrinol. 306:24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang T, Gao YK, Chen JY. 2002. KIAA0202, a human septin family member, interacting with hPFTAIRE1. Acta Biochim. Biophys. Sin. 34:520–525 [PubMed] [Google Scholar]
- 52. Zhang P, Li MZ, Elledge SJ. 2002. Towards genetic genome projects: genomic library screening and gene-targeting vector construction in a single step. Nat. Genet. 30:31–39 [DOI] [PubMed] [Google Scholar]
- 53. Zheng H, et al. 2007. Lack of Spem1 causes aberrant cytoplasm removal, sperm deformation, and male infertility. Proc. Natl. Acad. Sci. U. S. A. 104:6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]