Abstract
Myc family members are critical to maintain embryonic stem cells (ESC) in the undifferentiated state. However, the mechanism by which they perform this task has not yet been elucidated. Here we show that Myc directly upregulates the transcription of all core components of the Polycomb repressive complex 2 (PRC2) as well as the ESC-specific PRC2-associated factors. By expressing Myc protein fused with the estrogen receptor (Myc-ER) in fibroblasts, we observed that Myc, binding to the regulatory elements of Suz12, Ezh2, and Eed, induces the acetylation of histones H3 and H4 and the recruitment of elongating RNA polymerase II at their promoters. The silencing of both c-Myc and N-Myc in ESC results in reduced expression of PRC2 and H3K27me3 at Polycomb target developmental regulators and upregulation of genes involved in primitive endoderm differentiation. The ectopic expression of PRC2 in ESC, either silenced for c-Myc and N-Myc or induced to differentiate by leukemia inhibitory factor (LIF) withdrawal, is sufficient to maintain the H3K27me3 mark at genes with bivalent histone modifications and keep repressed the genes involved in ESC differentiation. Thus, Myc proteins control the expression of developmental regulators via the upregulation of the Polycomb PRC2 complex.
INTRODUCTION
Mouse embryonic stem cells (ESC) pluripotency and their self-renewing capabilities rely on independent regulatory networks (2, 7, 17). Recent reports have provided compelling evidence that Myc plays an important role in ESC homeostasis as well as in cell reprogramming toward the pluripotent state. The ectopic expression of Myc in ESC is able to promote their self-renewal and to maintain pluripotency also in the absence of the cytokine leukemia inhibitory factor (LIF) signaling (5) produced by feeder cells while inhibition of the expression of Myc proteins (c-Myc and N-Myc) induces loss of pluripotency and the spontaneous differentiation of ESC into primitive endoderm (41, 46). Myc overexpression in adult cells can block differentiation and cooperates with Oct3/4, Sox2, and Klf4 to reprogram adult differentiated cells into induced pluripotent stem cells (iPS), which are virtually indistinguishable from ESC (44). Genome-wide chromatin immunoprecipitation analyses of these factors both in ESC and during the reprogramming process (7, 15, 17, 42) pointed out that Myc is distinguished from Oct3/4, Sox2, and Nanog as it binds to a different subset of genes. These analyses showed that most of the Myc bound genes are involved in cell cycle progression and metabolism. However, Myc also binds to chromatin regulators, suggesting it might also indirectly regulate genes involved in cell differentiation. In agreement with this hypothesis, it has been shown that during the reprogramming process Myc promotes not only cell replication but also the repression of fibroblast-specific genes (42).
Myc is a master regulatory transcription factor that has been estimated to bind to over 10% of cellular promoters in different cellular types (10, 16, 22–24, 27, 49), modulating the expression of thousand genes. The mechanism by which Myc activates transcription has been studied in detail. Myc is a weak transcriptional activator that acts by recruiting to the chromatin modifier enzymes that open the chromatin or lead to the release of the RNA polymerase II by directly or indirectly recruiting to the promoters the transcription elongating factor b (P-TEFb) (8, 9, 26, 51). Much less is known about Myc-dependent transcription repression (13). Myc can negatively regulate transcription via its direct interaction with the transcription factors Myc interacting zinc protein 1 (Miz-1) (43) or SP1 (12), but a large number of developmental genes appear to be repressed by Myc independently from this mechanism.
Polycomb repressive complex 2 (PRC2) core complex is formed by three components: Suppressor of Zeste 12 (Suz12), Enhancer of Zeste Homolog 2 (Ezh2), and Embryonic Ectoderm Development (Eed) (39). Polycomb proteins in Drosophila have been shown to be required to maintain stem cell and differentiated cell identity (35). The PRC2 complex also contains several other subunits, including factors preferentially expressed in ESC like Jarid2, esPRC2p48, and Pcl2 (19, 21, 25, 31, 32, 37, 47, 50). In ESC, PRC2 catalyzes histone H3 methylation of lysine 27 at promoters of developmental regulators whose expression is required later in development, suggesting that PRC2 contributes to maintain ESC pluripotency by keeping repressed several developmental regulators (1, 3, 20, 25). The actual role of Polycomb in ESC differentiation has not been fully clarified. Suz12, Ezh2, or Eed null ESC can actually be established, demonstrating that these genes are dispensable for the establishment and maintenance of ESC (6, 30, 38). However, this appears to be accomplished either by direct complementation of Ezh2 enzymatic activity by Ezh1 (38) or by compensatory regulations that overcome PRC2 function in maintaining ES cells undifferentiated. In fact, Suz12 and Eed mutants ES cells have been shown to express higher levels of several ESC-specific genes, including Nanog, Oct 3/4, and Sox2, which could maintain the cells in the undifferentiated state trough a network of gene regulatory circuits (38, 45, 47).
Here we show that Myc contributes to maintain ESC undifferentiated by upregulating the transcription of PRC2 genes. We observed that Myc binds to the E box elements of PRC2 genes, where it recruits chromatin modifier enzymes, inducing the increase of active RNA polymerase II on their promoters. Silencing of Myc proteins in ESC leads to inhibition of the expression of all the components of the PRC2 complex, resulting in a global reduction of H3K27me3, derepression of developmentally regulated PRC2 target genes, and expression of regulators of primitive endoderm.
MATERIALS AND METHODS
Cell culture condition.
3T3 and 3T3-MycER fibroblast cells were cultured in growth medium (Dulbecco modified Eagle medium [DMEM] high glucose with 10% fetal calf serum [FCS]). For time course experiments, cells were starved for 36 h with 0.2% fetal bovine serum and induced with 10% of FCS and with or without 4-hydroxytamoxifen (OHT) (100 nM) for the indicated time. Cycloheximide was added 1 h before induction at a concentration of 10 μg/ml. R1 mouse ES cells, expressing MycT58AER (5) kindly provided by S. Dalton, and E14 mouse ES cells were cultured in ESC medium (DMEM high glucose with 15% fetal bovine serum [FBS], NNEA1x, NaPyr1x, 0.1 mM 2-mercaptoethanol, and 1,500 U/ml LIF).
Transfection.
Transfection of 3T3 cells and ES cells was performed using Lipofectamine 2000 transfection reagent according to the manufacturer's protocol using an equal amount of each plasmid in multiple transfections. To obtain a 3T3-MycER stable line, transfected cells were plated as single cells and cultured for 2 weeks in growth medium with 1 mg/ml puromycin and then drug-resistant clones were picked, grown, and analyzed.
DNA constructs and shRNA.
Suz12, Ezh2, and Eed expression vectors were purchased from AddGene (pCMVHA constructs deposited by K. Helin). Short hairpin RNA (shRNA) constructs were purchased by OpenBiosystems: c-Myc shRNA 1 (TRCN0000086913), c-Myc shRNA 2 (TRCN0000086916), N-Myc shRNA 1 (TRCN0000020694), N-Myc shRNA 2 (TRCN0000020696). MycER (Myc protein fused to estrogen receptor alpha) expression vector was purchased from ADDGENE (Plasmid 19128: pBabepuro-myc-ER). This Myc-ER-expressing retroviral vector was generated by inserting human c-Myc into the pBabepuro3:hbERTAM vector in frame with the tamoxifen-sensitive hormone binding domain of the estrogen receptor (34).
ChIP assay.
Each chromatin immunoprecipitation (ChIP) experiment was performed in at least three independent biological samples and performed as described previously (28). Briefly, 1 × 106 cells were cross-linked by adding formaldehyde 1% for 10 min at room temperature, quenched with 0.125 M Glycina for 5 min at room temperature, and then washed twice in phosphate-buffered saline (PBS) before freezing. Pellets were resuspended in 0.2 ml SDS lysis buffer, stored in ice for 10 min, sonicated for 15 min at 4°C, and then centrifuged at 14,000 rpm for 10 min at 4°C. Supernatants were diluted 10-fold with ChIP dilution buffer (1% were kept as input) and incubated overnight in gentle rotation at 4°C with 1 μg of antibody. Then 10 μl of preblocked protein A beads were added and incubated for 1 h in the same conditions. After five washes (one in low-salt wash buffer, one in high-salt wash buffer, one in LiCl salt wash buffer, and two in TE buffer), the complexes were eluted by adding 0.25 ml of elution buffer for 15 min in rotation at room temperature. After RNase and proteinase K treatment, DNA was purified by phenol-chloroform extraction followed by ethanol precipitation. Starting sites of genes coding for PRC2 subunits were designed according to previously published papers (28). DNA was analyzed by quantitative real-time PCR (RT-qPCR) by using the SYBR GreenER kit (Invitrogen). All experiment values were normalized to those obtained with a nonimmune serum (IgG) and divided by input (or by pan-H3 values), using the procedure previously described (18). The data are expressed as percentages of the DNA inputs or pan-H3. Oligonucleotide sequences are indicated in the supplemental material.
RNA analysis.
RNA samples were extracted directly from cultured cells using TRIzol reagent (Gibco) followed by isopropanol precipitation. RNA was analyzed by quantitative real-time PCR using the Superscript III platinum one-step qRT-PCR system kit (Invitrogen). For analysis of the heterogeneous nuclear RNA (hnRNA), RNA was treated with DNase I for 2 h and then analyzed with specific primers. Oligonucleotide sequences are indicated in the supplemental material.
FACS analysis.
Cultured cells were harvested after incubation with dissociation buffer (Gibco) under gentle agitation and incubated for 30 min in phosphate-buffered saline (PBS)-5% FBS on ice to block unspecific binding. Cells were then incubated with primary antibodies for 30 min in PBS-1% bovine serum albumin (BSA) and after three washes were stained with conjugated secondary antibodies. Cells were analyzed by fluorescence-activated cell scan (FACS).
Immunostaining.
Cells were plated on coated cover glasses, fixed with 4% paraformaldehyde in PBS for 20 min at room temperature, washed twice, and incubated with the blocking buffer (PBS, 1% BSA, 0.1% Triton X-100). The fixed cells were incubated with primary antibody in blocking buffer for 1 h at room temperature followed by three washes and incubation with conjugated secondary antibodies in blocking buffer for 1 h at room temperature. For nuclear staining, cells were incubated for 5 min at room temperature with ToPro reagent. Images were performed with Leica TCS SP2 confocal microscopy (Heidelberg, Germany).
Western blotting.
Cells were harvested, resuspended in F buffer, and sonicated for three pulses. Extracts were quantified by bicinchoninic acid (BCA) assay (BCA protein assay kit; catalog no. 23225; Pierce). Histone acidic extraction was performed as described previously (11). Samples were run in SDS-polyacrylamide gels at different percentages and then transferred to nitrocellulose membranes.
Antibodies.
The antibodies used in this work were purchased from Santa Cruz Biotechnology (anti-c-MYC sc-764 for ChIP experiments, anti-PIM1 sc-7856, anti-MAX sc-197, anti-Pol II sc-899, anti-HBO1 sc-25379, anti-TIP60 sc-5725, anti-OCT3/4 sc-5279, anti-N-MYC sc56729, anti-E2F1 sc-193, anti-GCN5 sc-6303, anti-P300 sc-584, anti-ER sc-544), from Covance (anti-Pol II, Ser2 H5, anti-polymerase II Ser5 H14), from Upstate (anti-histone H3 06-755, anti-acetyl histone H3 06-599, anti-phospho [Ser10]-histone H3 05-817, anti-acetyl histone H4 Lys16 06-762, anti-acetyl histone H4 Lys12 07-595, anti-acetyl histone H4 Lys8 07-328, anti-acetyl histone H4 Lys5 07-327, anti-acetyl histone H4 06-598, anti-trimethyl-histone H3 Lys4 07-473, anti-trimethyl-histone H3 Lys27 07-449, anti-EED clone AA19, 05-1320, anti SOX1 071673), from Abcam (anti-histone H4 ab7311, anti-acetyl histone H3 Lys9 ab10812, anti-acetyl histone H3 Lys14 ab52946, anti-c-MYC ab11917 for Western blotting experiments), from Cell Signaling (anti-SUZ12 3737, anti-EZH2 3147), from Bethyl Laboratories (anti-MOF A300-994A), from Sigma (anti-β-actin A5441), from Chemicon (anti-SSEA1 MAB4301), and from Zymed (anti-ECAD ECCD2).
Viability assay and cell cycle analysis.
A flow cytometry experiment was performed using LIVE/DEAD cell viability assays from Invitrogen according to the manufacturer's protocol. Two-dimensional cell cycle analysis of the mouse ESC was performed using Click-iT EdU assays from Invitrogen according to the manufacturer's protocol with 1 h of EdU pulse.
One-dimensional cell cycle analysis of the 3T3-MycER cells was performed by fixing cells in Et-OH 70% and staining with propidium iodide (PI) solution (0.1%Triton, 200 mg/ml RNase, 20 mg/ml PI in PBS) for 30 min at room temperature.
Both experiments were performed using Becton Dickinson FACSCalibur and FACScan and analyzed by FACS FlowJo Software.
Cell trace analysis.
Flow cytometry cell trace analysis was performed using a CellTrace CFSE cell proliferation kit from Invitrogen according to the manufacturer's protocol. The experiment was performed using Becton Dickinson FACSCalibur and FACScan and analyzed by FACS FlowJo Software.
AP staining.
Cells were plated as single cells and after 3 days were fixed with 4% paraformaldehyde for 2 min and then stained with alkaline phosphatase (AP) solution (Vector Red alkaline phosphatase substrate kit I; catalog no. SK-5100).
Histone methylation quantification.
Histones were extracted using an EpiQuik total histone extraction kit (Epigentek). Quantification of trimethylated H3K4 and H3K27 was performed using the EpiQuik global tri-methyl histone H3-K4 quantification kit (Colorimetric) and EpiQuik global tri-methyl histone H3-K27 quantification kit (Colorimetric), respectively (Epigentek). Data were normalized on total histone H3 quantified using the EpiQuik total histone H3 quantification kit (Colorimetric). Data are presented as the mean ± standard deviation (SD) of results from three independent experiments.
A histone methyltransferase assay (HMT) was performed on nuclear extracts as previously described (51). HMT assays of H3K4 and H3K27 were performed using, respectively, the EpiQuik histone methyltransferase activity/inhibition assay kit (H3K4) and the EpiQuik histone methyltransferase activity/inhibition assay kit (H3K27).
RESULTS
Myc regulates the transcription of the genes coding for the PRC2 complex.
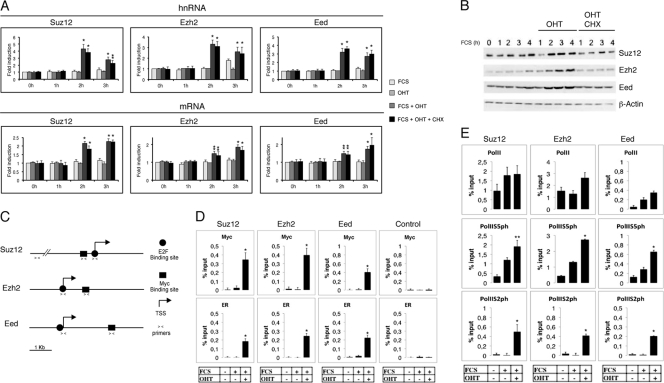
Analysis of Myc target genes by genome-wide chromatin immunoprecipitations (ChIP) in ESC reveals that, in addition to genes involved in metabolism and cell growth, Myc also binds to a number of genes coding for chromatin modifier enzymes, including some members of the Polycomb PRC2 complex (7, 14, 15). We therefore verified whether Myc regulates the expression of genes coding for the core components Suz12, Ezh2, and Eed of the PRC2 complex. To analyze the actual contribution of Myc in PRC2 regulation, we generated mouse fibroblast stable clones expressing Myc-ER fusion protein that could be activated by cell treatment with 4-hydroxytamoxifen (OHT). Activation of the Myc-ER fusion protein by nuclear translocation induced the expression of the unprocessed heterogeneous nuclear RNA (hnRNA) as well as the mature transcript of all three core components of PRC2 at 2 h and in the presence of the inhibitor of protein synthesis cycloheximide (CHX) (Fig. 1A), which inhibited cell cycle progression (see Fig. S1 in the supplemental material). Thus, excluding that the induction observed was an indirect effect due to the cell cycle progression. Western blot analysis showed a significant increase of Suz12, Ezh2, and Eed proteins in cells treated with OHT (Fig. 1B), confirming the induction at the protein level.
Fig 1.
Myc associates to regulatory elements of PRC2 core genes and activates their transcription. (A) Expression of PRC2 genes Suz12, Ezh2, and Eed hnRNA (top panel) and mRNA (bottom panel) in mouse NIH-3T3 fibroblasts expressing ectopic Myc-ER protein. Cells arrested by serum starvation were treated for the indicated time with 10% FCS with or without OHT in the absence or presence of cycloheximide. Data are presented as the mean ± SD of at least three independent experiments. Statistical analysis was performed with respect to uninduced samples. *, P value < 0.01. **, P value < 0.05. (B) Western blot analysis of mouse NIH-3T3 fibroblasts expressing exogenous Myc-ER protein after OHT treatment at the times indicated. Cells arrested by serum starvation were treated for the indicated times with 10% FCS with or without OHT in the absence or presence of cycloheximide. (C) Schematic representation of Suz12, Ezh2, and Eed promoter regions. Arrows indicate the transcription start sites (TSS). Squares represent the putative Myc binding sites (E boxes). Circles represent E2F binding sites (E2F). Convergent small arrows under each gene represent the positions of the regions analyzed by ChIP at the TSS, at Myc binding sites for each gene, or at 5 kb upstream of Suz12 as a Myc-negative control. (D) ChIP analysis on E-box regions of the PRC2 genes in mouse 3T3-MycER fibroblasts arrested by serum starvation and stimulated for 2 h with FCS with or without OHT using the anti-Myc (Myc) or anti-estrogen receptor (ER) antibodies to detect the binding of Myc-ER fusion protein. Purified rabbit IgG was used as a negative control. As a control for Myc-specific binding, we used a region that has been shown not to bind c-Myc or N-Myc by ChIP sequence analysis (7, 15); this region, located 5 kb upstream of the Suz12 gene, is shown. Statistical analysis was performed with respect to uninduced cells. *, P value < 0.01. **, P value < 0.05. (E) ChIP analysis on promoter regions (TSS) of the PRC2 genes as described for panel D. The antibody used is specified in each panel. Statistical analysis was performed with respect to uninduced samples. *, P value < 0.01. **, P value < 0.05.
To verify the direct binding of Myc to PRC2 genes, we performed chromatin immunoprecipitation (ChIP) assays. All three genes contain a conserved canonical E box element in proximity to the promoter (Fig. 1C), as well as an E2F binding site at their transcriptional start site (TSS) (4). Quantitative ChIP analysis revealed that within 2 h after OHT treatment Myc-ER was recruited at the E box of each of these genes, while no binding was observed in a negative-control region 5 kb upstream of the Suz12 gene (Fig. 1D). Two hours of OHT treatment also induced an increase of association to the promoter of the RNA polymerase II phosphorylated at Ser5 and Ser2 in its C-terminal domain (Fig. 1E). It is worth noting that, at 2 h after OHT treatment, we could not detect an increase in E2F1 binding at Suz12, Ezh2, or Eed TSS (Fig. S1B) and that at this time point the retinoblastoma protein (Rb) was not yet phosphorylated (see Fig. S1C in the supplemental material), confirming that we were observing a direct effect of Myc on these genes.
Taken together, these results demonstrate that Myc binding to the E box elements of Suz12, Ezh2, and Eed induces their transcriptional activation.
Myc activates the transcription of PRC2 genes by recruiting chromatin modifier enzymes to their E box elements.
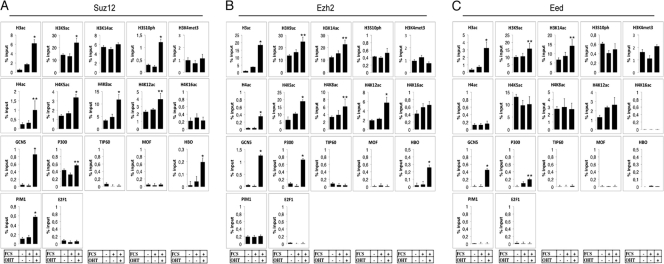
Next, we analyzed the mechanism by which Myc activates the transcription of Suz12, Ezh2, and Eed by the identification of histone modifier enzymes and the analysis of histone modifications that followed Myc binding to the E box elements. Following Myc binding to Suz12, Ezh2, and Eed E box elements, we observed an increase of H3 acetylation at K9 and K14, which correlated with the recruitment of the Myc cofactor GCN5 to these three genes (Fig. 2). Suz12 and Ezh2 E box elements also showed the recruitment of the acetyltransferase HBO1, which correlated with an increase of H4 acetylation at K5 and K12. At the E box element of Suz12 we also observed the recruitment of PIM1 and an increase of H3S10 phosphorylation. No difference was observed for E2F1, Tip60, or Mof on PRC2 E box elements (for the positive controls of these factors, see Fig. S1B and D in the supplemental material). Taken together, these data suggest that Myc activates the transcription of the genes coding for the core components of the PRC2 complex by Myc-dependent recruiting of chromatin modifier enzymes to their regulatory elements.
Fig 2.
Analysis of Myc-dependent chromatin modifications at E boxes of Polycomb PRC2 genes. (A to C) ChIP analysis of the E box elements of Suz12, Ezh2, and Eed in mouse 3T3-MycER fibroblasts arrested by serum starvation and then stimulated for 2 h with FCS with or without OHT using the antibodies indicated in each panel. Purified rabbit IgG were used as negative control. Statistical analysis was performed with respect to uninduced cells. *, P value < 0.01. **, P value < 0.05.
Myc maintains ES cells in the undifferentiated state.
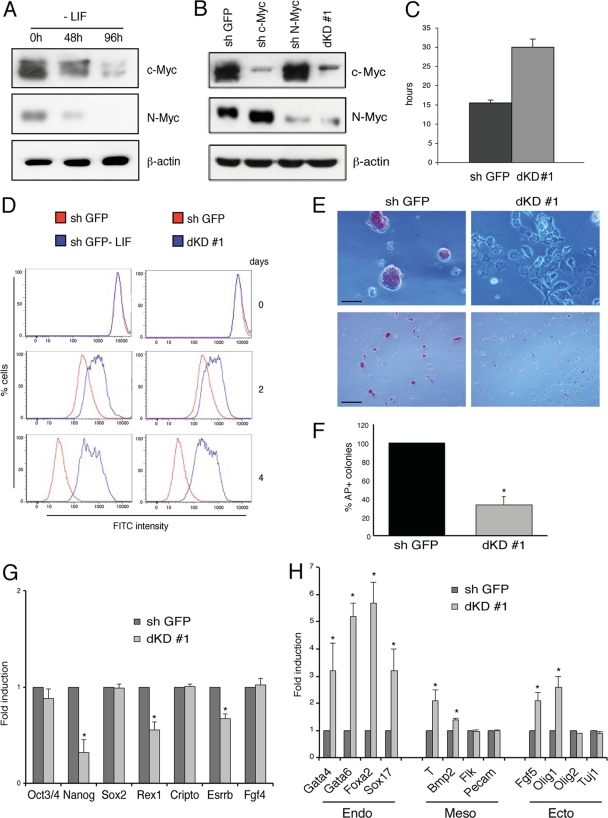
To analyze the role of Myc in the regulation of PRC2 in mouse ES cells, we performed Myc silencing experiments. In ESC, both c-Myc and N-Myc are highly expressed and LIF withdrawal determines the decline of both proteins (Fig. 3A), while the silencing of either one resulted in the increase of the transcriptional (not shown) and protein levels of the other (Fig. 3B). Therefore, to inhibit the expression of both c-Myc and N-Myc we performed a double knockdown (dKD) each with two independent shRNAs (Fig. 3B) (see Fig. S2A in the supplemental material).
Fig 3.
Role of c-Myc and N-Myc in mouse stem cell stemness. (A) Western blot analysis of c-Myc and N-Myc levels in mouse ESC E14 cultured without LIF for the time indicated. (B) Western blot analysis of c-Myc and N-Myc levels in control mouse ESC E14 or ESC silenced with the constructs indicated. (C) Mean time of cell replication in double c-Myc/N-Myc knockdown (dKD#1) ESC. The data are the mean results of three independent experiments. (D) Flow cytometry cell trace analysis using dye dilution. Red lines indicate the control (shGFP) stem cell proliferative tracing at the times indicated. The blue lines indicate stem cell proliferative tracing without LIF (left panels) or stem cell proliferative tracing c-Myc and N-Myc dKD (right panels). (E) Alkaline phosphatase (AP) staining of ESC colonies. The cells were plated as single cells to measure the capacity to form AP+ colonies in control cells (sh GFP) or in double knockdown (dKD #1). Two different magnifications are shown (scale bar: top panels, 50 μm; bottom panels, 200 μm). (F) Quantification of AP+ colonies in control cells (sh GFP) and in double silenced cells (dKD #1). *, P value < 0.01. (G) Expression analysis, by quantitative RT-PCR, of mRNAs in control ESC or in Myc double knockdown (dKD #1) cells as indicated. Data are presented as the means ± SD of results from three independent experiments. *, P value < 0.01. (H) Expression analysis, by quantitative RT-PCR, of developmental genes in control ESC or in Myc double knockdown (dKD #1) as indicated. Data are presented as the means ± SD of results from three independent experiments. *, P value < 0.01.
Myc dKD increased the cell doubling time with an expansion of G1 and early S phases (Fig. 3C) (see Fig. S2B to D in the supplemental material). We did not observe a reduction of cell viability by dKD (see Fig. S2E in the supplemental material). The downregulation of both Myc proteins led to a reduction of most characteristics of self-renewal, including ESC symmetric division, AP staining, and the ability to form single-cell colonies as dKD cells grew in a monolayer in a fibroblast-like manner (Fig. 3D to F) (see Fig. S2F to H in the supplemental material). In addition, dKD cells were larger than controls, mirroring the size increase obtained by LIF withdrawal, and showed a reduced expression of the stemness markers SSEA1 and E cadherin (see Fig. S3 in the supplemental material). We also observed a significant reduction of the key regulators of ESC Nanog, Rex1, and Esrrb but not of Oct3/4, Sox2, Cripto, and Fgf4 (Fig. 3G). Myc double-silenced ESC also showed an increased expression of early differentiation markers, including the endoderm markers Gata4, Gata6, FoxA2, and Sox17 and to a lesser extent the mesoderm marker Brachyury and the ectoderm markers Fgf5 and Olig1 (Fig. 3H). These findings suggest that Myc, in addition to its contribution to the enhancement of cell cycle progression, also contributes to the maintenance of ES cells in the undifferentiated state, as its expression is required to express key regulators of ES cell stemness and to keep repressed genes involved in ESC differentiation.
All of the genes coding for the PRC2 complex and its associated factors are bound and regulated by c-Myc and N-Myc in ESC.
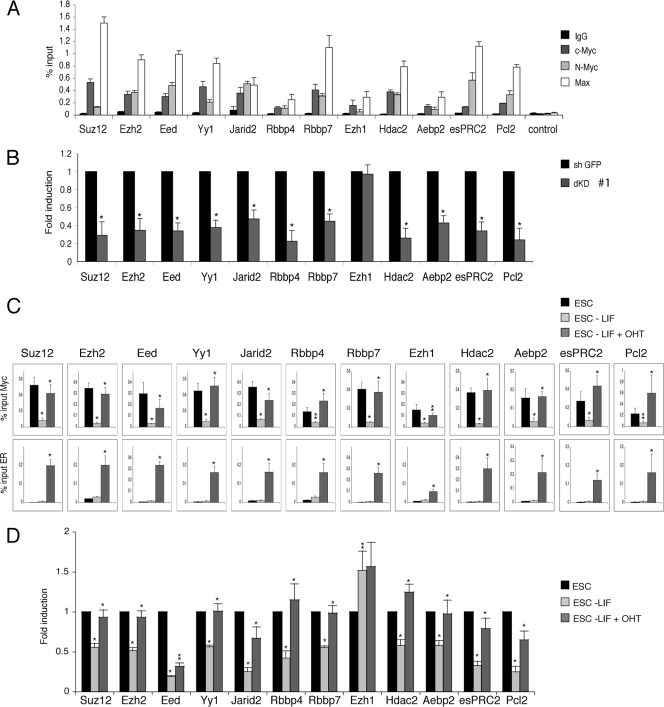
Next, we analyzed the role of Myc proteins in the regulation of the PRC2 complex and its associated factors in ESC. To this end, we measured by quantitative ChIP analysis the recruitment of endogenous c-Myc, N-Myc, and their dimerization partner Max to PRC2 genes. Both Myc proteins and Max associated to all three genes coding for the core PRC2 Suz12, Ezh2, and Eed as well as to the E box elements of the PRC2-associated factors YY1, Jarid2, Rbbp4, Rbbp7, Ezh1, Hdac2, Aebp2, esPRC2, and Pcl2 (Fig. 4A) known to be expressed in ESC (21, 31, 32, 37, 47, 48, 50). The specificity of c-Myc and N-Myc antibodies was confirmed by ChIP analysis in single and double KD (see Fig. S1E in the supplemental material). To verify whether Myc proteins contribute to the expression of these genes, we performed RT-qPCR in mock and dKD ES cells. The silencing of both c-Myc and N-Myc resulted in a significant reduction of the expression of all PRC2 components (Fig. 4B).
Fig 4.
All the components of the Polycomb complex are bound and regulated by c-Myc and N-Myc in embryonic stem cells. (A) ChIP analysis of Polycomb gene promoters in ESC using the antibodies indicated. Purified rabbit IgG (black columns) was used as a negative control. As a control for Myc specific binding was used a region that has been shown not to bind c-Myc or N-Myc by ChIP-sequence analysis (7, 15). (B) Expression analysis of mRNA of Polycomb genes in control ESC (sh GFP) or double silenced ESC (dKD #1). The levels of the transcripts were normalized against β-actin mRNA. Data are presented as the means ± SD of results from three independent experiments. *, P value < 0.01. (C) ChIP analysis of Polycomb gene E boxes in mouse ESC grown in the presence of LIF, in the absence of LIF for 72 h, or in the absence of LIF but treated with 4-OHT. ChIP experiments were performed using the indicated antibodies. Purified rabbit IgG was used as a negative control. Statistical analysis of ESC − LIF was performed with respect to control ESC, while statistical analysis of ESC − LIF + OHT was performed with respect to ESC − LIF. *, P value < 0.01. **, P value < 0.05. (D) Analysis of transcripts by quantitative PCR of Polycomb genes in ESC grown in the same conditions as above. The levels of each transcript were normalized against their levels in ESC. Data are presented as the means ± SD of results from three independent experiments. Statistical analysis was performed as in panel C.
As in the absence of LIF, Myc proteins are downregulated (Fig. 3A), we further analyzed whether the ectopic expression Myc proteins could upregulate the expression of PRC2 after LIF withdrawal. For this reason we used an ES cell line expressing a stable mutant of Myc-ER protein (5). LIF depletion determined a reduction of endogenous Myc binding to the E box elements and a reduction of the transcripts of all PRC2 genes analyzed but Ezh1 (Fig. 4C and D). The treatment of ESC with OHT, in the absence of LIF, induced the association of Myc-ER to the E boxes (Fig. 4C) and a significant increase in the expression of all mRNA subunits (Fig. 4D) but Ezh1. Taken together, these results suggest that Myc coordinately regulates the expression of all PCR2 subunits (yet known) expressed in ESC.
Myc proteins, by modulating PRC2 level, maintain H3K27 methylated at bivalent genes in ES cells grown in the absence of LIF.
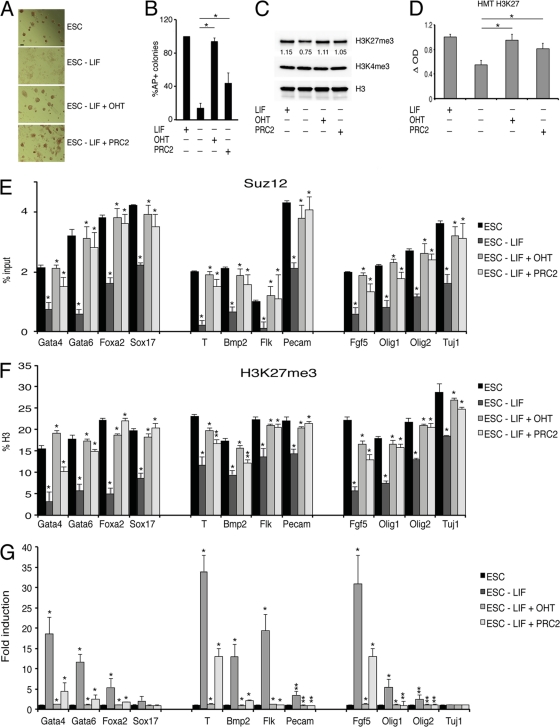
The results described above demonstrate that Myc directly controls the transcription of PRC2 in ESC. We then analyzed whether Myc-dependent regulation of PRC2 contributes to maintain ES cells in the undifferentiated state in the absence of LIF. We therefore activated Myc-ER by OHT treatment (which mediates the MycER translocation in the nuclei of ES cells) (see Fig. S4B in the supplemental material) in the absence of the LIF and analyzed ESC morphology. Active Myc-ER could maintain ES cells in their typical colony morphology and alkaline phosphatase activity (Fig. 5A and B) (see Fig. S4A and B). Significant inhibition of ES cells and a loss of colony formation and AP staining were also obtained by cotransfection of the constructs expressing the three PRC2 core proteins Suz12, Ezh2, and Eed (Fig. 5A and B) (see Fig. S4A and B).
Fig 5.
Myc or PRC2 maintain ESC undifferentiated and developmental genes in the OFF state in the absence of LIF. (A) Alkaline phosphatase (AP) staining of ESC colonies. The cells were plated as single cells to measure the capacity to form AP+ colonies of ESC grown in the presence of LIF, in the absence of LIF, or in the absence of LIF expressing either activated Myc-ER by OHT or ectopic PCR2 by transfection of CMV driven expression vectors (scale bar, 50 μm). (B) Quantification of AP+ colonies of ESC as in panel A. Data are presented as the means ± SD of results from three independent experiments. *, P value < 0.01. (C) Western blot analysis of ESC grown as in panel A. Histone H3 was used as a loading control. Numbers under first panel indicate the H3K27me3 blot quantification performed using ImageJ software. (D) H3K27 methyltransferase activity assays in ESC grown as in panel A. Data are presented as the means ± SD of results from three independent experiments. *, P value < 0.01. (E and F) ChIP analysis of the promoter regions of the indicated bivalent gene in ESC grown as in panel A using the antibodies indicated in each panel. Purified rabbit IgG were used as a negative control. Data are presented as the means ± SD of results from three independent experiments. Statistical analysis of ESC − LIF was performed with respect to ESC, while statistical analysis of ESC − LIF + OHT or ESC − LIF + PRC2 was performed with respect to ESC − LIF. *, P value < 0.01. **, P value < 0.05. (G) Expression analysis, by quantitative RT-PCR, of developmental genes in ESC grown as in panel A. Data are presented as the means ± SD of results from three independent experiments. Statistical analysis was performed as above.
The analysis of the levels of H3K27me3 and of H3K27 methyltransferase activity in ESC nuclei revealed a general reduction of H3K27me3 in ESC grown in the absence of LIF compared to control cells. The activation of Myc-ER by OHT treatment, or the ectopic expression of the core proteins of the PRC2 complex, significantly inhibited the reduction of the H3K27me3 mark as well as H3K27 methyltransferase activity in ESC nuclei grown in the absence of LIF (Fig. 5C and D). In contrast, we did not detect significant differences in the H3K4me3 mark under these conditions (see Fig. S4C and D in the supplemental material).
Next, we analyzed the binding of the PRC2 complex (measured as binding of the Suz12 subunit) and the H3K27me3 modification to the promoter regions of a subset of developmentally regulated “bivalent” genes that in ESC show H3K27me3 and H4K4me3 double marks. These experiments revealed that following LIF withdrawal, the binding of Suz12 was significantly reduced and increased again after Myc-ER activation by OHT treatment (Fig. 5E). The binding of Suz12 on these genes correlated with the level of H3K27me3 (Fig. 5F), whereas we could not observe a significant variation in the level of H3K4me3 (see Fig. S4D in the supplemental material). Similar results could be obtained by the coexpression of the core PRC2 proteins (Fig. 5E and F). Thus, modulation of PRC2 abundance in ESC, either by Myc or by ectopic expression of the core PRC2 proteins, is sufficient to reverse the chromatin status induced by LIF depletion at these Polycomb target genes. Analysis of the transcripts by RT-qPCR showed that the absence of LIF induced the upregulation of most genes analyzed (Fig. 5G). Thus, the activation of Myc-ER by OHT, or the constitutive expression of PRC2, was sufficient to keep in the OFF state the genes that are upregulated by LIF withdrawal. These results demonstrate that Myc-dependent regulation of PRC2 proteins maintains the silence of the bivalent genes involved in developmental programs in ESC.
Myc proteins are required to inhibit expression of bivalent developmental genes in ESC via PRC2 upregulation.
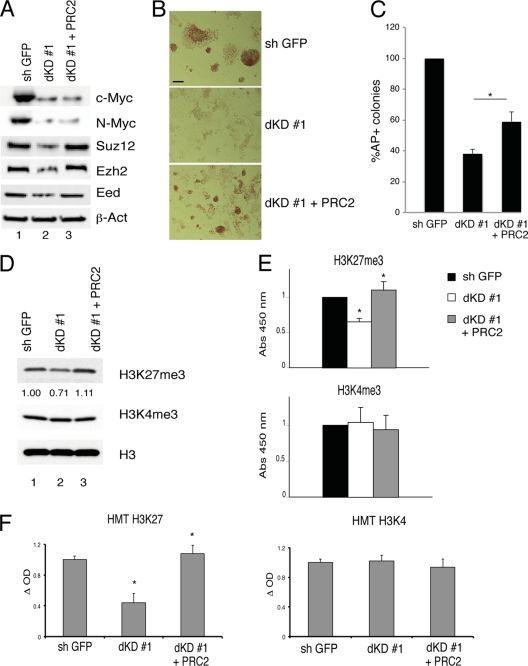
Next, we analyzed whether Myc expression is required to keep developmental genes in the OFF state via PRC2. ES cells silenced for the expression of Myc proteins showed, by Western blotting, a reduced expression of the core components of the PRC2 complex (Fig. 6A) (see Fig. S5A in the supplemental material). The ES cells subjected to dKD could no longer form AP-positive colonies and showed a significant reduction of H3K27me3 signal and H3K27me3 methyltransferase activity, but they did not show a significant reduction of H4K4me3 and H3K4me3 methyltransferase activity (Fig. 6B to F) (see Fig. S5). The ectopic expression of the PRC2 complex, by transfection of the expression vectors, partially inhibited the H3K27me3 reduction and loss of ESC morphology induced by Myc dKD (Fig. 6). In contrast, the ectopic expression of PRC2 could not rescue the cell cycle phenotype induced by Myc dKD (see Fig. S4F and G in the supplemental material). Thus, depletion of Myc activity in ESC affects the general level of H3K27me3 and ES cell morphology and these phenotypes can be counteracted, at least in part, by PRC2 constitutive expression in dKD cells.
Fig 6.
Ectopic PRC2 expression sustains the level of H3K27me3 that are downregulated by c-MYC and N-MYC double knockdown ESC. (A) Western blot analysis of Myc and PRC2 proteins in control ESC (sh GFP), in double knockdown cells (dKD #1), or in double knockdown cells expressing ectopic core PRC2 proteins. β-Actin was used as a loading control. (B) Alkaline phosphatase (AP) staining of ESC colonies as indicated. (C) Quantification of AP+ colonies of ESC as indicated. Data are presented as the means ± SD of results from three independent experiments. *, P value < 0.01. (D) Western blot analysis of ESC grown as in panel A. Histone H3 was used as a loading control. Numbers under first panel indicate the H3K27me3 blot quantification performed using ImageJ software. (E) H3K27 (top panel) and H3K4 (bottom panel) trimethylation quantification by colorimetric quantification assay (see Materials and Methods) in mouse ESC grown as indicated. Data are presented as means ± SD of results from three independent experiments. Statistical analysis of dKD #1 was performed with respect to control samples (sh GFP). dKD #1 + PRC2 was performed with respect to control samples (dKD #1). *, P value < 0.01. **, P value < 0.05. (F) H3K27 (left panel) and H3K4 (right panel) methyltransferase activity assays in ESC grown as in panel A. Data are presented as the means ± SD of results from three independent experiments. Statistical analysis was performed as above.
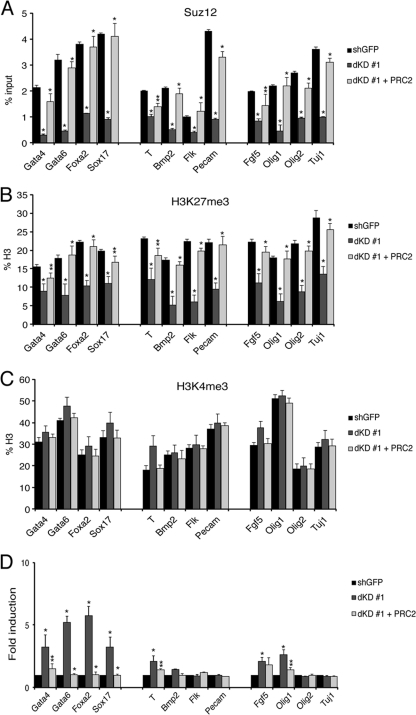
We then analyzed whether this effect could be seen at the level of single developmental bivalent genes. Analysis of Suz12 binding and H3K27me3 mark at a group of bivalent developmental genes involved in germ layer specification in dKD cells showed a significant reduction of PRC2 complex in these genes as well as a reduction of the H3K27me3 mark in all genes analyzed (Fig. 7A and B). The reduction of PRC2 association and H3K27me3 induced by the depletion of Myc proteins could be inhibited by the ectopic expression of PRC2 core proteins (Fig. 7A and B). No significant variations of H4K4me3 were observed by depletion of Myc proteins or ectopic expression of PRC2 in double-silenced cells (Fig. 7C). Analysis of the expression of these bivalent genes revealed that some were significantly upregulated. Importantly, the ectopic expression of PRC2 in the double-silenced cells was sufficient to keep all of these genes in the OFF state (Fig. 7D). It is worth to note that the depletion of Myc proteins in ESC resulted in the induction of expression of only a subset of the bivalent genes, although Myc depletion derepressed all bivalent genes (compare Fig. 7A, B and D). Taken together, these results strongly suggest that Myc proteins, by controlling the level of PRC2, regulate the level of H3K27me3 and the expression of PRC2 target genes, contributing to maintaining ESC in their undifferentiated state.
Fig 7.
Ectopic PRC2 expression maintains H3K27me3 at bivalent developmental genes in ESC keeping them in the repressed state. (A to C) ChIP analysis at the promoter regions of the indicated genes in control E14 ESC (sh GFP), in double-silenced cells (dKD #1), and in dKD cells expressing ectopic PRC2. The antibodies used are indicated above each panel. Purified rabbit IgG was used as a negative control. Data are presented as the means ± SD of results from three independent experiments. Statistical analysis was performed as Fig. 6E. (D) Transcriptional levels of the indicated genes in control ESC (sh GFP), in double-silenced cells (dKD #1), and in dKD cells expressing ectopic PRC2. The levels of transcripts were normalized against β-actin mRNA levels. Data are presented as the mean ± SD of results from three independent experiments. Statistical analysis was performed as in Fig. 6E.
DISCUSSION
Myc expression is required to maintain ESC in the undifferentiated state and significantly enhances the reprogramming of adult cells into induced pluripotent cells. By modulating the expression of Myc in fibroblasts and ESC, we here demonstrated that Myc upregulates the transcription of the Polycomb PRC2 complex both in adult and in stem cells. This regulation, contributing to keep silent bivalent genes, plays an important role in maintaining ES cells in the undifferentiated state.
Our experiments demonstrated that the direct binding of Myc to the PRC2 genes activates their expression by recruiting to their E box elements chromatin modifier enzymes, which results in the increase of active RNA polymerase II on their promoters. Previous analysis of PRC2 gene expression showed that these genes are regulated by the pRB-E2F pathway during the cell cycle progression (4). By analyzing PRC2 transcriptional regulation mediated by the Myc-ER chimera before E2F activation and by blocking the cell cycle progression, we could dissect the specific contribution of Myc in the transcription of these genes. Although we here demonstrated that Myc proteins directly upregulate the transcription of PRC2, it is worth mentioning that Myc can also regulate PRC2 indirectly (29, 36), thus, suggesting that Myc finely tunes PRC2 levels by positive and negative feedback mechanisms and corroborating the model by which the level of PRC2 is critical for ESC homeostasis. The PRC2 complex is formed by several subunits, some of which are preferentially expressed in ESC (25, 50).
Previous genome-wide ChIP analyses unveiled that Myc, besides binding to genes involved in cell cycle progression and metabolism, also binds to some members of the PcG family, although no functional studies were performed to confirm the actual binding and define the regulatory role of Myc on these genes. We have now demonstrated by ChIP analysis that Myc actually binds to all genes coding for the PRC2 complex and its binding results in their transcriptional activation both in ESC and in differentiated fibroblasts. This suggests that Myc acts as a regulator of the whole complex and not of some individual subunits, as suggested by the genome-wide analyses. Only Ezh1, which has been described to be more expressed in differentiated cells (50), was not upregulated by Myc in ESC, suggesting that Myc proteins not only regulate the levels of PRC2 but might also contribute to define the composition of the PRC2 complex in different cellular contexts.
In ES cells, Myc plays a dual role: it contributes to ESC self-renewal, by promoting cell cycle progression and metabolism, as well as to maintenance of ES cells in the undifferentiated state, as Myc-depleted ESC start to express developmental genes and spontaneously differentiate into primitive endoderm (5, 40, 41). We observed that, in the absence of LIF or in ESC silenced for Myc proteins, the downregulation of PRC2 results in the derepression of bivalent genes involved in ESC differentiation. Interestingly, the downregulation of PRC2 as a consequence of Myc reduction, differently from LIF withdrawal, did not result in the activation of all derepressed bivalent genes but induced mainly genes involved in primitive endoderm specification, suggesting that the derepression of developmental genes obtained by Myc depletion is not sufficient per se to activate positive regulators of mesoderm and ectoderm specification. Thus, the derepression of developmental genes does not induce the differentiation of ES cells as previously shown by Suz12−/−, and Eed mutant ES cells can still be maintained in culture, although they are impaired in their correct differentiation program (6, 30).
In ESC depleted for Myc proteins or grown in the absence of LIF, the ectopic expression of PRC2 is sufficient to keep the developmental genes in the OFF state and therefore able to inhibit the differentiation of the cells. Therefore, in ESC the level of PRC2 is critical and the reduced expression of Myc proteins, in response to differentiating signals, modulates this equilibrium, allowing cells to express developmental genes to start their differentiation programs. On the other hand, the ectopic expression of PRC2 alone could not restore the Myc-dependent regulation of the cell cycle progression, confirming that Myc is a pleiotropic factor that acts on independent pathways, one of which is inhibition of cell differentiation via PRC2.
A recent in-depth analysis proposed that three distinct subsets of regulated genes operate in ES cells; these include a core module determined by Oct3/4, Sox2, and Nanog, a Polycomb module, and a Myc module, which does not overlap with the Core or the Polycomb modules (17). Our data, showing that Myc actually regulates PRC2 in ESC, make a link between Myc and Polycomb modules, demonstrating that a major interconnection exists in ESC between these modules that increase the complexity of the networks in these cells.
The silencing of both c-Myc and N-Myc in ESC determines a general reduction of H3K27me3 mark, while we could not detect variations of the levels of H3K4me3 modification. These data differ from results of previous experiments in which Myc silencing was shown to induce a decrease of H3K4 but not H3K27 methylation (23). However, in the previous report only c-Myc was silenced while in our experiments we silenced both c-Myc and N-Myc, as in our cells, the silencing of c-Myc alone induced the overexpression of N-Myc.
It has been demonstrated, in ESC-mediated reprogramming experiments, that PcG-dependent repression is required for the conversion of adult differentiated B cells toward pluripotency (33) and that overexpression of PcG subunits, preferentially expressed in stem cells, enhances Oct3/4-, Sox2-, and Klf4-dependent cell reprogramming (50). Moreover, it was recently observed that in partially reprogrammed iPS a large number of developmental genes are repressed by methylation of H3K27 and almost none of these genes were direct targets of the four reprogramming factors (42). This finding supports the hypothesis that Myc contributes also to cell reprogramming by indirectly inhibiting the expression of genes involved in the differentiation via the upregulation of PRC2 complex.
As both Myc and PRC2 are both deregulated in a wide range of human cancers, our data might contribute to a better understandig of which regulatory programs are shared between stem cells and cancer cells.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Associazione Italiana Ricerca sul Cancro (AIRC), Regione Toscana programam salute, and Istituto Toscano Tumori (ITT).
We are indebted to Stephen Dalton for providing the R1 MycT58AER ES cell clone and to members of the laboratory for helpful suggestions and critical reading of the manuscript.
Footnotes
Published ahead of print 19 December 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Azuara V, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8:532–538 [DOI] [PubMed] [Google Scholar]
- 2. Boyer LA, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122:947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boyer LA, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353 [DOI] [PubMed] [Google Scholar]
- 4. Bracken AP, et al. 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22:5323–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cartwright P, et al. 2005. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 132:885–896 [DOI] [PubMed] [Google Scholar]
- 6. Chamberlain SJ, Yee D, Magnuson T. 2008. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26:1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen X, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133:1106–1117 [DOI] [PubMed] [Google Scholar]
- 8. Cowling VH, Cole MD. 2007. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol. Cell. Biol. 27:2059–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eilers M, Eisenman RN. 2008. Myc's broad reach. Genes Dev. 22:2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fernandez PC, et al. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fischle W. 2005. In nucleo enzymatic assays for the identification and characterization of histone modifying activities. Methods 36:362–367 [DOI] [PubMed] [Google Scholar]
- 12. Gartel AL, et al. 2001. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. U. S. A. 98:4510–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Herkert B, Eilers M. 2010. Transcriptional repression: the dark side of myc. Genes Cancer 1:580–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kidder BL, Yang J, Palmer S. 2008. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS One 3:e3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim J, Chu J, Shen X, Wang J, Orkin SH. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132:1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim J, Lee JH, Iyer VR. 2008. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One 3:e1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim J, et al. 2010. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143:313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kouskouti A, Talianidis I. 2005. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Landeira D, et al. 2010. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat. Cell Biol. 12:618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee TI, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125:301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li G, et al. 2010. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 24:368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Z, et al. 2003. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci. U. S. A. 100:8164–8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin CH, Lin C, Tanaka H, Fero ML, Eisenman RN. 2009. Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PLoS One 4:e7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mao DY, et al. 2003. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr. Biol. 13:882–886 [DOI] [PubMed] [Google Scholar]
- 25. Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469:343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McMahon SB, Wood MA, Cole MD. 2000. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 20:556–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meyer N, Penn LZ. 2008. Reflecting on 25 years with MYC. Nat. Rev. Cancer 8:976–990 [DOI] [PubMed] [Google Scholar]
- 28. Mikkelsen TS, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839–843 [DOI] [PubMed] [Google Scholar]
- 30. Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. 2007. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 27:3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pasini D, et al. 2010. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464:306–310 [DOI] [PubMed] [Google Scholar]
- 32. Peng JC, et al. 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139:1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pereira CF, et al. 2010. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell 6:547–556 [DOI] [PubMed] [Google Scholar]
- 34. Ricci MS, et al. 2004. Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol. Cell. Biol. 24:8541–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ringrose L, Paro R. 2007. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134:223–232 [DOI] [PubMed] [Google Scholar]
- 36. Sander S, et al. 2008. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 112:4202–4212 [DOI] [PubMed] [Google Scholar]
- 37. Shen X, et al. 2009. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139:1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen X, et al. 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32:491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Simon JA, Kingston RE. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10:697–708 [DOI] [PubMed] [Google Scholar]
- 40. Smith KN, Lim JM, Wells L, Dalton S. 2011. Myc orchestrates a regulatory network required for the establishment and maintenance of pluripotency. Cell Cycle 10:592–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith KN, Singh AM, Dalton S. 2010. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell 7:343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sridharan R, et al. 2009. Role of the murine reprogramming factors in the induction of pluripotency. Cell 136:364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Staller P, et al. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 3:392–399 [DOI] [PubMed] [Google Scholar]
- 44. Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 45. Ura H, et al. 2011. Eed/Sox2 regulatory loop controls ES cell self-renewal through histone methylation and acetylation. EMBO J. 30:2190–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Varlakhanova NV, et al. 2010. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation 80:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Walker E, et al. 2010. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell 6:153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Walker E, Manias JL, Chang WY, Stanford WL. 2011. PCL2 modulates gene regulatory networks controlling self-renewal and commitment in embryonic stem cells. Cell Cycle 10:45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zeller KI, et al. 2006. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci. U. S. A. 103:17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Z, et al. 2011. PRC2 complexes with JARID2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells 29:229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zippo A, et al. 2009. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138:1122–1136 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.