Abstract
The ETS transcription factor Elf-4 is an important regulator of hematopoietic stem cell (HSC) and T cell homeostasis. To gain insights into the transcriptional circuitry within which Elf-4 operates, we used comparative sequence analysis coupled with chromatin immunoprecipitation (ChIP) with microarray technology (ChIP-chip) assays for specific chromatin marks to identify three promoters and two enhancers active in hematopoietic and endothelial cell lines. Comprehensive functional validation of each of these regulatory regions in transgenic mouse embryos identified a tissue-specific enhancer (−10E) that displayed activity in fetal liver, dorsal aorta, vitelline vessels, yolk sac, and heart. Integration of a ChIP-sequencing (ChIP-Seq) data set for 10 key stem cell transcription factors showed Pu.1, Fli-1, and Erg were bound to the −10E element, and mutation of three highly conserved ETS sites within the enhancer abolished its activity. Finally, the transcriptional repressor Gfi1b was found to bind to and repress one of the Elf-4 promoters (−30P), and we show that this repression of Elf-4 is important for the maturation of primary fetal liver erythroid cells. Taken together, our results provide a comprehensive overview of the transcriptional control of Elf-4 within the hematopoietic system and, thus, integrate Elf-4 into the wider transcriptional regulatory networks that govern hematopoietic development.
INTRODUCTION
ETS transcription factors are known to be important for the proliferation and differentiation of cells within the hematopoietic system (reviewed in references 3 and 31). ETS proteins regulate transcription in concert with other transcription factors by binding a core ETS binding motif (GGAW) via their ETS domain. The ETS factor Elf-4 (also known as myeloid Elf-1-like factor, or MEF) was first cloned from the human megakaryocytic cell line CMK (36) and is expressed in hematopoietic stem cells (HSCs), myeloid and lymphoid lineages (24, 25, 36), ovary, placenta, and the colon (36). Elf-4−/− mice have a severe defect in NK and NK-T cell differentiation, implicating Elf-4 in the regulation of the innate immune system (24), and more recent studies have also shown that Elf-4 regulates the proliferation of both HSCs and CD8+ T cells (25, 58).
In addition to regulating the entry of HSCs into the cell cycle, Elf-4 also promotes the transition of cells from G1 to S phase (35) and has been shown to be a potent activator at the granulocyte-macrophage colony-stimulating factor, interleukin 3 (IL-3), IL-8, and lysozyme promoters (14, 18, 36). Moreover, Elf-4 has been implicated in the development of cancer, as it has been identified as a recurrent site of retroviral integration (30, 34) and has tumor suppressor activity in lung carcinoma cell lines (51). The AML-ETO and PML-retinoic acid receptor α (RARα) fusion oncogenes repress the expression of ELF-4, and analysis of acute myeloid leukemia (AML) patient samples with these translocations showed diminished levels of ELF-4 (1, 10). In addition, a chromosomal translocation involving ELF-4 and another ETS factor, ERG, has been described in a patient with AML (37). Elf-4 is therefore an important regulator of hematopoiesis with critical functions both in the stem cell compartment and mature lineages.
Despite the fact that biological functions of Elf-4 have been well characterized, very little work has focused on the transcriptional regulation of Elf-4. A minimal promoter region has been reported to be bound by the ubiquitous zinc finger transcription factor SP1 in human epithelial cells (21), but no upstream regulators responsible for tissue-specific expression of Elf-4 or comprehensive studies of Elf-4 regulation have been reported to date. Here we have taken a locus-wide approach to analyze the regulation of the mouse Elf-4 gene, which allowed us to identify 5 distinct regulatory regions, functionally validated using both in vitro and in vivo experiments. A tissue-specific enhancer active in blood progenitors and endothelium in transgenic mice was found to be dependent on ETS family transcription factors for its activity. Moreover, the transcriptional repressor Gfi1b bound to and repressed one of three Elf-4 promoters and, furthermore, we found that Elf-4 downregulation is important for normal erythropoiesis. Given the critical roles played by ETS factors in blood progenitors and endothelium and Gfi1b in erythroid cells, our study firmly integrates Elf-4 as an important component of regulatory networks controlling blood progenitor function and erythroid differentiation.
MATERIALS AND METHODS
Expression analysis.
RNA was isolated using TRI reagent (Sigma-Aldrich) according to the manufacturer's instructions. Prior to cDNA synthesis, genomic DNA was removed using Turbo DNA free (Ambion). cDNA was generated using oligo(dT) and Moloney murine leukemia virus reverse transcriptase (Sigma-Aldrich). Mouse Elf-4, Gfi1b, Gapdh, and 18S mRNA levels were measured using the primers listed in Table S1 at http:hscl.cimr.cam.ac.uk/genomic_supplementary.html and quantified using SYBR green (Stratagene) in a real-time PCR. Expression levels were calculated relative to that of Gapdh for cell lines and to 18S for the erythroid maturation analysis.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (27). Briefly, cells were treated with formaldehyde, and the cross-linked chromatin was sonicated (average size, 300 bp) prior to immunoprecipitation with anti-acetyl H3K9 antibody (06-599; Millipore) or anti-H3Me3K4 antibody (04-745; Millipore; a gift from L. O'Neill, Birmingham, United Kingdom). Oligonucleotides used to generate the Elf-4 tiling array were designed on a repeat masked sequence across the Elf-4 locus (ChrX:43492286-43571850, mm7). Oligonucleotides were spotted in triplicate by using the BioRobotics MicroGrid II total array system. The rolling average was calculated across each set of triplicates, and the resulting data were plotted using the variable-width bar graph drawer (http://hscl.cimr.cam.ac.uk/genomic_tools.html). For ChIP-sequencing (ChIP-Seq) analysis in J2E cells, 2 × 107 cells were cross-linked in 1% formaldehyde, lysed, and sonicated into fragments of about 150 to 400 bp prior to immunoprecipitation with anti-Gfi1b antibody (sc-8559X; Santa Cruz Biotechnology). Samples were amplified using the ChIP-Seq DNA sample prep kit from Illumina, following the manufacturer's instructions, and sequenced using the Illumina 2G genome analyzer. Sequencing reads were mapped to the mouse reference genome by using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) (28), converted to a density plot, and displayed with UCSC Genome Browser (http://genome.ucsc.edu/index.html) (19). The ChIP-Seq data set for Gfi1b is available at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html.
Reporter constructs and transfection.
Reporter constructs were generated by amplifying regions of the Elf-4 locus from bacterial artificial chromosome clone RP23-99C8 (BACPAC Resources, Oakland, CA) and cloning into the XhoI/HindIII sites of pGL2 basic or pGL2 promoter reporter constructs (Promega, United Kingdom). Primers are listed in Table S1 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html, and comparative sequence alignments are shown in Fig. S1 at the same website. Numbering of the cloned regions is relative to the ATG of ENSEMBL transcript Elf-4-001. Mutation constructs were made as previously described (13) and verified by sequencing. The primers used are listed in Table S1 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html. Cell lines were maintained as previously described (12, 27, 45). For transient transfections, cells were electroporated with 10 μg of plasmid and 2 μg or 5 μg (HPC7 and BW5147) of the control plasmid pEFBOS LacZ and assayed as previously described (12). Data were normalized to the control plasmid and are expressed relative to pGL2 basic expression. Stable transfections were performed as previously described (27).
Transgenic mouse analysis.
The transgenic beta-galactosidase reporter constructs were generated by cloning the Elf-4 −39P, −30P, and −2P fragments into the XhoI/HindIII site of the pGLac LacZ plasmid (GenBank accession number U19930). The Elf-4 −10E, −10EmETS1-2, −10EmETS3-5, and −16E reporter constructs were generated by cloning each of the fragments downstream of simian virus 40 (SV40) LacZ into the BamHI/SalI site of the pGLac promoter contains SV40 upstream of a LacZ reporter cassette (50). Embryonic day 11.5 (E11.5) F0 transgenic embryos were generated by pronuclear injection of the beta-galactosidase reporter constructs either in-house as previously described (52) or by Cyagen Biosciences (Guangzhou) Inc. (Guangzhou, China). Whole-mount embryos were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) for beta-galactosidase expression and photographed using a Nikon digital sight DS-FL1 camera attached to a Nikon SM7800 microscope for magnification. Images of sections were acquired with a Zeiss AxioCam MRc5 camera attached to a Zeiss Axioscope2plus microscope. All images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
Flow cytometry and in vitro erythroid differentiation assay.
Samples were stained with Ter119-allophycocyanin and CD71-phycoerythrin (BD, Oxford, United Kingdom) antibodies, and dead cells were excluded using 7-aminoactinoycin (7AAD). Flow cytometry analysis was performed using a FACSCalibur apparatus (BD), and cell sorting was performed using a Dakocytomation MoFlo cell sorter. In vitro erythroid differentiation assays were performed as previously described (57, 59). Briefly, fetal livers from E14.5 (C57BL/6 × CBA)F1 murine embryos were extracted by blunt dissection and homogenized. Lineage-negative hematopoietic progenitor cells were isolated using the magnetically activated cell sorting murine lineage cell depletion kit (Miltenyi Biotec GmbH, Gladbach, Germany) according to the manufacturer's instructions. A total of 2 × 105 lineage-negative cells were plated onto fibronectin-coated 12-well plates (BD Discovery Labware, Bedford, MA) and grown in erythroid differentiation medium (57, 59).
Retroviral transfection.
Elf-4 cDNA from Riken mouse FANTOM clone F630208F07 was cloned into a retroviral overexpression construct containing murine stem cell virus pGKprom-Puro-IRES-EGFP (PIG). Retrovirus production was carried out using the pCL-Eco retrovirus packaging vector (Imgenex, San Diego, CA) in the 293T cell line. Murine fetal liver cells were infected with retrovirus by centrifugation at 2,000 rpm at 30°C for 1.5 h with 4 μg/ml Polybrene (Sigma-Aldrich), after which the retroviral supernatant was replaced with erythroid differentiation medium (59).
Microarray sequence accession number.
The microarray sequence data derived from our experiments have been deposited in ArrayExpress under accession number E-MEXP-2908.
RESULTS
Locus-wide chromatin analysis across the Elf-4 locus revealed 5 candidate regulatory regions.
Comparative sequence analysis has been successfully used to predict functional regulatory elements for many genes (6, 26, 27, 43, 56). An 80-kb region encompassing the Elf-4 gene up to its flanking genes, Aifm1 and Bcorl1, contains multiple highly conserved noncoding regions (see Fig. S2A at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). To identify potential candidate promoter regions, we combined the sequence conservation analysis with Refseq (46) and ENSEMBL (17) gene structures based on experimentally observed transcripts, which showed transcripts arising from three distinct transcriptional start sites within the Elf-4 gene locus (see Fig. S2A at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). Locus-wide analysis of the percentage of GC bases also showed that these predicted start sites corresponded to regions of high GC content, a feature indicative of promoters.
Elf-4 has previously been shown to be expressed at different levels in a variety of hematopoietic and nonhematopoietic tissues (36). To identify experimental model systems for studying potential Elf-4 regulatory elements, levels of Elf-4 expression were assessed by quantitative real-time PCR in a panel of hematopoietic and endothelial cell lines (see Fig. S2B at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). Elf-4 expression levels relative to the housekeeping gene Gapdh were similar in the 416B (myeloid progenitor), HPC7 (multipotent hematopoietic progenitor), BW5147 (T-cell), and MS1 (endothelial) cell lines (see Fig. S2B at the URL above). In comparison, the expression levels of Elf-4 in the erythroid J2E cell line were virtually undetectable (see Fig. S2B at the URL above).
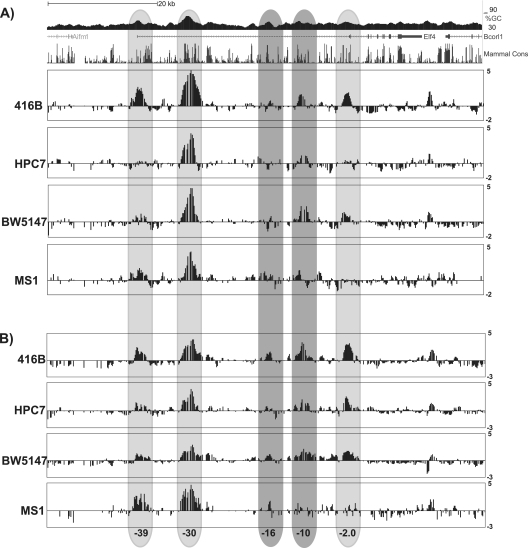
To identify candidate Elf-4 regulatory elements, we next analyzed chromatin modification profiles by ChIP with microarray technology (ChIP-chip). Chromatin was prepared from the four cell lines expressing Elf-4 (416B, HPC7, BW5147, and MS1) and immunoprecipitated with antibodies against the trimethylated form of lysine 4 of histone H3 (H3Me3K4) and acetylated lysine 9 of histone H3 (H3AcK9). Modification of chromatin by H3Me3K4 is generally thought to be a hallmark of active promoter regions, while H3AcK9 marks regions of accessible chromatin across both enhancers and promoters of active genes. However, a recent comprehensive analysis of multiple histone marks across a panel of human cell types demonstrated that the H3Me3K4 chromatin modification is not only found at active promoters but also characterizes regions with strong enhancer activity (9), and this notion was further supported by a recent functional analysis of T cell gene enhancers, where loss of H3K4me3 at enhancer regions was associated with downregulation of the associated genes during T cell development (42). Immunoprecipitated chromatin was interrogated using an 80-kb tiling array that encompassed the Elf-4 locus and reached into each of its flanking genes. Histone H3 trimethylation was observed at each of the three potential transcriptional start sites, but different patterns were observed in each of the cell lines tested (Fig. 1A). In all four cell lines, the highest level of enrichment was observed at the kb −30 region, which corresponds to the start of the predicted ensemble transcript Elf-4-001 and the region with the highest GC percentage across the Elf-4 locus (Fig. 1A). At the most-5′ region (kb −39), H3Me3K4 was enriched in the 416B myeloid progenitor cell line and in the MS1 endothelial cell line, with very low levels of enrichment in the BW5147 T cell line. No enrichment could be detected at this region for the HPC7 progenitor cell line (Fig. 1A). In comparison, low levels of enrichment for H3Me3K4 were detected at the kb −2 region in the 416B and BW5147 cell lines, with no enrichment observed in the HPC7 progenitor cells or the MS1 cell line (Fig. 1A).
Fig 1.
ChIP-chip analysis for histone H3 trimethylation and acetylation across the Elf-4 locus. Locus-wide ChIP-chip analysis of H3Me3K4 (A) and H3acK9 (B) was performed in hematopoietic and endothelial cell lines. The UCSC Genome Browser track (see also Fig. S2 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html) is shown for reference, and the y axis represents the fold enrichment over the median intensity across the whole locus and is expressed as log base 2 values. The light gray bars highlight regions that were strongly enriched for both H3Me3K4 and H3acK9 and correspond to potential transcriptional start sites. The dark gray bars highlight the regions that showed low-level enrichment for H3Me3K4 and were more highly enriched for H3acK9. Numbering reflects the distance (in kb) from the ATG of the ENSEMBL transcript Elf-4-001.
Histone H3 acetylation profiles across the Elf-4 locus largely confirmed the previous results obtained with the H3Me3K4 antibody, showing a highly enriched region of accessible chromatin at the kb −30 region in each of the four cell lines (Fig. 1). Similarly, the kb −39 region and the kb −2 regions were acetylated in the 416B, MS1, and BW5147 cell lines and displayed a pattern of enrichment consistent with the H3Me3K4 result (Fig. 1). In contrast to the H3Me3K4 profiles, low levels of acetylation were also detected at both the kb −39 and kb −2 regions in the HPC7 progenitor cells (Fig. 1B). In addition to the kb −39, −30, and −2 regions, the 416B, HPC7, and BW5147 cell lines were highly enriched at the kb −10 region and had low levels of acetylation at the kb −16 region (Fig. 1B). In comparison, the endothelial cell line MS1 showed a low level of acetylation at the kb −16 region, with an absence of acetylation at the kb −10 region, as seen in the other cell lines tested (Fig. 1).
To complement our ChIP-chip analysis, we also took advantage of publicly available ChIP-Seq data sets produced as part of the ENCODE project (48). Analysis of the Elf-4 locus showed H3K4me3 enrichment at each of the candidate Elf-4 regulatory regions in K562 cells (see Fig. S3 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html) and the −39P, −30P, −2P, and −10E regions in mouse bone marrow (see Fig. S4 at the URL above). The −30P promoter region was the most highly enriched in both K562 cells and mouse bone marrow, which was consistent with our ChIP-chip data in hematopoietic cell lines (see Fig. S3 and S4 the URL above). Moreover, RNA polymerase II (Pol II) was clearly enriched at each of the three candidate Elf-4 promoters in the murine erythroleukemia (MEL) cell line (see Fig. S5 at the URL above), confirming our ChIP-chip results. In addition, analysis of H3K4me1, a mark preferentially associated with enhancers, showed a broad peak of enrichment over the −16E and the −10E enhancer regions in the Elf-4 locus in both K562 and mouse bone marrow (see Fig. S3 and S4 the URL above). Importantly, two marks of active regulatory regions, H3K27ac and DNase I, both showed enrichment of each of the candidate Elf-4 regulatory regions identified by our ChIP-chip analysis (see Fig. S3 and S5 at the URL above). Together, our ChIP-chip analysis data combined with the data from the ENCODE project were used to generate epigenetic profiles for H3Me3K4, H3acK9, H3K4me1, H3K27ac, Pol II, and DNase I binding across the Elf-4 locus, which indicated that there are five potential regulatory regions for Elf-4 in hematopoietic and endothelial cell lines and in mouse bone marrow. The predicted gene structure for Elf-4 combined with the presence of H3Me3K4 and Pol II enrichment at the kb −39, −30, and −2 regions, is consistent with three promoter regions for Elf-4, with the kb −16 and −10 regions likely to represent transcriptional enhancers.
Transcriptional analysis of Elf-4 regulatory elements revealed three promoters and two enhancers active in hematopoietic and endothelial cell lines.
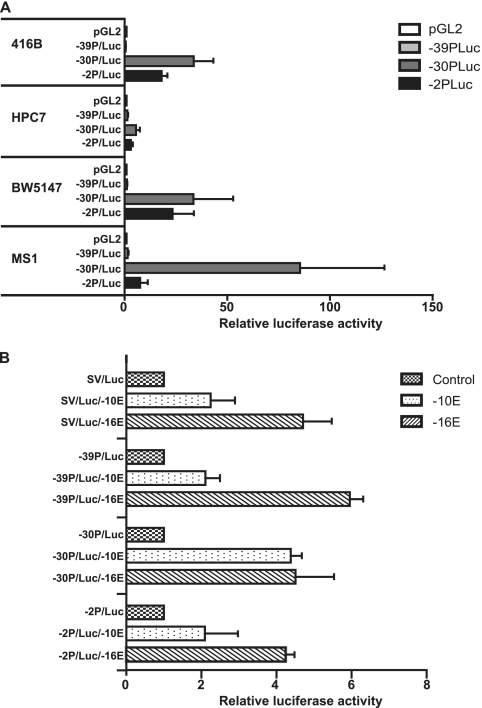
To functionally validate the candidate Elf-4 promoter regions identified by ChIP-chip, promoter elements were tested using luciferase transient-transfection assays. Each candidate promoter region (−39P, −30P, and −2P) was cloned upstream of luciferase in the pGL2 basic vector and tested relative to the empty vector control in the 416B, HPC7, BW5147, and the MS1 cell lines. In each of the four cell lines, the promoter region with the greatest activity was the Elf-4 −30P region, suggesting that this is the dominant promoter in hematopoietic and endothelial cell lines. Similar levels of activity for the Elf-4 −30P region were observed in the hematopoietic 416B and BW5147 cell lines, but the levels of activity were much lower in the HPC7 progenitor cell line (Fig. 2A). Interestingly, in the endothelial MS1 cell line Elf-4 −30P has particularly strong promoter activity (Fig. 2A), which is entirely consistent with our ChIP-chip analysis data, which showed the highest level of enrichment of H3Me3K4 and H3acK9 at this region in this cell line (Fig. 1). The Elf-4 −2P region also acts as a promoter in all four of the cell lines (Fig. 2A). The pattern of activity was similar to that seen for the −30P region, with the 416B and BW5147 cell lines showing similar levels of activity and the HPC7 cell line showing much lower activity. However, unlike the −30P region, the −2P region has very little activity in the MS1 cell line, again consistent with the absence of an H3Me3K4 peak at this region in MS1 cells (Fig. 2A). The Elf-4 −39P candidate promoter element showed no significant activity in any of the four cell lines tested (Fig. 2A).
Fig 2.
Transfection analysis of Elf-4 regulatory elements in hematopoietic and endothelial cell lines. (A) Candidate Elf-4 promoter elements (−39P, −30P, and −2P) were assayed by transient transfection in the 416B myeloid, HPC7 multipotent, BW5147 T cell, and MS1 endothelial cell lines. Promoter elements upstream of luciferase were tested, and the data were normalized relative to the pGL2 promoterless luciferase control vector. The means and the standard errors of the means are shown for three experiments performed in triplicate. (B) The candidate −10E and −16E Elf-4 enhancer elements were tested by stable transfection assays in the myeloid 416B cell line. Each enhancer was tested using either the heterologous SV40 promoter or the −39P, −30P, and −2P promoter elements, and results are plotted relative to the promoter-alone luciferase construct. The means and the standard errors of the means are shown for three experiments performed in triplicate.
In addition to the three promoter elements enriched for both H3Me3K4 and H3acK9, two other regulatory elements (−10E and −16E) were shown to be enriched predominantly for H3acK9 (Fig. 1B) and H3K4me1 (see Fig. S3 and S4 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html) and also did not correspond to any of the predicted transcriptional start sites. We therefore considered that these two elements may represent candidate enhancers for Elf-4. To assess enhancer activities, the −10E and −16E elements were cloned downstream of a luciferase reporter cassette driven by either the heterologous SV40 promoter or the −39P, −30P, and −2P promoter elements and tested in stable transfection assays using the 416B cell line (Fig. 2B). The −10E region displayed modest enhancer activity when tested with the SV40, the −39P, and the −2P promoters (Fig. 2B) and even stronger activity when tested in conjunction with the −30P promoter (Fig. 2B). The −16E region showed strong enhancer activity when tested with all four promoters. Overall, our analysis of the transcriptional activity of potential Elf-4 candidate regulatory elements has identified three promoters and two enhancers.
Conserved Ets binding motifs are required for the activity of the Elf-4 −10E enhancer.
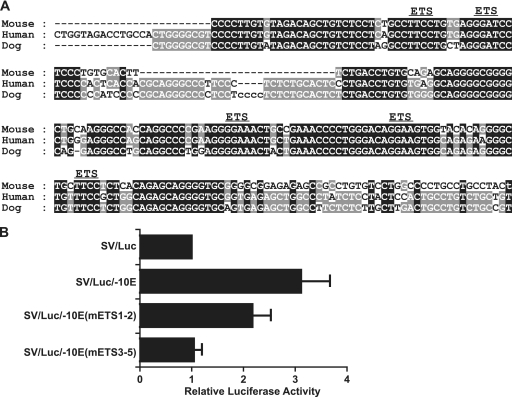
Our laboratory has recently generated a data set containing the genome-wide binding profiles for 10 different hematopoietic transcription factors in the multipotent hematopoietic progenitor cell line HPC7 (55). We therefore utilized this data set to gain insight into potential upstream regulators of Elf-4. There were no significant peaks for Scl/Tal-1, Lyl-1, and Lmo2 across the Elf-4 locus. However, ChIP-Seq analysis of the remaining 7 transcription factors (Gata2, Runx1, Meis1, Pu.1, Fli1, Erg, and Gfi1b) showed clear binding of each of the factors across the different Elf-4 regulatory elements identified in this study except for the −39P element, which did not bind any of the factors tested in the HPC7 cell line (see Fig. 5, below; see also Fig. S6 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). With the Elf-4 −10E enhancer being able to target expression in vivo to the fetal liver and dorsal aorta (see Fig. 4, below), we were particularly interested to use the ChIP-Seq data set to examine the binding profiles for hematopoietic transcription factors at this element. There was very little binding of Gata2 or Runx1, and Meis1 only showed weak binding (see Fig. S6 at the URL above). In contrast, the ETS transcription factors Pu.1, Fli1, and Erg showed much stronger binding to this element (see Fig. S6). Nucleotide sequence alignment of the Elf-4 −10E element in mouse, human, and dog revealed a highly conserved core region that contained an Ebox and 5 ETS factor binding sites (Fig. 3A; see also Fig. S1 at the URL above). Due to the strong binding of the ETS transcription factors shown by the ChIP-Seq data, we hypothesized that the ETS binding sites present in the conserved core of the −10E enhancer may be responsible for its activity. To investigate the importance of the ETS motifs, mutations were introduced into the −10E enhancer and tested in stable transfection assays in the 416B cell line. Mutation of the first and second ETS motifs resulted in a partial decrease in the activity of the −10E enhancer (Fig. 3B), whereas mutation of the third, fourth, and fifth ETS motifs was sufficient to abolish the activity of the −10E enhancer completely (Fig. 3B). Taken together, these results show that activity of the −10E enhancer critically depends on ETS motifs present in the conserved region of the −10E enhancer, with Pu.1, Fli1, and Erg being likely activators of Elf-4 expression.
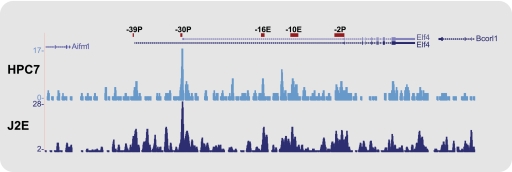
Fig 5.
Gfi1b binds to the Elf-4 −30 promoter in HPC7 and J2E cell lines. Density plots obtained from ChIP-Seq reads for Gfi1b in the HPC7 and J2E cell lines are displayed as UCSC Genome Browser tracks for the Elf-4 locus. Elf-4 candidate regulatory elements are shown as a reference.
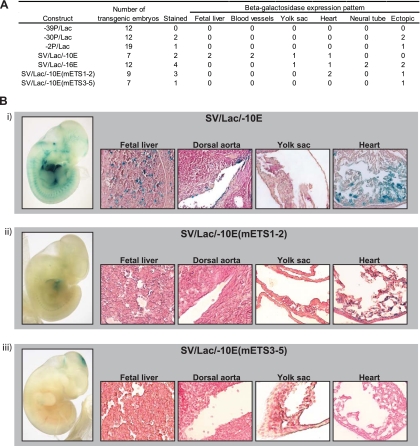
Fig 4.
The Elf-4 −10E enhancer specifically targets blood and endothelium in the developing embryo. Elf-4 regulatory elements were validated in vivo using F0 transgenic embryos. (A) Table summarizing the expression patterns of Elf-4 regulatory elements analyzed by beta-galactosidase staining of E11.5 transgenic embryos. (B) Representative whole-mount E11.5 embryos stained with X-Gal to determine beta-galactosidase expression and tissue sections of the fetal liver, dorsal aorta, yolk sac, and the heart are shown for SV/Lac/−10E (i), SV/Lac/−10E(mETS1-2) (ii), and SV/Lac/−10E(mETS3-5) (iii). Whole-mount images were taken at 2× magnification, and the tissue section images were taken at 20× magnification.
Fig 3.
Conserved ETS binding sites are required for the activity of the Elf-4 −10E enhancer. (A) Nucleotide sequence alignment of the conserved region of the Elf-4 −10E enhancer. The conserved ETS binding sites are marked. (B) Stable transfection assay results in 416B cells of luciferase reporter constructs containing the wild-type −10E enhancer and the −10E enhancer with different combinations of the ETS binding sites mutated. Luciferase values were normalized relative to the SV40-luciferase control vector. The means and the standard errors of the means are shown for three experiments performed in triplicate.
The Elf-4 −10E enhancer specifically directs expression to blood and endothelium in transgenic mouse E11.5 embryos.
To functionally validate the in vivo activity of the Elf-4 regulatory elements, we generated beta-galactosidase reporter constructs for all five regulatory elements and tested them by generating multiple F0 transgenic embryos for each of the five constructs. The expression pattern for each Elf-4 regulatory element was assessed by X-Gal staining of whole-mount E11.5 embryos. No specific staining patterns were observed for any of the three promoter regions (Fig. 4A). In contrast, the Elf-4 enhancer elements showed more consistent staining patterns. The Elf-4 −16E enhancer was expressed in the developing neural tube and one embryo showed staining in the yolk sac and in the heart (Fig. 4A), but when analyzed in histological sections we could not detect any consistent hematopoietic staining (Fig. 4A and data not shown). In contrast, the Elf-4 −10E enhancer showed strong staining in hematopoietic and endothelial tissues in whole-mount embryos (SV/Lac/−10E) (Fig. 4A and B, panel i). Analysis of histological sections confirmed that the −10E enhancer directs expression to a small proportion of cells in the fetal liver, the endothelial lining of the dorsal aorta, the endothelial lining of vitelline vessels (data not shown), the yolk sac, and the endocardium of the heart (SV/Lac/−10E) (Fig. 4B, panel i). To determine whether the ETS motifs present in the Elf-4 −10E enhancer were also critical for the in vivo activity of the enhancer in hematopoietic and endothelial tissues, we also tested the mutated −10E enhancer constructs in F0 transgenic embryos (Fig. 4A). Mutation of the first and second ETS motifs of the Elf-4 −10E enhancer showed staining in the endothelial lining of the heart, but at lower levels than seen with the full −10E enhancer, and resulted in a complete loss of activity in the fetal liver, the dorsal aorta, and the yolk sac [SV/Lac/−10E(mETS1-2)] (Fig. 4A and B, panel ii). Mutation of the third, fourth, and fifth ETS motifs dramatically reduced the in vivo activity of the Elf-4 −10E enhancer, with no detectable staining in hematopoietic or endothelial tissues in the whole-mount embryos or tissue sections [SV/Lac/−10E(mETS3-5)] (Fig. 4A and B, panel iii). In summary, our transgenic analysis of Elf-4 regulatory elements identified a tissue-specific enhancer for Elf-4 that specifically directs Elf-4 expression in the blood and endothelium, and it confirmed that the ETS motifs found in the enhancer are critical for its activity.
Gfi1b represses Elf-4 expression by binding at the −30P promoter element, and Elf-4 repression is important for erythroid maturation.
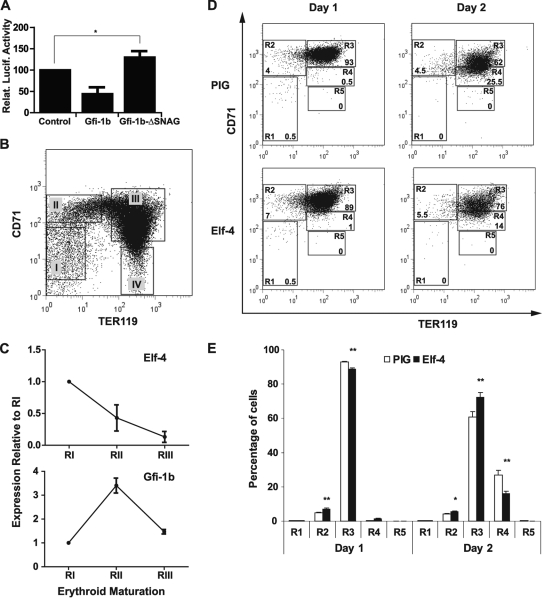
Gfi1b is a transcriptional repressor that is an essential regulator of erythroid and megakaryocyte development (11, 49, 54). Further interrogation of our ChIP-Seq data set revealed specific binding of Gfi1b at the Elf-4 −30P promoter in the HPC7 cell line (Fig. 5). Following the generation of a new ChIP-Seq data set for Gfi1b in J2E cells (see Materials and Methods), specific binding of Gfi1b to the Elf-4 −30P promoter was also observed in this erythroid cell model (Fig. 5). Moreover, analysis of ELF-4 and GFI1B expression across an array of human cell types showed contrasting expression patterns, particularly in CD71-positive erythroid progenitors, in which GFI1B expression is very high compared to ELF-4 (see Fig. S7 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). To determine the relationship between Gfi1b and the Elf-4 −30P promoter, transient cotransfections were performed in the J2E cell line, which expresses very low levels of endogenous Elf-4 (see Fig. S2 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). J2E cells were transfected with the Elf-4 −30P luciferase reporter plasmid and either the empty expression vector (MigRI), full-length Gfi1b (MigRI-Gfi1b), or a truncated version of the protein lacking the SNAG repressor domain (MigRI-Gfi1b-ΔSNAG) (Fig. 6A). Overexpression of Gfi1b repressed the activity of the Elf-4 −30P promoter by 50% relative to the control (Fig. 6A). In contrast overexpression of Gfi1b lacking the SNAG repressor domain resulted in a significant (P ≤ 0.05) 30% increase in activity of the Elf-4 −30P promoter relative to the control, signifying that repression of the Elf-4 −30P promoter is mediated through the SNAG domain of Gfi1b (Fig. 6A). Taken together, these results suggest that Gfi1b acts upstream of Elf-4 to downregulate its expression.
Fig 6.
Overexpression of Elf-4 delays erythroid maturation during in vitro differentiation of E14.5 fetal liver cells. (A) Repression of the Elf-4 −30P promoter by overexpression of Gfi1b in the J2E cell line. Cotransfection of the Elf-4 −30P luciferase reporter construct with either MigRI (control), MigRI-Gfi1b, or MigRI-Gfi1b-ΔSNAG expression plasmids was analyzed in the J2E cell line. Luciferase values are expressed relative to the control, and the means from four independent transfections performed in triplicate are shown. Error bars indicate the standard errors of the means. Statistically significant differences (based on Student's t test) are indicated with an asterisk (P ≤ 0.05). (B) Four populations of murine E14.5 fetal liver cells isolated by flow cytometry and representing sequential stages of erythroid differentiation: (I) CD71−/low TER119−; (II) CD71+ TER119−; (III) CD71+ TER119+; (IV) CD71− TER119+. (C) Expression of Elf-4 and Gfi1b during in vivo terminal erythroid differentiation in the fetal liver. Real-time PCR analysis for the expression of Elf-4 and Gfi1b was performed on E14.5 fetal liver cells sorted by flow cytometry (regions I to III). The gene expression levels of each population were normalized to mouse 18s, and the data are presented as the expression level relative to that at stage I. Error bars indicate standard deviations. (D) Representative FACS plots for TER119 and CD71 expression during in vitro differentiation in lineage-negative E14.5 fetal liver cells overexpressing Elf-4 or a control (PIG) vector after 24 h (day 1) or 48 h (day 2). Five populations corresponding to stages of erythroid differentiation from progenitors to enucleated cells were gated: R1, CD71−/med TER119−; R2, CD71high TER119low; R3, CD71high TER1119high; R4, CD71med TER1119high; R5, CD71low TER1119high. Percentages of GFP+ cells in each of these stages are indicated. (E) Bar charts indicating the proportions of cells at each of the five stages of erythroid differentiation (R1 to R5) after 24 h (day 1) and 48 h (day 2) of in vitro culture of lineage-negative E14.5 fetal liver cells retrovirally transfected with an overexpressing Elf-4 or control (PIG) vector. A representative plot (with means and standard deviations) for one experiment, performed in triplicate, is shown. Statistically significant differences (based on Student's t test) are indicated with asterisks: *, P ≤ 0.05; **, P ≤ 0.01.
Gfi1b−/− embryos die at E15 due to a delay in the maturation of primitive erythrocytes and a failure to produce definitive enucleated erythrocytes (49). Downregulation of ETS factors is also known to be important for terminal erythroid differentiation (2, 6, 47). Since Gfi1b is able to repress the activity of the main promoter element in the Elf-4 locus and because Elf-4 expression is almost absent in the erythroid cell line J2E, we next explored the possibility of a relationship between these two factors. First, we analyzed the expression of Elf-4 and Gfi1b during fetal liver erythropoiesis. Expression levels of Elf-4 and Gfi1b in E14.5 fetal liver cells were measured by real-time PCR in CD71−/low TER119−, CD71+ TER119−, and CD71+ TER119+ cells, corresponding to populations of increasing maturity along the erythroid differentiation pathway (59) (Fig. 6B, regions I to III, respectively). The most mature CD71− TER119+ population (region IV) consists largely of enucleated cells (59) and was therefore excluded from the real-time PCR analysis. During erythroid maturation, the expression of Elf-4 decreased, while Gfi1b showed a marked increase in expression (Fig. 6C).
Given the known importance of Gfi1b for erythroid development, repression of the Elf-4 promoter by Gfi1b, and reciprocal expression of Gfi1b and Elf-4 during erythroid maturation, we next investigated whether downregulation of Elf-4 in itself constituted an important step during erythroid differentiation. To this end, we ectopically expressed murine Elf-4 in primary E14.5 fetal liver erythroblasts by using a retroviral vector (MSCV-Elf-4-PGKprom-puro-IRES-GFP), which also encodes green fluorescent protein (GFP) (see Fig. S8 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html) and differentiated transduced cells in vitro (59). Differentiation of transduced cells was monitored by flow cytometry, and Elf-4-overexpressing cells were compared to cells transduced with the empty retroviral vector. Untransduced GFP− cells served as an internal control to ensure that erythroid differentiation was unaffected by retroviral infection. Analysis was performed on five populations representing the sequential stages of erythroid differentiation as defined by their surface expression of CD71 and TER119 (Fig. 6D, regions R1 to R5). Figure 6E shows the results of a representative experiment. Overexpression of Elf-4 caused significant changes in fluorescence-activated cell sorting profiles after just 24 h of differentiation, and those changes were more dramatic after 48 h. Overexpression of Elf-4 resulted in a substantial delay in terminal erythroid differentiation, as evidenced by the highly significant (Student's t test, P ≤ 0.01) accumulation of more immature cells in R2 and R3 at the expense of more mature cells in R4 compared to the empty vector control (Fig. 6E). Together with the transfection and expression data, these results suggest that Elf-4 constitutes a key downstream target gene of Gfi1b during its control of erythroid maturation.
DISCUSSION
The capability of hematopoietic stem cells to remain quiescent, self-renew, or differentiate is a highly regulated process dependent on a number of different transcription factors. To understand how these factors work in concert with each other to form the regulatory networks that orchestrate stem cell fate decisions, it will be essential to identify both the transcription factors involved as well as their upstream regulators.
The Elf-4 gene encodes an ETS family transcription factor that is widely expressed in hematopoietic tissues and is an important regulator of HSC and CD8+ T cell proliferation and the differentiation of NK and NK-T cells (24, 25, 58). Using a locus-wide ChIP-chip array, we have identified five conserved regulatory regions across the Elf-4 locus. Functional analysis of these regions showed two promoters and two enhancers to be active in hematopoietic and endothelial cell lines. Further analysis of these conserved regions in transgenic mouse embryos identified a tissue-specific enhancer that specifically directs Elf-4 expression in the blood and endothelium. By integration with a genome-wide ChIP-Seq data set, we identified a number of upstream regulators for Elf-4, and through functional mutation analysis we identify ETS family transcription factors Fli1, Erg, and Pu.1 as key mediators of the activity of the tissue-specific enhancer. Finally, we demonstrated that the transcriptional repressor Gfi-1b specifically represses the most potent Elf-4 promoter and that Elf-4 repression is critical for the normal maturation of primary erythroid cells.
Comparative sequence analysis has been widely used to identify key regulatory regions for a number of major hematopoietic regulators (7, 26, 40, 44, 56). However, in the case of Elf-4, it was only through combining a computational approach with ChIP-chip analysis for the presence of specific histone marks that we were able to more accurately predict those regions within the Elf-4 gene locus that warranted functional validation. Comparative sequence analysis still remains a powerful tool for analyzing gene regulation, but it is becoming increasingly superceded by the current advances in experimental techniques such as ChIP-chip and ChIP-Seq. As it becomes easier to rapidly perform these genome-scale studies for a number of different histone markers and/or transcription factors across several different cell types, the identification of gene regulatory regions and the tissues they may be active in will undoubtedly be simplified. Moreover, a recent study aiming to identify enhancers active during embryonic heart development found that ChIP-Seq for the enhancer-associated protein p300 identified many heart-specific enhancers that have poor evolutionary conservation and may otherwise be overlooked (5), thus further emphasizing the potential advantages of genome-scale experimental approaches over comparative genomic sequence analysis.
While comparative genomics and/or ChIP-chip/ChIP-Seq have proven to be extremely accurate methods for the identification of candidate regulatory regions, the true functional activity of any given regulatory element can only be determined through in vivo validation using transgenic embryos. One potential drawback of analyzing regulatory function in vivo is that a single region alone may not be enough to produce a specific staining pattern for the gene of interest. This was indeed the case with Elf-4, for which the three promoters (−39P, −30P, and −2P) and the −16E enhancer failed to show reproducible staining in E11.5 embryos. This in itself was not unsurprising, as we and others previously observed that the promoters alone of other key hematopoietic regulators, such as Scl, Lmo2, Gfi1, Gata1, Runx1, and endoglin (4, 27, 32, 43, 50, 52), have very weak or no activity in transgenic embryos. Moreover, there is increasing evidence that it is the pattern of histone markers and transcription factor binding across enhancers rather than promoters that are responsible for cell-type-specific gene expression patterns (15, 16). Nevertheless, additional explanations for the lack of staining may be that combinations of promoters and enhancers are required along with testing at other time points of development. However, a comprehensive analysis of all combinations of promoters and enhancers at different time points would become ineffective in terms of cost and time. Importantly, our in vivo analysis of the Elf-4 regulatory regions provided definitive proof that the −10E region has enhancer activity with a tissue-specific pattern that is restricted to the blood and endothelium and thus consistent with the major domains of endogenous Elf-4 expression. Moreover, analysis of histological sections from embryos expressing the Elf-4 −10E region suggests that the enhancer is only active in a very small proportion of circulating primitive red bloods cells. Transcriptional programs in blood stem/progenitor and endothelial cells have long been recognized to be highly similar, with many regulatory elements showing activity in both lineages (13, 26, 27, 38, 39, 44, 50). In view of the endothelial staining pattern observed for the −10E region, we propose that the hematopoietic activity seen for this element is most likely in progenitor cells rather than mature blood cells. However, analysis of the Elf-4 −10E region in transgenic mouse lines is required to determine exactly which hematopoietic subsets express the Elf-4 −10E region.
Potential upstream regulators of the −10E enhancer element were identified by integrating a recently published 10-factor ChIP-Seq data set to our analysis of the −10E enhancer. Three members of the ETS family of transcription factors, Pu.1, Fli1, and Erg were found to be highly enriched at this region, and we demonstrated that the functionally important motifs in the conserved region of the −10E enhancer are indeed ETS motifs. The ETS family of transcription factors have previously been shown to be important during hematopoietic and endothelial development. Two of the ETS transcription factors binding the −10E enhancer, Fli1 and Erg, have greater homology to each other than to any of the other ETS family members. In mouse embryos, Fli1 is expressed in hematopoietic cells and in the developing vasculature (33), and Fli−/− embryos die around E12.5 from severe hemorrhaging (53). Unlike Fli1, loss of Erg has little effect on the vasculature, but embryos show a failure of definitive hematopoiesis (29). Moreover, in a mouse model using double heterozygotes for Fli1 and Erg, both transcription factors have been shown to be indispensable for normal HSC and megakaryocytic homeostasis (22). Pu.1, the third ETS family member that binds the −10E enhancer, has a critical role in the maintenance and expansion of HSCs in the fetal liver, and Pu.1−/− mice die at E18.5 from complete hematopoietic failure (20). It is also important to consider that as a member of the ETS family of transcription factors, Elf-4 may autoregulate its own expression. However, we have tested all commercial Elf-4 antibodies available to us and have so far failed to detect any enrichment on candidate target regions, including the −10E enhancer region, and therefore at this time we cannot definitively state whether Elf-4 autoregulation occurs.
ETS factor binding, along with Gata and Scl, has been shown to be critical for driving the hematopoietic specificity of several enhancers known to be important in hematopoiesis such as Scl +19, Lmo2 −75, and the Runx1 +23 (13, 26, 40). Surprisingly, the −10E enhancer does not contain any conserved Gata binding sites and shows no binding of Gata 2 in ChIP-Seq analysis. In addition, Scl does not bind significantly anywhere across the Elf-4 locus (data not shown). Recently it has been shown that many endothelium-specific enhancers contain a FOX:ETS motif that is synergistically activated by Forkhead and ETS transcription factors (8). Given that the −10E enhancer is active in the endothelium of the dorsal aorta and the vitelline vessels, we looked for the presence of the FOX:ETS motif in the −10E enhancer, but again, this motif was absent. Therefore, it appears that the ETS motifs present in the −10E enhancer are sufficient to drive the hematopoietic/endothelium-specific activity of Elf-4, although we cannot exclude the possibility that an as-yet-unidentified motif works in concert with the ETS factors to produce this tissue specificity.
Analysis of comprehensive ChIP-Seq data allowed us to identify not only transcription factors, such as the Ets factors Pu.1, Erg, and Fli1, that activate expression of Elf-4, but also Gfi1b as a factor that downregulates Elf-4 expression. The transcriptional repressor Gfi1b has been shown to be a positive regulator of erythroid differentiation and is critical for production of definitive enucleated erythrocytes in the developing embryo (11, 49). Moreover, forced expression of Gfi1b in the erythroid cell line K562 induces erythroid differentiation that, upon deletion of the SNAG repressor domain, is abolished (11). Although transcriptional repression by Gfi1b during erythropoiesis (11) is widely accepted, very little is known about the Gfi1b target genes that need to be switched off to allow erythroid differentiation to proceed. Our retroviral expression experiments now suggest that Elf-4 represents a key Gfi1b target gene, and it will be interesting to investigate whether Elf-4 also functions as an important target of Gfi1b or its close relative, Gfi1, in other hematopoietic cell types. Interestingly, following repression of Elf-4 during the RII (CD71high/TER119low proerythroblast) stage of fetal liver erythropoiesis, Gfi1b expression itself decreased as the cells matured toward the RIII (CD71high/TER119high basophilic erythroblast) stage of erythroid differentiation, which is consistent with previous observations that Gfi1b needs to be downregulated for the final maturation of erythroid cells beyond proerythroblasts (23, 41). In addition to providing mechanistic insights into Gfi1b control of erythropoiesis, our results also significantly advance our understanding of Elf-4 function. Based on the finding that Elf-4−/− HSCs are more quiescent than their wild-type counterparts (25), one might have concluded that Elf-4 functions in a rather generic way to promote proliferation. However, we did not see any evidence for increased proliferation. Instead, cell numbers were actually reduced in Elf-4-expressing cultures (data not shown) and showed a pronounced defect in maturation. Of note, a recent study of T lymphocytes suggested that Elf-4 suppresses both homeostatic and antigen-driven proliferation of CD8+ T cells (58). Taken together, these studies highlight the notion that the influence of Elf-4 on the balance between cell proliferation and quiescence is highly context dependent. It is likely that these diverse functions of Elf-4 are at least in part mediated through context-specific protein-protein interactions and involve the control of distinct sets of downstream target genes. Elucidation of cell-type-specific Elf-4 interaction partners and of target genes therefore represent promising areas for future investigation.
Taken together, the data presented here represent not only the first comprehensive analysis of Elf-4 regulation but also provide new insights into Elf-4 function within wider hematopoietic regulatory networks. Using a combination of large-scale experimental approaches and in vivo functional validation, we have identified key regulatory sequences and upstream regulators responsible for Elf-4 expression in hematopoietic progenitor cells and Elf-4 downregulation during erythroid maturation. Moreover, we identified a previously unrecognized role for Elf-4 in the control of erythroid differentiation, where it could function as a key target of the major erythroid regulator Gfi1b. Collectively these results have allowed us for the first time to firmly integrate the key hematopoietic regulator Elf-4 within the emerging transcriptional networks that control not only blood stem cells and progenitor cells but also some of their more differentiated progeny.
ACKNOWLEDGMENTS
This work was supported by the Leukaemia and Lymphoma Research Fund (LLR), Cancer Research UK, and the Leukemia and Lymphoma Society.
We thank Richard Auburn at Flychip for printing the custom arrays and our departmental transgenic team and Cyagen Biosciences for generating transgenic embryos. We also thank B. Kee (University of Chicago) for providing MigRI-Gfi1b and MigRI-Gfi1b-ΔSnag vectors. The ENCODE data used in this publication would not have been available without the work of the members of the ENCODE consortium, and we thank the laboratories of Bradley Bernstein (Broad Institute) for the K562 data, Michael Snyder (Stanford University), Sherman Weissman (Yale University) for the RNA polymerase II data, John Stamatoyannopoulos (University of Washington) for the DNase I data, and Bing Ren (Ludwig Institute for Cancer Research) for the bone marrow data.
A.M.S., F.J.C.-N., J.S., S.K., R.T.T., N.K.W., R.L.H., and J.-R.L. performed the experiments for our study; A.M.S. and F.J.C-N. analyzed the results and created the figures; A.M.S., F.J.C.-N., and B.G. designed the research and wrote the paper.
We declare no competing financial interests.
Footnotes
Published ahead of print 12 December 2011
REFERENCES
- 1. Alcalay M, et al. 2003. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Invest. 112:1751–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Athanasiou M, Mavrothalassitis G, Sun-Hoffman L, Blair DG. 2000. FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia 14:439–445 [DOI] [PubMed] [Google Scholar]
- 3. Bartel FO, Higuchi T, Spyropoulos DD. 2000. Mouse models in the study of the Ets family of transcription factors. Oncogene 19:6443–6454 [DOI] [PubMed] [Google Scholar]
- 4. Bee T, et al. 2009. The mouse Runx1 +23 hematopoietic stem cell enhancer confers hematopoietic specificity to both Runx1 promoters. Blood 113:5121–5124 [DOI] [PubMed] [Google Scholar]
- 5. Blow MJ, et al. 2010. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 42:806–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calero-Nieto FJ, et al. 2010. Transcriptional regulation of Elf-1: locus-wide analysis reveals four distinct promoters, a tissue-specific enhancer, control by PU.1 and the importance of Elf-1 downregulation for erythroid maturation. Nucleic Acids Res. 38:6363–6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chapman MA, et al. 2004. Analysis of multiple genomic sequence alignments: a web resource, online tools, and lessons learned from analysis of mammalian SCL loci. Genome Res. 14:313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Val S, et al. 2008. Combinatorial regulation of endothelial gene expression by Ets and Forkhead transcription factors. Cell 135:1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ernst J, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fukushima T, et al. 2003. The level of MEF but not ELF-1 correlates with FAB subtype of acute myeloid leukemia and is low in good prognosis cases. Leuk. Res. 27:387–392 [DOI] [PubMed] [Google Scholar]
- 11. Garcon L, et al. 2005. Gfi-1B plays a critical role in terminal differentiation of normal and transformed erythroid progenitor cells. Blood 105:1448–1455 [DOI] [PubMed] [Google Scholar]
- 12. Gottgens B, et al. 1997. Transcription of the SCL gene in erythroid and CD34 positive primitive myeloid cells is controlled by a complex network of lineage-restricted chromatin-dependent and chromatin-independent regulatory elements. Oncogene 15:2419–2428 [DOI] [PubMed] [Google Scholar]
- 13. Gottgens B, et al. 2002. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 21:3039–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hedvat CV, Yao J, Sokolic RA, Nimer SD. 2004. Myeloid ELF1-like factor is a potent activator of interleukin-8 expression in hematopoietic cells. J. Biol. Chem. 279:6395–6400 [DOI] [PubMed] [Google Scholar]
- 15. Heintzman ND, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heinz S, et al. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38:576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hubbard TJ, et al. 2009. ENSEMBL 2009. Nucleic Acids Res. 37:D690–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kai H, et al. 1999. Myeloid ELF-1-like factor up-regulates lysozyme transcription in epithelial cells. J. Biol. Chem. 274:20098–20102 [DOI] [PubMed] [Google Scholar]
- 19. Kent WJ, et al. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim HG, et al. 2004. The ETS family transcription factor PU.1 is necessary for the maintenance of fetal liver hematopoietic stem cells. Blood 104:3894–3900 [DOI] [PubMed] [Google Scholar]
- 21. Koga T, et al. 2005. Sp1-dependent regulation of myeloid Elf-1 like factor in human epithelial cells. FEBS Lett. 579:2811–2816 [DOI] [PubMed] [Google Scholar]
- 22. Kruse EA, et al. 2009. Dual requirement for the ETS transcription factors Fli-1 and Erg in hematopoietic stem cells and the megakaryocyte lineage. Proc. Natl. Acad. Sci. U. S. A. 106:13814–13819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuo YY, Chang ZF. 2007. GATA-1 and Gfi-1B interplay to regulate Bcl-xL transcription. Mol. Cell. Biol. 27:4261–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lacorazza HD, et al. 2002. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity 17:437–449 [DOI] [PubMed] [Google Scholar]
- 25. Lacorazza HD, et al. 2006. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell 9:175–187 [DOI] [PubMed] [Google Scholar]
- 26. Landry JR, et al. 2009. Expression of the leukemia oncogene Lmo2 is controlled by an array of tissue-specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors. Blood 113:5783–5792 [DOI] [PubMed] [Google Scholar]
- 27. Landry JR, et al. 2005. Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells. Blood 106:2680–2687 [DOI] [PubMed] [Google Scholar]
- 28. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loughran SJ, et al. 2008. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat. Immunol. 9:810–819 [DOI] [PubMed] [Google Scholar]
- 30. Lund AH, et al. 2002. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat. Genet. 32:160–165 [DOI] [PubMed] [Google Scholar]
- 31. Maroulakou IG, Bowe DB. 2000. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene 19:6432–6442 [DOI] [PubMed] [Google Scholar]
- 32. McDevitt MA, Fujiwara Y, Shivdasani RA, Orkin SH. 1997. An upstream, DNase I hypersensitive region of the hematopoietic-expressed transcription factor GATA-1 gene confers developmental specificity in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 94:7976–7981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Melet F, Motro B, Rossi DJ, Zhang L, Bernstein A. 1996. Generation of a novel Fli-1 protein by gene targeting leads to a defect in thymus development and a delay in Friend virus-induced erythroleukemia. Mol. Cell. Biol. 16:2708–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mikkers H, et al. 2002. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat. Genet. 32:153–159 [DOI] [PubMed] [Google Scholar]
- 35. Miyazaki Y, et al. 2001. Cyclin A-dependent phosphorylation of the ETS-related protein, MEF, restricts its activity to the G1 phase of the cell cycle. J. Biol. Chem. 276:40528–40536 [DOI] [PubMed] [Google Scholar]
- 36. Miyazaki Y, Sun X, Uchida H, Zhang J, Nimer S. 1996. MEF, a novel transcription factor with an Elf-1 like DNA binding domain but distinct transcriptional activating properties. Oncogene 13:1721–1729 [PubMed] [Google Scholar]
- 37. Moore SD, et al. 2006. ELF4 is fused to ERG in a case of acute myeloid leukemia with a t(X;21)(q25-26;q22). Leuk. Res. 30:1037–1042 [DOI] [PubMed] [Google Scholar]
- 38. Ng CE, et al. 2010. A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. Stem Cells 28:1869–1881 [DOI] [PubMed] [Google Scholar]
- 39. North T, et al. 1999. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126:2563–2575 [DOI] [PubMed] [Google Scholar]
- 40. Nottingham WT, et al. 2007. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 110:4188–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Osawa M, et al. 2002. Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood 100:2769–2777 [DOI] [PubMed] [Google Scholar]
- 42. Pekowska A, et al. 2011. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 30:4198–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pimanda JE, et al. 2006. Endoglin expression in the endothelium is regulated by Fli-1, Erg, and Elf-1 acting on the promoter and a −8-kb enhancer. Blood 107:4737–4745 [DOI] [PubMed] [Google Scholar]
- 44. Pimanda JE, et al. 2008. Endoglin expression in blood and endothelium is differentially regulated by modular assembly of the Ets/Gata hemangioblast code. Blood 112:4512–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pinto do O P, Kolterud A, Carlsson L. 1998. Expression of the LIM-homeobox gene LH2 generates immortalized steel factor-dependent multipotent hematopoietic precursors. EMBO J. 17:5744–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rao G, Rekhtman N, Cheng G, Krasikov T, Skoultchi AI. 1997. Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene 14:123–131 [DOI] [PubMed] [Google Scholar]
- 48. Rosenbloom KR, et al. 2010. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 38:D620–D625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saleque S, Cameron S, Orkin SH. 2002. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 16:301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sanchez M, et al. 1999. An SCL 3′ enhancer targets developing endothelium together with embryonic and adult haematopoietic progenitors. Development 126:3891–3904 [DOI] [PubMed] [Google Scholar]
- 51. Seki Y, et al. 2002. The ETS transcription factor MEF is a candidate tumor suppressor gene on the X chromosome. Cancer Res. 62:6579–6586 [PubMed] [Google Scholar]
- 52. Sinclair AM, et al. 1999. Distinct 5′ SCL enhancers direct transcription to developing brain, spinal cord, and endothelium: neural expression is mediated by GATA factor binding sites. Dev. Biol. 209:128–142 [DOI] [PubMed] [Google Scholar]
- 53. Spyropoulos DD, et al. 2000. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell. Biol. 20:5643–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vassen L, Okayama T, Moroy T. 2007. Gfi1b:green fluorescent protein knock-in mice reveal a dynamic expression pattern of Gfi1b during hematopoiesis that is largely complementary to Gfi1. Blood 109:2356–2364 [DOI] [PubMed] [Google Scholar]
- 55. Wilson NK, et al. 2010. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7:532–544 [DOI] [PubMed] [Google Scholar]
- 56. Wilson NK, et al. 2010. Gfi1 expression is controlled by five distinct regulatory regions spread over 100 kilobases, with Scl/Tal1, Gata2, PU.1, Erg, Meis1, and Runx1 acting as upstream regulators in early hematopoietic cells. Mol. Cell. Biol. 30:3853–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wood AD, et al. 2009. ID1 promotes expansion and survival of primary erythroid cells and is a target of JAK2V617F-STAT5 signaling. Blood 114:1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yamada T, Park CS, Mamonkin M, Lacorazza HD. 2009. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Kruppel-like factors KLF4 and KLF2. Nat. Immunol. 10:618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang J, Socolovsky M, Gross AW, Lodish HF. 2003. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood 102:3938–3946 [DOI] [PubMed] [Google Scholar]