Abstract
In this report, we describe the phosphorylation of Yin Yang 1 (YY1) in vitro and in vivo by CK2α (casein kinase II), a multifunctional serine/threonine protein kinase. YY1 is a ubiquitously expressed multifunctional zinc finger transcription factor implicated in regulation of many cellular and viral genes. The products of these genes are associated with cell growth, the cell cycle, development, and differentiation. Numerous studies have linked YY1 to tumorigenesis and apoptosis. YY1 is a target for cleavage by caspases in vitro and in vivo as well, but very little is known about the mechanisms that regulate its cleavage during apoptosis. Here, we identify serine 118 in the transactivation domain of YY1 as the site of CK2α phosphorylation, proximal to a caspase 7 cleavage site. CK2α inhibitors, as well as knockdown of CK2α by small interfering RNA, reduce S118 phosphorylation in vivo and enhance YY1 cleavage under apoptotic conditions, whereas increased CK2α activity by overexpression in vivo elevates S118 phosphorylation. A serine-to-alanine substitution at serine 118 also increases the cleavage of YY1 during apoptosis compared to wild-type YY1. Taken together, we have discovered a regulatory link between YY1 phosphorylation at serine 118 and regulation of its cleavage during programmed cell death.
INTRODUCTION
Yin Yang 1 (YY1) is a ubiquitously expressed multifunctional transcription factor that can act either as an activator or a repressor of gene expression (6). It is involved in different cellular processes, such as proliferation, embryogenesis, differentiation, development, tumorigenesis, and apoptosis (22, 54). YY1 was originally cloned because of its interaction with an element in the adeno-associated virus (AAV) P5 promoter. It was shown to either repress or activate transcription of the AAV gene, depending on the absence or presence of the adenovirus E1A oncoprotein (E1A). This factor was thus named Yin Yang 1, referring to its dual transcriptional activity, or “yin-yang” behavior (55). Total ablation of the YY1 gene in mice caused embryonic lethality at the peri-implantation stage (15), while disruption of one YY1 allele resulted in significant growth retardation and developmental abnormalities (15), reflecting the essential role of the YY1 gene in embryogenesis, growth, and differentiation.
A wealth of experimental data has linked YY1 to cell cycle control. YY1 has been functionally associated with multiple components of cell cycle signaling pathways, such as c-Myc (5, 49, 56), retinoblastoma protein (Rb) (12, 46), and p53 (57), implicating YY1 in tumor development (19, 23). Previous studies from our laboratory have also reported that YY1 is involved in cell cycle regulation, showing differential cellular localization and complex formation with DNA across the cell cycle (18, 45). Also, at the onset of the G1/S phase, YY1 is involved in upregulation of replication-dependent histone genes through its interaction with the alpha element (18, 45), a 7-bp element in the coding region of all the replication-dependent core histone genes (9, 26).
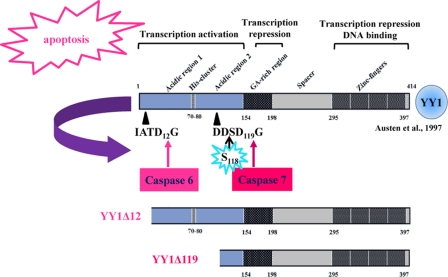
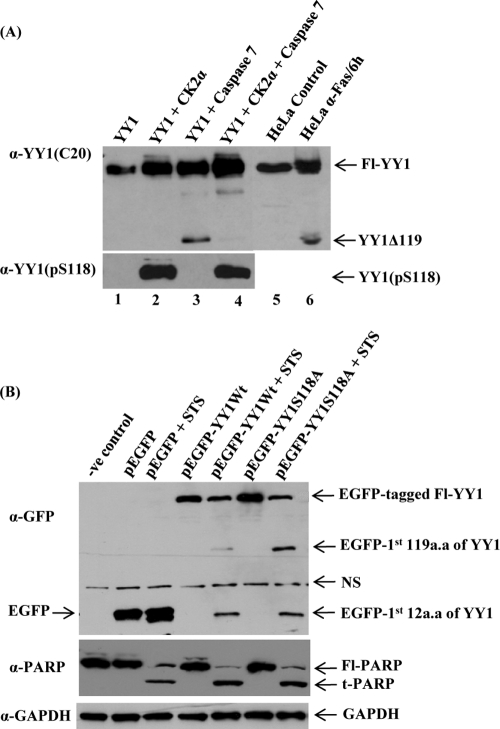
An important role for YY1 in tumorigenesis is further supported by its link to apoptosis. In response to DNA damage, YY1 acts as a negative regulator of p53. Sui and coworkers (57) demonstrated that YY1 controls endogenous p53 levels through regulation of Hdm2-mediated p53 ubiquitination via a direct physical interaction mechanism. Moreover, Gronroos et al. proposed that YY1 disrupts the p53-p300 interaction and blocks p300-dependent p53 acetylation and stabilization (23). Knockdown of endogenous YY1 sensitizes HeLa cells to DNA-damaging and other apoptotic stimuli (1, 23). Another study suggested that YY1 negatively regulates the expression of the Fas receptor, causing resistance to Fas-induced apoptosis (21). YY1 knockdown or inhibition results in upregulation of Fas expression and sensitization of tumor cells to Fas-induced apoptosis (25). We analyzed the fate of YY1 during apoptosis. We showed that YY1 is cleaved by caspases both in vitro and in vivo in response to apoptotic stimuli (29) (summarized in Fig. 1). Two distinct caspase cleavage sites were identified in the transactivation domain of YY1. The two sites, IATD12G and DDSD119G, are cleaved by caspase 6 and caspase 7, respectively. This process generates two N-terminally truncated fragments, YY1Δ12 and YY1Δ119, which have lost their first 12 and 119 amino acids (Fig. 1) (29). The N-terminal caspase 7 cleavage fragment (YY1Δ119) lacks its transactivation domain and is no longer able to stimulate gene transcription. Its DNA binding domain and two repressor domains remain intact (Fig. 1). Interestingly, YY1Δ119 but not the wild-type protein or the caspase-resistant protein YY1D12A/D119A can modify the apoptotic response to anti-Fas, suggesting that cleaved YY1 plays a positive feedback role during later stages of apoptosis (29).
Fig 1.
The functional domains and caspase cleavage sites of YY1. YY1 is cleaved by caspases in vitro and in vivo in response to apoptotic stimuli. This process generates two N-terminal cleaved fragments: YY1Δ12 and YY1Δ119, cleaved by caspases 6 and 7, respectively (29). The functions of the different domains of human YY1 are indicated (6).
Numerous residues on YY1 have been reported to be targets of posttranslational modifications, such as S-nitrosation (25), acetylation (58, 65), O-linked glycosylation (24), sumoylation (13), and poly(ADP-ribosyl)ation (41, 42). YY1 has been shown by others to be a phospho-protein (8, 54). More recently, we have been able to map distinct phosphorylation sites in YY1 (50, 51). We have identified kinases that phosphorylate YY1 in vitro, and we mapped several phosphorylation sites on YY1 in vivo (50, 51). We provide evidence here that one of these kinases is casein kinase 2α (CK2α), a serine/threonine protein kinase that phosphorylates and regulates many cellular substrates involved in cell growth, proliferation, differentiation, and tumorigenesis (2, 10, 16, 17, 32, 37). CK2 is ubiquitously present in all eukaryotic cells, highly conserved from yeast to humans, and is constitutively active in cells. It is normally present as a tetrameric complex and consists of two catalytic subunits (α and/or α′) in a homozygous or heterozygous composition and two noncatalytic regulatory (β) subunits (16, 32, 37, 47, 60). There is high CK2 constitutive activity in all cancer types examined. This includes kidney, mammary gland, head and neck, prostate, and lung cancers, linking CK2 to tumorigenesis (16).
CK2 recognizes consensus sequences that include Ser/Thr residues specified by clusters of acidic negatively charged amino acids located (+1 to +3) C-terminal to CK2 phosphorylation sites (27) in hundreds of cellular substrates, including caspase target proteins, such as connexin 45.6 (66), hematopoietic lineage cell-specific protein 1 (HS1) (53), presenilin 2 (62), IκB-α (IκB) (7), phosphatase and tensin homolog (PTEN) (39), apoptosis repressor with caspase recruitment domain (ARC) (31), Bid (14), caspase 9 (36), and Max (28). CK2 phosphorylation of these proteins occurs at sites flanking their caspase consensus regions, resulting in their protection from caspase-mediated cleavage and apoptosis (14, 17, 28, 31, 36, 39, 53, 59, 62, 66).
Even though YY1 has been extensively implicated in cancer biology and in programmed cell death, the regulation of its cleavage during apoptosis by caspases and the function of this cleavage in tumor progression are unclear. YY1 contains a potential CK2 phosphorylation site located within the caspase 7 recognition motif and directly adjacent to the negatively charged caspase 7 cleavage residue D119, suggesting that YY1 phosphorylation at S118 may protect YY1 from cleavage during programmed cell death. Here, we report that CK2α phosphorylates YY1 at serine 118 in vitro and in vivo, implicating this kinase in the regulation of apoptotic cleavage of YY1 by caspases.
MATERIALS AND METHODS
Cell culture and reagents.
HEK293 and HeLa S3 and HeLa-Flag-YY1 cells, a stable cell line that was generated as described previously (51), were grown in Dulbecco's modified Eagle's medium (Cellgro, Herndon, VA) supplemented with 10% fetal bovine serum (FBS; Sigma, St. Louis, MO), 1% nonessential amino acids (Sigma, St. Louis, MO), and 1% penicillin-streptomycin (Mediatech). U2OS cells were cultured in McCoy's 5A medium (Cellgro, Herndon, VA) supplemented with 10% FBS and 1% penicillin-streptomycin. All cells were grown at 37°C in 5% CO2. Cells were trypsinized and split into new plates at subconfluency.
The CK2 chemical inhibitors TBCA (tetrabromocinnamic acid) and DMAT (2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole) were obtained from Calbiochem and Sigma, respectively, and dissolved in dimethyl sulfoxide (DMSO). For the induction of apoptosis, asynchronously growing cells were either treated with staurosporine (STS; 1 μM; Sigma) or anti-Fas antibody (100 ng/ml; Upstate, Lake Placid, NY). The cells were preincubated with cycloheximide (CHX; 2.5 μg/ml; Sigma) for 30 min before the addition of anti-Fas antibody (64).
Mutagenesis.
Generation of a YY1 point mutation at position 118 from serine to alanine was performed using the pET-20b(+)-YY1 plasmid and subcloning into PGEX-2T-YY1 (50, 51). Mutagenesis was performed using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The serine-to-alanine mutation was confirmed by sequencing. The primers used for the mutagenesis of serine 118-to-alanine substitution were as follows: sense, 5′-GCGGCGACGACGCGGACGGGCTG-3′; antisense, 5′-CAGCCCGTCCGCGTCGTCGCCGC-3′.
Bacterial expression of GST-YY1 and deletion mutants.
Glutathione S-transferase (GST)–YY1 constructs, both in full length and as various deletion mutants, were overexpressed in bacterial cells as described previously (51).
Cold in vitro kinase assay.
GST-YY1 attached to glutathione beads was used in cold in vitro kinase assays with the purified catalytic subunits of CK2 (CK2α or CK2α′), which were purchased from SignalChem (British Columbia, Canada), or with casein kinase 1 (CK1; NEB). Kinase reactions were performed in kinase buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 10 mM MgCl2, 5 mM cold ATP) for 30 min at 30°C, with shaking. Reactions were then stopped by the addition of SDS-PAGE buffer and loaded for separation on a 10% SDS-PAGE gel.
Radioactive in vitro kinase assay.
Kinase reactions were performed in kinase buffer (50 mM Tris [pH 7.4], 10 mM MgCl2, 50 μM ATP, 0.25 μM [γ-32P]ATP, 5 mM β-glycerophosphate, 10 mM NaF, 1 mM dithiothreitol [DTT]) for 30 min at 30°C, with shaking. Reactions were stopped by the addition of SDS-PAGE buffer and separated on a 10% SDS-PAGE gel. After staining with Coomassie brilliant blue R-250 (Sigma) to visualize the protein bands, gels were dried and exposed to a phosphorimager screen at room temperature overnight. The screen was then scanned on a Typhoon 9410 imager (GE Healthcare, Waukesha, WI) for analysis.
Whole-cell extract preparation.
After washing cells three times with cold phosphate-buffered saline (PBS) on ice, cells were scraped in freshly prepared ice-cold lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 2 mM EDTA, 1 mM DTT, 10 mM NaF, 25 mM β-glycerophosphate, and a cocktail of protease inhibitors [Sigma]). Cells were lysed on ice for 15 min. Lysates were pipetted up and down several times to shear DNA, followed by centrifugation at 18,000 × g for 15 min at 4°C.
Immunodepletion of CK2α from cellular extracts.
HEK293 whole-cell extract (WCE) was prepared as previously described. A 540-μg aliquot of WCE was subjected to three rounds of immunodepletion with either an antibody raised against the α-catalytic subunit of CK2 or an antibody to green fluorescent protein (GFP) as a control. The first two immunodepletion rounds included 4 h of incubation at 4°C with the antibodies and then 1 h of incubation at 4°C with protein A/G Plus-agarose beads (Santa Cruz Biotechnology). Each cleared supernatant was used for the next round of immunodepletion. After two rounds, the cleared supernatants were subjected to a final overnight incubation with CK2α or GFP antibody at 4°C, and then the protein A/G slurry was added and incubated with the mixture for an additional hour. Equal amounts (40 μg) of the control or CK2α-immunodepleted cellular extracts from all the immunodepletion rounds were then used as the kinase source in a cold in vitro kinase assay (as described earlier).
Western blotting.
Protein samples were separated on SDS-PAGE gels and then transferred by electroblotting onto a Trans-Blot transfer membrane (Bio-Rad Laboratories). After blotting, the transfer of proteins was inspected by quickly staining and destaining the membrane with Ponceau S solution (Sigma). Afterwards, the membrane was blocked for 30 min at room temperature (RT) in blocking solution (PBS, 0.5% Tween 20, 5% nonfat dry milk) and then incubated with primary antibodies in blocking solution overnight at 4°C. The membrane was washed 3 times for 10 min with PBST (PBS with 0.5% Tween 20). Horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit (GE Healthcare, Waukesha, WI) or anti-goat (Santa Cruz Biotechnology) secondary antibodies were added to the membrane in blocking solution and incubated for an hour at RT, after which it was washed as above. Specific protein bands were detected by the addition of SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL) for 5 min and exposure to X-ray film (Fuji Medical Systems, Stamford, CT). Three anti-YY1 antibodies from Santa Cruz Biotechnology were used for Western blot analyses. Anti-YY1 (C20), a rabbit polyclonal antibody, recognizes the last 20 amino acids at the C-terminal end of YY1. Anti-YY1 (H-10) is a mouse monoclonal antibody (MAb) raised against the full-length protein, while anti-YY1 (H414) is a rabbit polyclonal antibody raised against the full-length protein. The rabbit polyclonal anti-pS118 was generated by New England Peptide using a synthesized phosphopeptide corresponding to amino acids 113 to 124 of YY1 [Ac-CVGGDD(pS)DGLRAE-amide]. Antibodies specific for poly(ADP-ribose) polymerase 1 (PARP-1; mouse monoclonal) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; rabbit polyclonal) and CK2α (goat polyclonal) were purchased from Santa Cruz Biotechnology. Antibody specific for GFP (goat polyclonal) was purchased from Rockland Immunochemicals (Gilbertsville, PA).
In vitro caspase 7 cleavage assay of YY1.
Purified nontagged YY1 as described previously (50) was first phosphorylated in vitro. One microgram of YY1 was incubated for 90 min at 30°C in 20 μl of kinase reaction buffer in the absence or presence of CK2α. After phosphorylation, each kinase reaction mix was diluted to a final volume of 28 μl with caspase 7 buffer (50 mM HEPES [pH 7.5], 50 mM NaCl, 10 mM EDTA, 0.1% 3-[(3-choladmidopropyl)-dimethylammonio]-1-propanesulfonate, 5% glycerol, and 10 mM DTT) and incubated further for 40 min at 37°C in the absence or presence of 1U of active recombinant human caspase 7 (Millipore). Reactions were stopped by the addition of SDS-PAGE buffer, separated on a 10% SDS-PAGE gel, and analyzed by Western blotting using anti-YY1 (C20) and anti-YY1 (pS118) antibodies.
Immunoprecipitation and λ-phosphatase assay.
Immunoprecipitation (IP) of Flag-YY1 from HeLa–Flag-YY1 stable cells was performed using the anti-Flag mouse MAb cross-linked to resin beads (resin M2; Sigma). WCEs were prepared and incubated with the antibody overnight, with rotation, at 4°C. Resin M2-Flag-YY1 complex was collected by centrifugation at 500 × g at 4°C for 2 min and then washed three times with lysis buffer and one additional time with lysis buffer without phosphatase inhibitors. Equal aliquots of the beads were then resuspended in phosphatase buffer (New England BioLabs, Beverly, MA) in the presence of 2 mM MnCl2 and incubated at 30°C, with or without λ-phosphatase (New England BioLabs, Beverly, MA) for 30 min. Reactions were then stopped by the addition of 4× SDS-PAGE buffer and loaded for separation on a 10% SDS-PAGE gel.
Plasmid constructions: pEGFP-YY1.
To construct pEGFP-tagged YY1 (WT) and (S118A) mammalian expression plasmids, an NcoI/EcoI fragment encompassing the open reading frame of human YY1 was digested from pET-20b-YY1, gel purified, and inserted into a BglII/EcoRI digest of the pEGFP-C1 vector (Clontech), after blunting the NcoI and BglII sites of the insert and vector, respectively. The NcoI and BglII sites were blunted using the Klenow fragment of polymerase I (NEB).
Transfections of small interfering RNA and plasmid constructs.
The two bidirectional epitope-tagged constructs used for CK2α overexpression, pRS3 and pGV13 (61), were a gift from David W. Litchfield, Department of Biochemistry, University of Western Ontario, London, Ontario, Canada. The plasmids were transiently cotransfected into cells along with the reverse tetracycline transactivator advanced vector (rtTA; a gift from Choogan Lee, College of Medicine, Florida State University). In the presence of 1 μg/ml of doxycycline (Dox; Sigma), pRS3 or pGV13 coordinately express the catalytic isoform of CK2 (CK2α) along with the regulatory subunit (CK2β), or the kinase-inactive CK2α mutant (K68M) together with (CK2β) subunit. The genes of interest were transfected into cells by using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. The small interfering RNAs (siRNAs) were transfected into cells (grown as previously described) by using DharmaFECT 1 transfection reagent from Dharmacon (Lafayette, CO) according to the manufacturer's instructions. The control On-Target plus nontargeting pool siRNA (catalog number D-001810-10-05]) and CK2α On-Target plus SMARTpool siRNA (catalog number L-003475-00]) were purchased from Dharmacon (Lafayette, CO). The CK2α SMARTpool siRNA contained four different siRNA duplexes that targeted the 3′-untranslated region (UTR) of the CK2α gene.
CK2α cDNA rescue assay.
A CK2α cDNA rescue experiment was performed to verify that the observed decrease in S118 phosphorylation was due to CK2α knockdown. HEK293 cells were plated, cultured overnight, and then transfected with control siRNA or CK2α siRNA. After 48 h of knockdown, the siRNA-resistant CK2α cDNA construct (pRS3) (61) was transfected into the cells as described previously and allowed to express for an additional 48 h in culture in the presence of doxycycline. The cells were then lysed, and WCEs were analyzed by Western blotting.
Densitometric quantitation and statistical analysis.
Relative intensities of autoradiogram bands were quantitated using NIH ImageJ software (http://rsbweb.nih.gov/ij/). The two-tailed Student t test analysis was used to test for significance where indicated. Differences were considered significant when P was <0.05.
RESULTS
Mapping of the CK2α phosphorylation site in vitro.
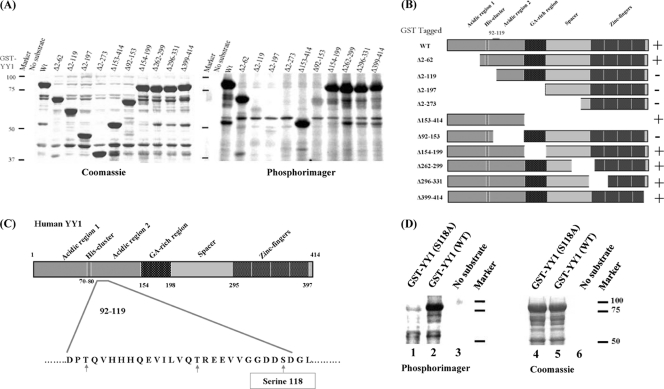
CK2α showed very high activity for phosphorylating YY1 in an in vitro assay. A two-step procedure was used for mapping the CK2α phosphorylation site on YY1. First, N-terminal GST-tagged YY1 (WT) and YY1 deletion mutants (Fig. 2B) were overexpressed in Rosetta cells (DE3), and then bacterial lysates were used in a radioactive in vitro kinase assay with purified CK2α. All reactions were performed in kinase buffer for 30 min at 30°C, stopped with sample buffer, and analyzed as described in Materials and Methods (Fig. 2A). As shown in Fig. 2A, CK2α phosphorylated GST-YY1 (WT) and GST-YY1(Δ2–62), while the deletion of the first (N-terminal) 119 amino acids [GST-YY1(Δ2–119), (Δ2–197), and (Δ2–273)] abolished phosphorylation. This indicated that the region of CK2α phosphorylation of YY1 lies between residues 62 and 119. As additional evidence, deletion clones GST-YY1(Δ153–414), (Δ154–199), (Δ262–299), (Δ296–331), and (Δ399–414) were positive for phosphorylation, while deletion of amino acids 92 to 153 abolished phosphorylation (Fig. 2A). The latter result narrowed down the site of phosphorylation to a region between amino acids 92 and 119 (Fig. 2B).
Fig 2.
CK2α phosphorylation of YY1 in its transactivation domain in vitro. (A) GST-tagged YY1 deletion mutants used in the radioactive in vitro kinase reactions are indicated above the lanes. The no-substrate kinase reaction served as a negative control to eliminate the possibility of CK2α autophosphorylation. Kinase reaction mixtures were separated on a 10% SDS-PAGE gel, and the gel was stained with Coomassie blue (left) to visualize the protein bands, dried, and incubated overnight with a phosphorimager screen (right) at room temperature. The screen was then scanned on a Typhoon 9410 imager (Amersham Biosciences). (B) Diagram of GST-YY1 (WT) and GST-YY1 deletion mutants (6) used in the kinase reactions shown in panel A. The region between amino acids 92 and 119, shown on the full-length YY1, is the region identified as the site of CK2α phosphorylation. + and − indicate the presence or absence of phosphorylation. (C) Diagram of the different domains of the YY1 protein. Amino acid residues 92 to 119 are shown; serine and threonine residues within this amino acid sequence are indicated by arrows. Serine 118, marked in a rectangle, is the best candidate residue for CK2α phosphorylation. (D) Radioactive in vitro kinase assay results with GST-YY1 WT or S118A with CK2α, as described for panel A.
Next, we set out to determine the specific residue phosphorylated by CK2α. Examining residues 92 to 119 of YY1 revealed the presence of one serine and two threonine residues (Fig. 2C). The serine 118 residue was judged to be the most favorable CK2α phosphorylation site of the three, since it is surrounded by two negatively charged aspartic amino acids. CK2 specificity involves acidic amino acids adjacent to the phosphorylation site (27). To test whether this residue is the phosphorylation site for CK2α, a mutation of S118 to an alanine residue, which cannot be phosphorylated, was generated and subsequently used as substrate for CK2α in a radioactive in vitro kinase assay. Substitution of serine with alanine at this residue drastically reduced phosphorylation on YY1, as observed in lane 1 compared to the wild-type YY1 in lane 2 (Fig. 2D). Equal loading of GST-YY1 and the mutant YY1 is shown in lanes 4 and 5. Lanes 3 and 6 are negative-control lanes, to check for CK2α autophosphorylation activity (Fig. 2D). These data are consistent with the interpretation that serine 118 is the site for phosphorylation of YY1 in vitro. Also, high-throughput phospho-proteomic studies have shown that serine 118 is phosphorylated in vivo (11, 33, 35, 40, 43).
Use of anti-YY1 (pS118) antibody to detect specific YY1 phosphorylation at serine 118.
To monitor YY1 phosphorylation at S118 in vivo as well as in vitro and to investigate the biological functional consequences of YY1 phosphorylation at this site, a phospho-specific antibody was generated. Anti-YY1 (pS118), a rabbit polyclonal antibody against a synthetic peptide phosphorylated on serine 118, was produced. Three independent approaches were performed to validate the specificity of the anti-YY1 (pS118) antibody.
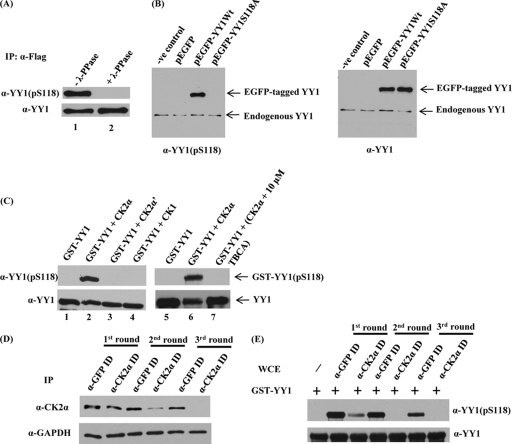
The first step to determine the specificity of our antibody was to verify that it did not recognize dephosphorylated YY1. WCEs from the HeLa-Flag-YY1 stable cell line were prepared, and immunoprecipitation of Flag-YY1 was performed. The immune complexes were incubated at 30°C, with or without λ-phosphatases. Reactions were stopped by adding sample buffer, and the phosphorylation status of YY1 at S118 was assessed by Western blotting with anti-YY1 (pS118) antibody (Fig. 3A). The addition of λ-phosphatase (lane 2), a general protein phosphatase, abolished the reactivity of the antibody with YY1 compared to the untreated sample (lane 1). Reprobing the blot with YY1-specific antibody (Fig. 3A) showed Flag-YY1 loading and confirmed the specificity of the antibody for the phosphorylated S118; the complete loss of the Western blot signal was shown to be due to YY1 dephosphorylation at S118 by λ-phosphatase treatment.
Fig 3.
YY1 is phosphorylated in vivo at S118 and by CK2α in vitro, and immunodepletion of CK2α abolishes S118 phosphorylation in vitro. (A) Flag-YY1 was immunoprecipitated from a HeLa cell line stably overexpressing Flag-YY1 by using anti-Flag mouse MAb cross-linked to resin beads. Immune complexes bound to the beads were washed with lysis buffer, resuspended in phosphatase buffer, and incubated at 30°C, with (lane 2) or without (lane 1) λ-phosphatase for 30 min. Samples were separated on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane was examined for YY1 phosphorylated at serine 118 by using anti-YY1 (pS118) antibody. The blot was stripped and hybridized with anti-YY1 (H10) to verify equal amounts of Flag-YY1. (B) pEGFP-YY1 (WT), the nonphosphorylatable mutant pEGFP-YY1 (S118A), empty pEGFP vector, and the transfection control were expressed transiently in HeLa cells. At 24 h posttransfection, WCEs were prepared, separated by SDS-PAGE, and transferred to a membrane for analysis with anti-YY1 (pS118) antibody and anti-YY1 (H10). (C) Cold in vitro kinase assay of GST-YY1 (WT) with CK2α, CK2α′, or CK1. A CK2-specific inhibitor, TBCA, was added to the kinase reaction mixture (lane 7) at the indicated concentration. All lanes contained purified CK2α as identified, except for lane 1 and lane 5, which served as negative controls. Kinase reaction mixtures were separated on 10% SDS-PAGE gels and transferred to nitrocellulose membranes. The membranes were examined first for YY1 phosphorylation at serine 118 by using anti-YY1 (pS118) antibody. GST-YY1 loading was examined by stripping and hybridizing the blot with anti-YY1 (H10). (D) Whole HEK293 cell extracts were subjected to three sequential rounds of immunodepletion (ID) with GFP antibody as a control or antibody to the α-catalytic subunit of CK2. Following ID, CK2α expression levels were assessed by separating cell lysates by SDS-PAGE and analyzing by Western blotting using anti-CK2α antibody. Also, the blot was stripped and hybridized with anti-GAPDH as a loading control. (E) Western blot analysis was performed after cold in vitro kinase assay reactions with both the control and CK2α-immunodepleted HEK293 WCEs from all the immunodepletion rounds, as the sources for kinase activity, and bacterially expressed GST-YY1 bound to glutathione beads as substrate. Reaction mixtures were separated on SDS-PAGE, transferred to nitrocellulose membranes, probed with anti-YY1 (pS118), and then stripped and reprobed with anti-YY1 (H10) antibody.
Next, we used the site-directed S118 mutation to alanine. The YY1S118A mutant was subcloned into an enhanced green fluorescent protein (EGFP)-tagged mammalian expression plasmid. WCEs were prepared from HeLa cells transiently transfected with pEGFP-YY1 (WT) or pEGFP-YY1 (S118A) constructs and analyzed by Western blotting. The anti-YY1 (pS118) antibody detected the endogenous YY1 protein, as well as EGFP-tagged YY1 from cells that were transfected with wild-type YY1 plasmid, but not the S118A YY1 mutant (Fig. 3B). Immunoblot analysis measuring total YY1 revealed that EGFP-YY1 (S118A) and EGFP-YY1 (WT) proteins were expressed at comparable levels (Fig. 3B).
We then performed a cold in vitro kinase assay to check the immunoreactivity of the phospho-specific antibody (Fig. 3C). GST-YY1 (WT) was used as a substrate for purified CK2α, CK2α′, or CK1. All kinase reactions were assessed by Western blotting with anti-YY1 (pS118). In this assay, GST-YY1 (WT) was highly phosphorylated by CK2α (lanes 2 and 6). Neither CK2α′ (lane 3) nor CK1 (lane 4) phosphorylated S118. The addition of the specific CK2 inhibitor TBCA (tetrabromocinnamic acid) (44) (lane 7) abolished the CK2α phosphorylation at S118. As expected in the absence of CK2α (lanes 1 and 5), incubation of GST-YY1 (WT) with kinase buffer and ATP exhibited no reactivity with anti-YY1 (pS118). Equal loading in all lanes was verified by stripping and reprobing the blot with anti-YY1 (H10). These results indicated the following: (i) anti-YY1 (pS118) antibody is very specific for the phosphorylated form of YY1 at serine 118, (ii) YY1 is phosphorylated in vivo at S118, and (iii) CK2α phosphorylates YY1 at S118 in vitro.
Immunodepletion of CK2α from WCE abolishes in vitro phosphorylation of YY1.
Next, we wanted to provide in vivo evidence that serine 118 on YY1 is indeed a substrate for CK2α. For this purpose, CK2α was immunodepleted from HEK293 cellular extracts. After three sequential rounds of immunodepletion, the levels of CK2α were greatly diminished compared to control levels as well as levels observed after the first and second rounds of CK2α immunodepletion. GAPDH was used to show equal loading (Fig. 3D). Equal amounts of control and CK2α-depleted extracts from all the immunodepletion rounds were then used in a cold in vitro kinase assay with GST-YY1. The kinase reaction mixtures were analyzed by Western blotting, using anti-YY1 (pS118) antibody and then anti-YY1 (H10) antibody. Figure 3E shows that incubation of GST-YY1 with depleted extracts from the second and third rounds of immunodepletion with CK2α abolished phosphorylation of YY1 at S118 relative to the antibody controls (GFP-depleted WCEs) and to those incubated with the extracts from the first round of CK2α immunodepletion, indicating very low or missing CK2α activity in the depleted extracts. In the absence of extracts, incubation of GST-YY1 with kinase buffer and ATP exhibited no reactivity with anti-YY1 (pS118). Equal loading in all lanes was verified by stripping and reprobing the blot with anti-YY1 (H10). These results confirm that CK2α is the kinase responsible for YY1 phosphorylation at S118.
CK2α overexpression increases S118 phosphorylation while CK2α knockdown decreases S118 phosphorylation in vivo.
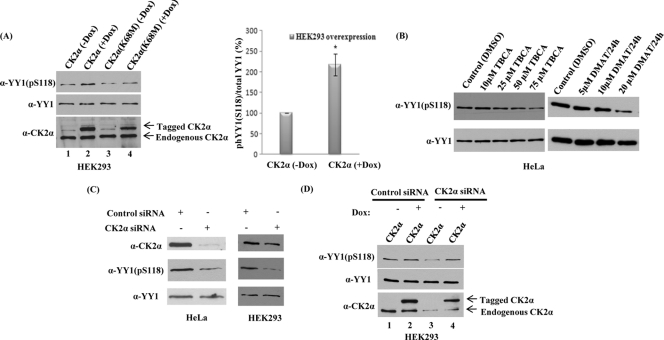
We then examined whether endogenous YY1 could be phosphorylated by CK2α in vivo. CK2α overexpression was used to examine the role of CK2α on YY1 S118 phosphorylation. Using the Tet-On gene expression system, two CK2α bidirectional constructs, CK2α and the kinase-inactive CK2α mutant (K68M) (61), were transiently cotransfected into HEK293 cells along with the rtTA vector to regulate the expression of CK2α. The constructs were expressed in the presence of Dox and repressed in the absence of Dox. To examine whether the regulated CK2α expression altered YY1 S118 phosphorylation, cells were lysed 24 h after transfection and analyzed by Western blotting. Figure 4A shows that overexpression of CK2α in the presence of doxycycline induced a significant increase in phosphorylation of S118 in vivo (lane 2). Conversely, overexpression of a catalytically inactive form of CK2α (lane 4) did not show an increase in phosphorylation of YY1. The same blot was stripped and reanalyzed with anti-YY1 (H10) antibody (Fig. 4A) and showed that the level of endogenous YY1 protein in all lanes was equal and unaffected by CK2α overexpression. In the absence of Dox, expression levels of CK2α and the kinase-inactive CK2α mutant (K68M) plasmids were repressed and served as negative controls (lanes 1 and 3). The expression of both the active and the catalytically inactive forms of CK2α was confirmed by reprobing with anti-CK2α antibody (Fig. 4A, lanes 2 and 4). These data showed that increased expression of only the active form of CK2α resulted in a significant increase in S118 phosphorylation of YY1 in vivo.
Fig 4.
CK2α overexpression increases S118 phosphorylation while CK2α knockdown decreases S118 phosphorylation in vivo. (A) CK2α and the kinase-inactive CK2α mutant (K68M) constructs were transiently cotransfected into HEK293 cells with the tetracycline transactivator advanced vector (rtTA) in the presence (+) or absence (-) of 1 μg/ml of Dox. The constructs were expressed in the presence of Dox and repressed in the absence of Dox. Twenty-four hours later, WCEs were prepared and proteins were separated on an SDS-PAGE gel. Levels of protein expression were analyzed on Western blots with anti-YY1 (pS118) antibody, anti-YY1 (H10) antibody, or anti-CK2α antibody. As shown in panel A, data from three repeats of the HEK293 CK2α overexpression experiments were quantitated. Here, the y axis represents the percent phosphorylation of YY1 at S118 relative to the total amount of YY1 protein and normalized to the control. Percentages are presented as means ± standard errors of the means of three independent experiments. *, P < 0.05 as determined by two-tailed Student's t test analysis. (B) Asynchronous HeLa cells were grown in medium containing either TBCA or DMAT for 24 h at the indicated concentrations. Cells were collected, WCEs were prepared, and samples were analyzed on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane was examined for YY1 phosphorylation at serine 118 by using anti-YY1 (pS118) antibody. The blot was stripped and hybridized with anti-YY1 (H10) antibody to verify YY1 levels. (C) HeLa and HEK293 cells were transiently transfected for 72 h with control siRNA or CK2α-specific siRNA to knock down CK2α. CK2α expression levels following knockdown as well as the phosphorylation status of S118 on YY1 were assessed by analyzing cell lysates by Western blotting using anti-CK2α antibody and anti-YY1 (pS118) antibody, respectively. To rule out possible effects of CK2α on endogenous YY1 protein levels, the blot was stripped and reanalyzed with anti-YY1 (H10) antibody. (D) Control and CK2α siRNA transfections were carried out in HEK293 cells as described previously. After 48 h of siRNA transfection, HEK293 cells were transfected with an siRNA-resistant CK2α cDNA construct as mentioned above for 48 h. The CK2α expression levels, the phosphorylation status of S118 on YY1, and also the endogenous YY1 protein levels were assessed by analyzing cell lysates with Western blotting.
After overexpression of CK2α caused an increase in S118 phosphorylation, we investigated the effects of two different specific CK2 inhibitors (TBCA and DMAT) and CK2α siRNA knockdown on YY1 S118 phosphorylation. Asynchronous HeLa cells were incubated with either TBCA or DMAT or the solvent, DMSO, as control, for the concentrations and times indicated in Fig. 4B. At the end of the incubation period, WCEs were prepared, and samples were analyzed. With the anti-YY1 (pS118) antibody, we found that incubation of cells with TBCA or DMAT for 24 h significantly reduced in vivo phosphorylation of YY1 (Fig. 4B). The total amount of YY1 protein loaded in all lanes was comparable, as shown by reprobing the same Western blots, which were stripped and reanalyzed with anti-YY1 (H10) antibody (Fig. 4B).
Next, CK2α levels were significantly reduced in vivo by siRNA knockdown when we used a pool of four different siRNA duplexes which target the CK2α 3′-UTR. HeLa and HEK293 cells were transiently transfected for 72 h with control siRNA or CK2α siRNA (Fig. 4C). Following knockdown, both CK2α levels and the phosphorylation of YY1 at S118 were significantly reduced compared to YY1 levels (Fig. 4C).
Importantly, the decrease in YY1 S118 phosphorylation and CK2α protein levels observed upon CK2α knockdown in Fig. 4C were effectively reversed when endogenous CK2α was rescued by overexpression (Fig. 4D). After 48 h of CK2α knockdown in HEK293 cells, the siRNA-resistant CK2α cDNA construct was transfected into the cells and allowed to express for an additional 48 h of culture in the presence of doxycycline. WCEs were analyzed by Western blotting with anti-CK2α, anti-YY1 (pS118), and anti-YY1 (H10) antibodies. Endogenous CK2α protein levels were significantly reduced in HEK293 cells transfected with CK2α siRNA (Fig. 4D, lanes 3 and 4) compared with cells transfected with control siRNA (Fig. 4D, lanes 1 and 2). However, CK2α overexpression in control siRNA- or CK2α siRNA-transfected cells rescued CK2α expression, resulting in phosphorylation of S118 in vivo (Fig. 4D, lanes 2 and 4). The total amounts of endogenous YY1 protein in all lanes were equal and unaffected by the transfections. Collectively, the results demonstrated that YY1 S118 phosphorylation is due to CK2α and that YY1 is a substrate for CK2α in vivo.
Phosphorylation of YY1 on S118 interferes with its cleavage by caspase 7 in vitro.
We next examined whether the presence of a phosphate group on S118 on YY1 could inhibit its cleavage by caspase 7 in vitro. Bacterially expressed and purified YY1 was incubated in kinase buffer in the presence or absence of CK2α and then subjected to an in vitro caspase 7 cleavage assay as described in Materials and Methods. The full-length YY1 and its cleavage product were assessed by Western blotting with anti-YY1 (C20) antibody (Fig. 5A, upper panel). Lane 4 shows that YY1 phosphorylation on S118 by CK2α prior to cleavage resulted in a significant reduction in the levels of YY1Δ119 cleavage product, compared to lane 3, where kinase was absent and YY1 was not phosphorylated. The mobility of the YY1Δ119 fragment seen in lane 3 was identical to that of the fragment generated within apoptotic HeLa WCEs (lane 6). Phosphorylation of YY1 at S118 by CK2α was confirmed by probing with anti-YY1 (pS118) antibody (Fig. 5A, lower panel). As expected in the absence of CK2α (lanes 1 and 3), incubation of YY1 with kinase buffer and ATP exhibited no reactivity with the phospho-specific antibody. Therefore, these data reveal that phosphorylation of YY1 at S118 prevents cleavage by caspase 7 in vitro, while unphosphorylated YY1 is susceptible to caspase 7 cleavage.
Fig 5.
Phosphorylation of YY1 at S118 decreases its susceptibility to cleavage by caspase 7. (A) Bacterially expressed and purified YY1 was incubated with nonradioactive cold ATP in kinase buffer in the presence or absence of purified CK2α. The kinase reaction mixtures were then used as substrate sources for an in vitro cleavage assay with active recombinant caspase 7 protein. Reaction mixtures were then resolved on SDS-PAGE gels and transferred to a nitrocellulose membrane. Reaction mixtures containing full-length YY1 and YY1Δ119 cleavage product were analyzed by Western blotting using anti-YY1 C20 (upper panel). HeLa control (lane 5) and apoptotic (lane 6) WCEs were loaded on the same gel as positive controls. The faint additional fragment in lanes 3 and 4 is an artifact, since it was not observed in apoptotic HeLa WCEs (lane 6). The in vitro samples analyzed in the upper panel (lanes 1 to 4) were separated by PAGE and blotted with the anti-YY1 (pS118) to distinguish the phosphorylated form of YY1 at S118 from the unphosphorylated YY1, as shown in the lower panel. (B) pEGFP-YY1 (WT), nonphosphorylatable mutant pEGFP-YY1 (S118A), empty pEGFP vector, and the transfection control were expressed transiently in HeLa cells. At 24 h posttransfection, cells were either left untreated or were treated with 1 μM STS for 5 h. Samples were separated by PAGE for Western analysis using the anti-GFP antibody to distinguish the recombinant YY1 from endogenous proteins. The antibody detected the Fl-tagged YY1 as well as the N-terminally EGFP-tagged YY1 apoptotic fragments cleaved from the tagged overexpressed YY1 protein. Fl, full length. The blot shown was stripped and analyzed for PARP cleavage, a well-known caspase substrate, using an anti-PARP antibody. Fl-PARP, full-length PARP; t-PARP, truncated PARP; NS, nonspecific. Also, the blot was stripped and hybridized with anti-GAPDH as a loading control.
Role of serine 118 phosphorylation during apoptosis.
Since the phosphorylated S118 residue is adjacent to the caspase 7 cleavage site (D119) on YY1 (29) (Fig. 1), we reasoned that phosphorylation at S118 might influence susceptibility of YY1 to cleavage by caspase 7 during apoptosis.
To investigate this hypothesis, we analyzed the extent of cleavage of the YY1S118A mutant protein during apoptosis. This mutant construct was subcloned into an EGFP-tagged mammalian expression plasmid. When pEGFP-YY1 (WT) and pEGFP-YY1 (S118A) were expressed in mammalian cells, these constructs produced YY1 fusion proteins with an N-terminal EGFP tag.
The constructs were expressed transiently in asynchronous HeLa cells. Twenty-four hours posttransfection, cells were either left untreated or were treated with the apoptotis-inducing agent staurosporine (STS) at 1 μM for 5 h. STS, a broad-spectrum protein kinase inhibitor, has been reported to induce the intrinsic (stress) pathway of apoptosis (4).
To distinguish N-terminal YY1 peptides produced by cleavage of the tagged YY1 proteins from those of endogenous YY1 during apoptosis, WCEs were analyzed by Western blotting using anti-GFP antibody (Fig. 5B). This antibody recognizes only the overexpressed EGFP-tagged form of YY1. It also detects the N-terminal YY1 fragments cleaved from the tagged overexpressed YY1 protein, consisting of the first 12 and/or 119 amino acids that still have the N-terminal EGFP tag and which the YY1 C20 antibody does not recognize. As observed in Fig. 5B, the overexpressed EGFP-YY1 (S118A) mutant protein was more sensitive to caspase cleavage than the overexpressed YY1 wild-type protein. Also, we analyzed caspase-mediated cleavage of PARP (Fig. 5B), a marker of apoptosis that was found to occur in parallel to that of YY1 cleavage, as expected. GAPDH was used to show equal loading. We conclude that prevention of phosphorylation at serine 118 enhances YY1 cleavage by caspase 7.
CK2α knockdown in vivo enhances YY1 cleavage during apoptosis.
To determine whether a similar resistance to cleavage by caspase 7 is conferred by YY1 phosphorylation in living cells, asynchronous HeLa cells were grown in medium containing TBCA or the solvent, DMSO, as control for 24 h at the concentrations indicated in Fig. 6A. After 24 h, STS was added to cells for 5 h in the absence or presence of the CK2 inhibitor. Cell lysates were harvested and analyzed by immunoblotting. Using anti-YY1 (C20) antibody, we found that inhibiting endogenous CK2α with TBCA enhanced YY1 caspase cleavage when exposed to STS (Fig. 6A). PARP cleavage is similar to YY1, as shown by the same Western blot after stripping and hybridizing with anti-PARP antibody. TBCA by itself at the doses used in this study could not induce either YY1 cleavage or apoptosis (Fig. 6A).
Fig 6.
CK2α knockdown in vivo enhances YY1 cleavage in the presence of apoptotic stimuli. (A and B) Asynchronous HeLa cells were preincubated with TBCA (a specific CK2 inhibitor), or with DMSO as a control, for 24 h at the indicated concentrations. After 24 h, either STS (A) or anti-Fas (B) anitbody was added to cells at 1 μM for 5 h or 100 ng/ml for 6 h, respectively, in the absence or presence of TBCA. WCEs were prepared, and proteins were separated on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. Cleavage of full-length and endogenous YY1 was detected by hybridizing the blot with anti-YY1 antibody (C20; rabbit polyclonal). Two exposures of the blot showed the apoptotic YY1Δ119 and YY1Δ12 cleavage products. The blots were stripped and reprobed by Western analysis using anti-PARP antibody. (C) U2OS cells were transiently transfected with control siRNA or siRNA to knock down CK2α. Following siRNA transfections for 48 h, cells were either left untreated or treated with the apoptotic stimulus STS at 1 μM for 5 h. CK2α expression levels, as well as the phosphorylation status of S118 on YY1, were assessed by separating cell lysates by SDS-PAGE and analyzing by Western blotting using anti-CK2α antibody and anti-YY1 (pS118) antibody, respectively. Also, the same samples were analyzed with anti-YY1 (H414) antibody (to detect endogenous YY1 protein levels and cleavage) and anti-PARP antibody (a marker of apoptosis).
To test whether YY1 cleavage is similarly enhanced by a CK2 inhibitor in response to other apoptotic inducers, we used anti-Fas antibody instead of STS (Fig. 6B) to induce apoptosis. Anti-Fas antibody stimulates the extrinsic (death receptor-dependent) apoptotic pathways after binding to Fas (20, 48). HeLa cells were exposed to anti-Fas antibody in the absence or presence of TBCA (Fig. 6B), as described for the experiment shown in Fig. 6A. The cleavage of endogenous YY1 and induction of apoptosis were analyzed by immunoblotting as described previously. The enhancement of YY1 cleavage seen in Fig. 6A is also apparent in Fig. 6B. Consistent with the results of the experiment discussed in Fig. 5B, for which the YY1 S118 mutant was used, inhibition of CK2α significantly increased the amount of YY1 cleavage in the presence of apoptotic stimuli.
To further address the effect of serine 118 phosphorylation by CK2α on YY1 cleavage during apoptosis, endogenous CK2α was depleted in vivo by siRNA knockdown. U2OS cells were transiently transfected with control siRNA or CK2α siRNA. CK2α expression levels following knockdown were markedly reduced, as was the phosphorylation of S118 on YY1, as shown by immunoblot analysis using anti-CK2α and anti-YY1 (pS118) antibodies (Fig. 6C). The total amounts of YY1 protein loaded in lane 1 and lane 3 were comparable, ruling out any possible effect of CK2α knockdown on endogenous YY1 protein levels. To investigate the effect of CK2α kinase activity on YY1 cleavage by caspases, cells were either left untreated or treated with STS following transfection with CK2α siRNA. Whole-cell lysates prepared from transfected cells were analyzed with anti-YY1 (H414) antibody, which detected endogenous YY1 cleavage, similar to the anti-YY1 (C20) antibody. As shown in Fig. 6C, reduction of endogenous CK2α, this time with siRNA, enhanced YY1 cleavage in U2OS cells exposed to STS. Under these conditions, PARP was also cleaved, with kinetics similar to YY1. Interestingly, the phosphorylation of YY1 at S118 was reduced upon STS treatment (Fig. 6C, lanes 2 and 4). Also, after incubation with STS, an accelerated cleavage of YY1 induced by CK2α knockdown was observed in HeLa cells (data not shown).
DISCUSSION
A large amount of experimental data has led to the hypothesis that YY1 is a factor that can lead to cell survival and thus transformation by enhancing cellular resistance and insensitivity to apoptotic stimuli (1, 21, 23, 25). These data also suggest that this is the reason YY1 is targeted for destruction by the caspases. However, the mechanisms regulating YY1 cleavage and the relevance of its destruction during apoptosis are still not clear. In this work, we have shown that the caspase-dependent cleavage of YY1 is regulated by posttranslational modification, phosphorylation, during programmed cell death.
The fact that aspartic acid is the frequent residue in the CK2 phospho-acceptor sites as well as the target site of caspases (37, 47) suggests that CK2-mediated phosphorylation regulates the cleavage of several caspase substrates and protects them from caspase-mediated proteolysis (66). Caspase 7 cleaves YY1 immediately C-terminal of aspartic acid 119 (D119) during apoptosis (29). Since the S118 residue is located in the transcription activation region of YY1, proximal to the caspase 7 consensus cleavage site (Fig. 1), we predicted that the phosphate group added to S118 by CK2α might regulate the accessibility of caspase 7 to its recognition site (Asp119) and thus affect the cleavage of YY1. To test this hypothesis, we replaced the CK2α phosphorylation site with an alanine residue and observed that the YY1S118A mutant protein, which could not be phosphorylated, was more sensitive to caspase-mediated cleavage in vivo (Fig. 5B). Also, the in vitro data presented here clearly demonstrate that phosphorylation of YY1 at S118 by CK2α renders YY1 resistant to cleavage by caspase 7 (Fig. 5A). Moreover, we showed that CK2α overexpression increased phosphorylation of S118 in vivo (Fig. 4A), while knockdown of intracellular CK2α or loss of its kinase activity by CK2 inhibitors and siRNA decreased S118 phosphorylation of YY1 (Fig. 4B and C) and enhanced its cleavage in response to both extrinsic and intrinsic apoptotic stimuli (Fig. 6). This demonstrated that CK2α-mediated phosphorylation of YY1 likely protects it from caspase cleavage during cell death. Since TBCA and DMAT, as well as CK2α knockdown with siRNA, significantly reduced phosphorylation at S118 but did not completely abolish it, it is possible that another kinase(s) may phosphorylate S118 in vivo in other contexts or in response to other signals. We checked whether other acidophilic kinases, such as CK2α′ or CK1, cooperate with CK2α to phosphorylate YY1 at S118. None of these kinases was shown to phosphorylate S118 in an in vitro cold kinase assay (Fig. 3C). A more likely explanation is that this serine 118 phosphorylation is long lived (low turnover rate) in the absence of apoptotic induction signals.
In our examination of YY1 phosphorylation at S118, we wanted to investigate whether phosphorylation at this site has any effect on the biochemical activities of YY1. Phosphorylation at S118 did not affect either the cellular localization of YY1 or its DNA binding activity (data not shown). YY1 is a transcriptional regulator known to have an antiapoptotic role. It is involved in controlling the expression of numerous genes, including several directly associated with apoptosis (1). We speculate that since S118 is located in the transactivation domain of YY1, protection of the transcriptional activation activity of this factor by phosphorylation, particularly for the expression of specific antiapoptotic genes, including Bcl-2 family members or inhibitors of caspases (3, 38, 52, 63), is responsible for the enhanced sensitivity of the S118A mutant to apoptotic stimuli. It is also possible that this phosphorylation has no effect other than to protect YY1 from cleavage during apoptosis.
We checked if phosphorylation at S118 is cell cycle regulated. We synchronized HeLa cells by using an automated mitotic shake-off machine to collect mitotic cells. Also, cells synchronized by double-thymidine blocks were used as an alternate method to examine cells in mid-S-phase through mitosis. Using the two methods to synchronize cell populations, we prepared WCEs and performed Western blot analysis with our phospho-specific antibody to phosphorylated serine 118 (data not shown). We found that YY1 was constitutively phosphorylated under normal growth conditions at all points during the cell cycle. There was no fluctuation in S118 phosphorylation (data not shown). This was not surprising in light of the constitutive activity of CK2α throughout the cell cycle (60). Our data indicate that serine 118 is dephosphorylated in response to signals such as apoptotic stimuli (e.g., STS and Fas) for subsequent cleavage by caspase 7 (Fig. 6C, lanes 2 and 4). The putative phosphatase(s) involved in dephosphorylating YY1 at serine 118 is unknown.
The consensus CK2 phosphorylation sequence, S/T-X-X-E/D, involves acidic residues (aspartic or glutamic acid) or a phospho-serine/threonine at position n + 3 relative to the phospho-acceptor target (17, 27, 30, 47). It is interesting that the S118 site we mapped in the YY1 caspase 7 recognition motif does not perfectly match this consensus. YY1 lacks the determinant at n + 3 (Leu in YY1 [Fig. 2C]), as does another proven caspase target protein, Bid (14). However, YY1 is surrounded by negatively charged aspartic amino acids, a dominant specificity determinant for CK2, upstream of the phospho-acceptor site and at position n + 1, as do a number of other known CK2 substrates, such as Fragmin, NDPK A, EBV (Epstein-Barr virus) ZEBRA, CUT, ER, and RAD proteins (37). Thr108 (T108REE111), a residue also located in the region where CK2α phosphorylates YY1 (Fig. 2C), conforms perfectly with the bioinformatics-generated CK2 consensus sequence. However, we did not test this residue, since several pioneer studies on sites phosphorylated by CK2 revealed the negative role that basic residues exert (in this case R) at any position close to Ser/Thr, especially at positions n + 1 and n + 3, where the frequency of the presence of an acidic residue predominates (34). Also, our in vitro and in vivo findings showed clearly that S118 is indeed a site for CK2α phosphorylation.
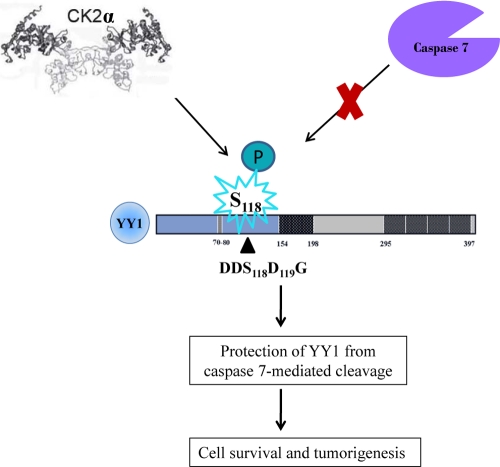
Our study here describes an in vivo regulatory mechanism of the transcriptional regulator YY1 by which its sensitivity to caspase cleavage is prevented by CK2α phosphorylation (Fig. 7). Dephosphorylation/phosphorylation of YY1 and its precise role in the apoptotic process remain to be determined.
Fig 7.
Regulation of caspase 7 cleavage of YY1 by CK2α. The model represents how YY1, a caspase substrate, is protected from caspase 7 cleavage when it is phosphorylated at S118 by CK2α. S118 is a residue adjacent to the caspase 7 recognition site. Failure of YY1 to undergo caspase cleavage in response to apoptotic signals could contribute to tumorigenesis.
Footnotes
Published ahead of print 19 December 2011
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Affar el B, et al. 2006. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 26:3565–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahmad KA, Wang G, Unger G, Slaton J, Ahmed K. 2008. Protein kinase CK2: a key suppressor of apoptosis. Adv. Enzyme Regul. 48:179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andera L, Wasylyk B. 1997. Transcription abnormalities potentiate apoptosis of normal human fibroblasts. Mol. Med. 3:852–863 [PMC free article] [PubMed] [Google Scholar]
- 4. Andreau K, et al. 2004. Preapoptotic chromatin condensation upstream of the mitochondrial checkpoint. J. Biol. Chem. 279:55937–55945 [DOI] [PubMed] [Google Scholar]
- 5. Austen M, Cerni C, Lüscher-Firzlaff JM, Lüscher B. 1998. YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene 17:511–520 [DOI] [PubMed] [Google Scholar]
- 6. Austen M, Lüscher B, Lüscher-Firzlaff JM. 1997. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 272:1709–1717 [DOI] [PubMed] [Google Scholar]
- 7. Barkett M, Xue D, Horvitz HR, Gilmore TD. 1997. Phosphorylation of IκB-α inhibits its cleavage by caspase CPP32 in vitro. J. Biol. Chem. 272:29419–29422 [DOI] [PubMed] [Google Scholar]
- 8. Becker KG, Jedlicka P, Templeton NS, Liotta L, Ozato K. 1994. Characterization of hUCRBP (YY1, NF-E1, delta): a transcription factor that binds the regulatory regions of many viral and cellular genes. Gene 150:259–266 [DOI] [PubMed] [Google Scholar]
- 9. Bowman TL, Kaludov NK, Klein M, Hurt MM. 1996. An H3 coding region regulatory element is common to all four nucleosomal classes of mouse histone-encoding genes. Gene 176:1–8 [DOI] [PubMed] [Google Scholar]
- 10. Canton DA, Litchfield DW. 2006. The shape of things to come: an emerging role for protein kinase CK2 in the regulation of cell morphology and the cytoskeleton. Cell. Signal. 18:267–275 [DOI] [PubMed] [Google Scholar]
- 11. Chen L, Giorgianni F, Beranova-Giorgianni S. 2010. Characterization of the phosphoproteome in LNCaP prostate cancer cells by in-gel isoelectric focusing and tandem mass spectrometry. J. Proteome Res. 9:174–178 [DOI] [PubMed] [Google Scholar]
- 12. Delehouzee S, et al. 2005. GABP, HCF-1 and YY1 are involved in Rb gene expression during myogenesis. Genes Cells 10:717–731 [DOI] [PubMed] [Google Scholar]
- 13. Deng Z, Wan M, Sui G. 2007. PIASy-mediated sumoylation of Yin Yang 1 depends on their interaction but not the RING finger. Mol. Cell. Biol. 27:3780–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desagher S, et al. 2001. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol. Cell 8:601–611 [DOI] [PubMed] [Google Scholar]
- 15. Donohoe ME, et al. 1999. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 19:7237–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duncan JS, Litchfield DW. 2008. Too much of a good thing: the role of protein kinase CK2 in tumorigenesis and prospects for therapeutic inhibition of CK2. Biochim. Biophys. Acta 1784:33–47 [DOI] [PubMed] [Google Scholar]
- 17. Duncan JS, et al. 2010. Regulation of cell proliferation and survival: convergence of protein kinases and caspases. Biochim. Biophys. Acta 1804:505–510 [DOI] [PubMed] [Google Scholar]
- 18. Eliassen KA, Baldwin A, Sikorski EM, Hurt MM. 1998. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol. Cell. Biol. 18:7106–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ericsson J, Usheva A, Edwards PA. 1999. YY1 is a negative regulator of transcription of three sterol regulatory element-binding protein-responsive genes. J. Biol. Chem. 274:14508–14513 [DOI] [PubMed] [Google Scholar]
- 20. Fesik SW. 2005. Promoting apoptosis as a strategy for cancer drug discovery. Nat. Rev. Cancer 5:876–885 [DOI] [PubMed] [Google Scholar]
- 21. Garban HJ, Bonavida B. 2001. Nitric oxide inhibits the transcription repressor Yin-Yang 1 binding activity at the silencer region of the Fas promoter: a pivotal role for nitric oxide in the up-regulation of Fas gene expression in human tumor cells. J. Immunol. 167:75–81 [DOI] [PubMed] [Google Scholar]
- 22. Gordon S, Akopyan G, Garban H, Bonavida B. 2006. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25:1125–1142 [DOI] [PubMed] [Google Scholar]
- 23. Gronroos E, Terentiev AA, Punga T, Ericsson J. 2004. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. U. S. A. 101:12165–12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hiromura M, et al. 2003. YY1 is regulated by O-linked N-acetylglucosaminylation (O-glcNAcylation). J. Biol. Chem. 278:14046–14052 [DOI] [PubMed] [Google Scholar]
- 25. Hongo F, et al. 2005. Inhibition of the transcription factor Yin Yang 1 activity by S-nitrosation. Biochem. Biophys. Res. Commun. 336:692–701 [DOI] [PubMed] [Google Scholar]
- 26. Hurt MM, Bowman TL, Marzluff WF. 1991. A common transcriptional activator is located in the coding region of two replication-dependent mouse histone genes. Mol. Cell. Biol. 11:2929–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kennelly PJ, Krebs EG. 1991. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 266:15555–15558 [PubMed] [Google Scholar]
- 28. Krippner-Heidenreich A, et al. 2001. Targeting of the transcription factor Max during apoptosis: phosphorylation-regulated cleavage by caspase-5 at an unusual glutamic acid residue in position P1. Biochem. J. 358:705–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krippner-Heidenreich A, et al. 2005. Caspase-dependent regulation and subcellular redistribution of the transcriptional modulator YY1 during apoptosis. Mol. Cell. Biol. 25:3704–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuenzel EA, Mulligan JA, Sommercorn J, Krebs EG. 1987. Substrate specificity determinants for casein kinase II as deduced from studies with synthetic peptides. J. Biol. Chem. 262:9136–9140 [PubMed] [Google Scholar]
- 31. Li PF, et al. 2002. Phosphorylation by protein kinase CK2: a signaling switch for the caspase-inhibiting protein ARC. Mol. Cell 10:247–258 [DOI] [PubMed] [Google Scholar]
- 32. Litchfield DW. 2003. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 369:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malik R, et al. 2009. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J. Proteome Res. 8:4553–4563 [DOI] [PubMed] [Google Scholar]
- 34. Marin O, Meggio F, Draetta G, Pinna LA. 1992. The consensus sequences for cdc2 kinase and for casein kinase-2 are mutually incompatible. A study with peptides derived from the beta-subunit of casein kinase-2. FEBS Lett. 301:111–114 [DOI] [PubMed] [Google Scholar]
- 35. Mayya V, et al. 2009. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2:ra46. [DOI] [PubMed] [Google Scholar]
- 36. McDonnell MA, et al. 2008. Phosphorylation of murine caspase-9 by the protein kinase casein kinase 2 regulates its cleavage by caspase-8. J. Biol. Chem. 283:20149–20158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meggio F, Pinna LA. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349–368 [DOI] [PubMed] [Google Scholar]
- 38. Mok CL, et al. 1999. BAD can act as a key regulator of T cell apoptosis and T cell development. J. Exp. Med. 189:575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Montiel-Duarte C, et al. 2008. Resistance to Imatinib Mesylate-induced apoptosis in acute lymphoblastic leukemia is associated with PTEN down-regulation due to promoter hypermethylation. Leuk. Res. 32:709–716 [DOI] [PubMed] [Google Scholar]
- 40. Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. 2006. Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. U. S. A. 103:5391–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oei SL, et al. 1997. Interaction of the transcription factor YY1 with human poly(ADP-ribosyl) transferase. Biochem. Biophys. Res. Commun. 240:108–111 [DOI] [PubMed] [Google Scholar]
- 42. Oei SL, Griesenbeck J, Schweiger M, Ziegler M. 1998. Regulation of RNA polymerase II-dependent transcription by poly(ADP-ribosyl)ation of transcription factors. J. Biol. Chem. 273:31644–31647 [DOI] [PubMed] [Google Scholar]
- 43. Olsen JV, et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3:ra3. [DOI] [PubMed] [Google Scholar]
- 44. Pagano MA, et al. 2007. Tetrabromocinnamic acid (TBCA) and related compounds represent a new class of specific protein kinase CK2 inhibitors. Chembiochem 8:129–139 [DOI] [PubMed] [Google Scholar]
- 45. Palko L, Bass HW, Beyrouthy MJ, Hurt MM. 2004. The Yin Yang-1 (YY1) protein undergoes a DNA-replication-associated switch in localization from the cytoplasm to the nucleus at the onset of S phase. J. Cell Sci. 117:465–476 [DOI] [PubMed] [Google Scholar]
- 46. Petkova V, et al. 2001. Interaction between YY1 and the retinoblastoma protein. Regulation of cell cycle progression in differentiated cells. J. Biol. Chem. 276:7932–7936 [DOI] [PubMed] [Google Scholar]
- 47. Pinna LA. 2002. Protein kinase CK2: a challenge to canons. J. Cell Sci. 115:3873–3878 [DOI] [PubMed] [Google Scholar]
- 48. Plati J, Bucur O, Khosravi-Far R. 2008. Dysregulation of apoptotic signaling in cancer: molecular mechanisms and therapeutic opportunities. J. Cell Biochem. 104:1124–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Riggs KJ, et al. 1993. Yin-yang 1 activates the c-myc promoter. Mol. Cell. Biol. 13:7487–7495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rizkallah R, Alexander KE, Kassardjian A, Lüscher B, Hurt MM. 2011. The transcription factor YY1 is a substrate for Polo-like kinase 1 at the G2/M transition of the cell cycle. PLoS One 6:e15928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rizkallah R, Hurt MM. 2009. Regulation of the transcription factor YY1 in mitosis through phosphorylation of its DNA-binding domain. Mol. Biol. Cell 20:4766–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rong P, Bennie AM, Epa WR, Barrett GL. 1999. Nerve growth factor determines survival and death of PC12 cells by regulation of the bcl-x, bax, and caspase-3 genes. J. Neurochem. 72:2294–2300 [DOI] [PubMed] [Google Scholar]
- 53. Ruzzene M, Penzo D, Pinna LA. 2002. Protein kinase CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole (TBB) induces apoptosis and caspase-dependent degradation of haematopoietic lineage cell-specific protein 1 (HS1) in Jurkat cells. Biochem. J. 364:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shi Y, Lee JS, Galvin KM. 1997. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332:F49–F66 [DOI] [PubMed] [Google Scholar]
- 55. Shi Y, Seto E, Chang LS, Shenk T. 1991. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67:377–388 [DOI] [PubMed] [Google Scholar]
- 56. Shrivastava A, Yu J, Artandi S, Calame K. 1996. YY1 and c-Myc associate in vivo in a manner that depends on c-Myc levels. Proc. Natl. Acad. Sci. U. S. A. 93:10638–10641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sui G, et al. 2004. Yin Yang 1 is a negative regulator of p53. Cell 117:859–872 [DOI] [PubMed] [Google Scholar]
- 58. Takasaki N, Kurokawa D, Nakayama R, Nakayama J, Aizawa S. 2007. Acetylated YY1 regulates Otx2 expression in anterior neuroectoderm at two cis-sites 90 kb apart. EMBO J. 26:1649–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tozser J, et al. 2003. Effect of caspase cleavage-site phosphorylation on proteolysis. Biochem. J. 372:137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Turowec JP, et al. 2010. Protein kinase CK2 is a constitutively active enzyme that promotes cell survival: strategies to identify CK2 substrates and manipulate its activity in mammalian cells. Methods Enzymol. 484:471–493 [DOI] [PubMed] [Google Scholar]
- 61. Vilk G, Saulnier RB, St Pierre R, Litchfield DW. 1999. Inducible expression of protein kinase CK2 in mammalian cells. Evidence for functional specialization of CK2 isoforms. J. Biol. Chem. 274:14406–14414 [DOI] [PubMed] [Google Scholar]
- 62. Walter J, Schindzielorz A, Grunberg J, Haass C. 1999. Phosphorylation of presenilin-2 regulates its cleavage by caspases and retards progression of apoptosis. Proc. Natl. Acad. Sci. U. S. A. 96:1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weinmann P, Gaehtgens P, Walzog B. 1999. Bcl-Xl- and Bax-alpha-mediated regulation of apoptosis of human neutrophils via caspase-3. Blood 93:3106–3115 [PubMed] [Google Scholar]
- 64. Weiss T, et al. 1998. TNFR80-dependent enhancement of TNFR60-induced cell death is mediated by TNFR-associated factor 2 and is specific for TNFR60. J. Immunol. 161:3136–3142 [PubMed] [Google Scholar]
- 65. Yao YL, Yang WM, Seto E. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yin X, Gu S, Jiang JX. 2001. The development-associated cleavage of lens connexin 45.6 by caspase-3-like protease is regulated by casein kinase II-mediated phosphorylation. J. Biol. Chem. 276:34567–34572 [DOI] [PubMed] [Google Scholar]