Abstract
Studies of small noncoding RNAs (sRNAs) have been conducted predominantly using culturable organisms, and the acquisition of further information about sRNAs from global environments containing uncultured organisms now is very important. In this study, hot spring water (57°C, pH 8.1) was collected directly from the underground environment at depths of 250 to 1,000 m in Yunohama, Japan, and small RNA sequences obtained from the environment were analyzed. A phylogenetic analysis of both archaeal and bacterial 16S rRNA gene sequences was conducted, and the results suggested the presence of unique species in the environment, corresponding to the Archaeal Richmond Mine Acidophilic Nanoorganisms (ARMAN) group and three new Betaproteobacteria. A metatranscriptomic analysis identified 64,194 (20,057 nonredundant) cDNA sequences. Of these cDNAs, 90% were either tRNAs, tRNA fragments, rRNAs, or rRNA fragments, whereas 2,181 reads (10%) were classified as previously uncharacterized putative candidate sRNAs. Among these, 15 were particularly abundant, 14 of which showed no sequence similarity to any known noncoding RNA, and at least six of which form very stable RNA secondary structures. The analysis of a large number of tRNA fragments suggested that unique relationships exist between the anticodons of the tRNAs and the sites of tRNA degradation. Previous bacterial tRNA degradation studies have been limited to specific organisms, such as Escherichia coli and Streptomyces coelicolor, and the current results suggest that specific tRNA decay occurs more frequently than previously expected.
INTRODUCTION
Small noncoding RNAs (sRNAs) are known to be one of the major players in the regulation of gene expression in the Prokaryota in both Bacteria and Archaea (20, 25, 55). A number of sRNA studies, particularly of Bacteria, have been undertaken in the last decade (1, 20). For example, about 80 sRNAs from Escherichia coli have been registered in RegulonDB (17), and these have been identified with several methods: high-throughput computational searches (2, 10, 47, 69), shotgun cloning (29, 60), and tiling array analyses (58, 70). In recent years, a new powerful method, next-generation sequencing technology, has been used to discover new sRNAs in the Bacteria, and the results have shown that hundreds of still-undiscovered sRNA genes might exist in the E. coli genome (45, 51). It has been reported that the majority of bacterial sRNAs are regulators of gene expression in response to environmental stresses (38) and typically are involved in the modulation of translation efficiency and the stability of mRNA molecules (1).
Interestingly, many tRNA fragments are observed in the small RNA fraction prepared from E. coli. (29, 51). The developmentally regulated cleavage of tRNAs also has been observed in the bacterium Streptomyces coelicolor (22). Plasmid-encoded enzymes, the colicins, are involved in the cleavage of the anticodons of specific tRNAs in E. coli (36, 43), resulting in the production of tRNA fragments. Similarly, it is reported that tRNA cleavage is a response to stresses in the Eukaryota (15, 56), and cancer- or age-associated changes in fragmented tRNAs have been observed (27, 32). Recently, rRNA fragmentation has become another topic of interest (16, 39), although the functions of fragmented rRNAs remain unclear. Papers that report unique RNA regulatory sequences, called riboswitches, in prokaryotic cells also are accumulating (49). These regulatory RNA sequences are predominantly located in the 5′ untranslated regions of some mRNAs and control target translation (49). Taken together, these findings confirm that sRNAs are important regulators in the Prokaryota.
However, previous sRNA studies usually were conducted in model organisms, so the knowledge of environmental sRNAs derived from uncultured microbes still is sparse. Metagenomics is a powerful technique with which to establish the population dynamics and phylogenies of uncultured microbes to overcome this problem. Previous metagenomic research has revealed the diversity of microbial species and their genomic sequences in various environments, including the ocean (59), hot springs (6, 23, 24, 30), hydrothermal vents (68), gold mines (41, 42), human skin (21), the human gut (3), and so on. However, the metagenomic approach does not identify the kinds of genes that are actually expressed in certain environments, which reflect the major functional activities and adaptations of the microorganisms that express them. Therefore, the metatranscriptomic approach has become increasingly important in defining these overall environmental gene expression profiles (7, 50, 61) and in identifying both the precursor and processing status of such transcripts. Therefore, a combination of metagenomic and metatranscriptomic approaches should allow an overview to be established of the diversity of microbial communities and the dynamics of their gene expression. Like other sRNA studies, recent reports have revealed the presence of large structured noncoding RNAs (64) and depth-specific novel sRNAs in the ocean (50), suggesting that these sRNAs play important roles in the adaptations required to live in this environment.
In the current study, we used both metagenomic and metatranscriptomic approaches to collect more information about sRNAs from microorganisms in hot spring water, which have minimal exposure to the external environment. The unique archaeal and bacterial species and novel candidate sRNAs found in the environment, including huge amounts of tRNA fragments, are discussed.
MATERIALS AND METHODS
Sample collection and DNA/RNA extraction.
The sample was obtained from the Yunohama hot spring (Tsuruoka, Yamagata, Japan; 38°78′04.57″N, 139°75′21.18″E), which has a temperature of 57°C and a pH of 8.1, on 4 August 2009. Unfiltered Yunohama hot spring water (20 liters) was bottled from a tank into which the hot water had been pumped (by eight pumps) from depths ranging from 250 to 1,000 m for mixing and storage. For genomic DNA preparation, the sample water was concentrated by filtration using an UltraClean water DNA isolation kit (MO BIO Laboratories, Carlsbad, CA) according to the manufacturer's protocol. The genomic DNA (14.3 μg) then was extracted from 3.5 liters of the sampled water using the same kit. For sRNA preparation, 3 liters of the sampled water was centrifuged (333 ml; 9 times) at 11,000 × g for 10 min at 4°C. The pellet was stored at −80°C until analysis. The sRNA was prepared with the mirVana miRNA isolation kit (Life Technologies, Carlsbad, CA). The stored pellet was initially resuspended in 1 ml of cold phosphate-buffered saline, to which 600 μl of lysis/binding buffer (mirVana miRNA isolation kit) was added. Finally, the sRNA fraction (approximately <200 bp in size; 650 ng) was isolated from 3 liters of the sample water.
PCR amplification and determination of 16S rRNA gene sequences.
16S rRNA genes were amplified from the genomic DNA isolated from the Yunohama sample using an archaeal primer set recently described by Gantner et al. (18) (340F, 5′-CCCTAYGGGGYGCASCAG-3′; and 1000R, 5′-GGCCATGCACYWCYTCTC-3′) and a commonly used bacterial universal primer set described by Wilms et al. (66) (341F, 5′-CCTACGGGAGGCAGCAG-3′; and 907R, 5′-CCGTCAATTCCTTTGAGTTT-3′). PCR was performed with TaKaRa Ex Taq DNA polymerase (Takara Bio Inc., Shiga, Japan). For the archaeal 16S rRNA sequences, amplification proceeded with one denaturation step at 98°C for 2 min, followed by 40 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 1 min 30 s, with a final extension step at 72°C for 7 min. The bacterial 16S rRNA sequences were amplified with one denaturation step at 95°C for 5 min, followed by 30 cycles at 95°C for 30 s, 50°C for 30 s, and 72°C for 2 min, with a final extension step at 72°C for 2 min. To amplify as many unknown archaeal species as possible, another set of primers (340F2, 5′-CCCTAYGGGGYGCASCAGGC-3′; and 932R, 5′-GCYCYCCCGCCAATTCMTTTA-3′) was designed for the archaeal 16S rRNA genes. The PCR amplification with these primers proceeded with one denaturation step at 95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 50°C for 15 s, and 72°C for 1 min, with a final extension step at 72°C for 2 min.
The amplified PCR products (see Fig. S1 in the supplemental material) then were cloned into the pGEM-T Easy vector (Promega Corporation, Madison, WI). The resulting plasmids were transformed into competent Escherichia coli DH5α cells (Takara Bio Inc.). The colonies were selected by blue-white screening on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and isopropyl-β-d-thiogalactopyranoside plates. After the preparation of the plasmid DNAs, their inserts were sequenced on both the sense and antisense strands with an ABI PRISM 3100 DNA sequencer (Life Technologies) using either the T7 primer (5′-TAATACGACTCACTATAGGG-3′) or the SP6 primer (5′-ATTTAGGTGACACTATAGAA-3′).
Construction of a cDNA library from the sRNA fraction and 454 pyrosequencing.
The sRNAs initially were treated with tobacco acid pyrophosphatase (Nippon Gene, Tokyo, Japan) and then used to construct a cDNA library. The cDNA library was constructed with the small RNA cloning kit (Takara Bio Inc.) with the following procedures: (i) 3′ adapters (5′-CATCGATCCTGCAGGCTAGAGAC-3′) and 5′ adapters (5′-AAAGATCCTGCAGGTGCGTCA-3′) were ligated with T4 RNA ligase; (ii) the transcripts then were reverse transcribed to cDNAs with the reverse transcription (RT) primer 5′-GTCTCTAGCCTGCAGGATCGATG-3′, which is the antisense sequence of the 3′ adapter; (iii) the cDNAs were amplified with the F primer (the same DNA sequence as that of the 5′ adapter) and the RT primer for 15 cycles, followed by a second PCR for 18 cycles with the same primer set. The amplified cDNAs were separated on a 10% polyacrylamide gel, and the fragments were cut from the gel. The excised gel fragments were incubated at 37°C for 4 h in a buffer containing 0.5 M ammonium acetate and 1 mM EDTA. The eluted cDNAs were purified by ethanol precipitation and dissolved in TE buffer. The final preparation of the sample was confirmed by 10 to 20% polyacrylamide gel electrophoresis (see Fig. S2 in the supplemental material), and the samples were used for pyrosequencing with GS FLX (Roche Diagnostics, Indianapolis, IN).
Bioinformatic analysis.
The following software and databases were used for the analysis of the pyrosequencing data: tRNA sequences were detected with tRNAscan-SE (34), and the microbial taxon corresponding to each tRNA sequence was predicted based on the genomic tRNA database, GtRNAdb (http://gtrnadb.ucsc.edu/) (9); 5S rRNA sequences were detected with a BLAST search (ftp://ftp.ncbi.nih.gov/blast/executables/) of the 5S rRNA database (52); fragment sequences for tRNA and 5S rRNA were detected with a BLAST search of the mature tRNA and 5S rRNA sequences detected in this study, respectively; 16S rRNA and 23S rRNA partial sequences were detected with a BLAST search of the RDP (http://rdp.cme.msu.edu/) (12) and the SILVA database (44); and other noncoding RNAs were detected by comparison to the Rfam database (http://rfam.sanger.ac.uk/) (19). We have summarized the numbers of redundant/nonredundant reads and the proportions of different types of sRNAs in Table 1. Sfold (http://sfold.wadsworth.org) (13, 14) was used to predict RNA secondary structures and to calculate the free energies of the candidate sRNAs.
Table 1.
Numbers of sequences and the percentage of each type of sRNA in the cDNA libraryh
| Small RNA type | Sequence type |
|||
|---|---|---|---|---|
| Nonredundant |
Redundant |
|||
| No. | % | No. | % | |
| tRNAa | ||||
| Mature | 8,069 | 40.2 | 43,092 | 67.1 |
| Fragment | 2,063 | 10.3 | 4,484 | 7.0 |
| rRNAb | ||||
| Mature 5S | 532 | 2.7 | 2,036 | 3.2 |
| 5S fragment | 320 | 1.6 | 673 | 1.0 |
| 16S fragment | 3,499 | 17.4 | 5,640 | 8.8 |
| 23S fragment | 3,348 | 16.7 | 5,133 | 8.0 |
| Other noncoding RNAc | ||||
| SRP bacte | 36 | 0.18 | 66 | 0.10 |
| TPPf | 4 | 0.02 | 4 | 0.01 |
| 6S RNA | 4 | 0.02 | 4 | 0.01 |
| Alpha RBSg | 1 | 0 | 2 | 0 |
| Unclassifiedd | ||||
| SURFY (≥10 reads) | 50 | 0.25 | 186 | 0.29 |
| Others (<10 reads) | 2,131 | 10.6 | 2,874 | 4.5 |
Total number of nonredundant sequences, 10,132 (50.5%). Total number of redundant sequences, 47,576 (74.1%).
Total number of nonredundant sequences, 7,699 (38.4%). Total number of redundant sequences, 13,482 (21.0%).
Total number of nonredundant sequences, 45 (0.22%). Total number of redundant sequences, 76 (0.12%).
Total number of nonredundant sequences, 2,181 (10.9%). Total number of redundant sequences, 3,060 (4.8%).
Bacterial signal recognition particle RNA.
TPP riboswitch (THI element).
Alpha operon ribosome binding site.
A total of 20,057 and 64,194 nonredundant and redundant sequences were analyzed, respectively.
For the phylogenetic analysis, representative 16S rRNA gene sequences (either 13 archaeal or 15 bacterial sequences, respectively) obtained from the Yunohama PCR clones were combined with known 16S rRNA gene sequences registered in the RDP (either 305 archaeal or 316 bacterial sequences), and then each phylogenetic tree was constructed using ClustalW 2.0 (31) with 100 bootstrap iterations. The tree was visualized with iTOL (33).
Nucleotide sequence accession numbers.
The pyrosequencing data have been submitted to the DDBJ Sequence Read Archive (http://trace.ddbj.nig.ac.jp/dra/index_e.shtml) under accession number DRA000364. The 16S rRNA gene sequences determined in this study have been deposited in the DDBJ database (http://getentry.ddbj.nig.ac.jp/) under accession numbers AB665420 to AB665440, AB679512 to AB679519, and AB683043 (see Table S1 in the supplemental material). The small unique RNAs from Yunohama (SURFYs) also were deposited in the DDBJ database under accession numbers AB665588 to AB665602 (see Table S2).
RESULTS AND DISCUSSION
Diversity of archaeal and bacterial communities in the Yunohama hot spring.
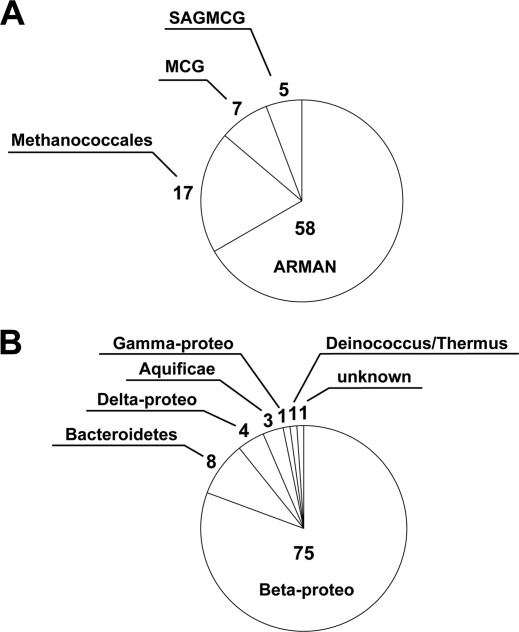
To clarify the phylogeny of the microbes in the deep subsurface water of the Yunohama hot spring, 16S rRNA genes from both archaeal and bacterial species were amplified by PCR using domain-specific primer sets. Recently, Gantner et al. reported a new archaeal primer set (340F and 1000R) for 16S rRNA genes (18). The authors validated these primers using three diverse environmental samples. Therefore, in the current study, this primer set was used to amplify the archaeal 16S rRNA gene sequences from the genomic DNA prepared from the Yunohama hot spring water. Two bands were amplified (approximately 680 and 820 nucleotides [nt] in size; see Fig. S1A in the supplemental material). After the PCR products were subcloned, 87 clones were randomly selected and their nucleotide sequences determined. DNA sequencing and phylogenetic analysis (see Fig. S3 in the supplemental material) of the archaeal 16S rRNA clones revealed that the longer band (58/87 clones) corresponded to only one taxon, the Archaeal Richmond Mine Acidophilic Nanoorganisms (ARMAN) (5), whereas the shorter band (29/58 clones) corresponded to the following three taxa: (i) Methanococcales, (ii) a miscellaneous crenarchaeotic group (MCG) (41), and (iii) the South African Gold Mine Crenarchaeotic Group (SAGMCG) (54). Therefore, archaeal species belonging to ARMAN comprised 67% (58/87) of the archaeal clones derived from the Yunohama hot spring (Fig. 1A). Previously, ARMAN have been found only in acidic environments throughout the world (4), so the Yunohama ARMAN could be the first example of ARMAN living in an alkaline environment (pH 8.1).
Fig 1.
Archaeal and bacterial species found in the Yunohama hot spring. The pie charts represent the closest known taxa and their read numbers for (A) 87 archaeal 16S rRNA gene sequences and (B) 93 bacterial 16S rRNA gene sequences. These taxa were estimated based on the phylogenetic analysis shown in Fig. S3 and S5 in the supplemental material.
Interestingly, intervening repeat sequences were present in two distinct regions of the 16S rRNA gene sequences of the Yunohama ARMAN (see Fig. S4A and S4B in the supplemental material), whereas these repeat sequences were not observed in the 16S rRNA gene sequence of the candidate Micrarchaeum acidiphilum (ARMAN-2) (4), which is the species phylogenetically closest to the Yunohama ARMAN (see Fig. S3). Moreover, the Yunohama ARMAN were classifiable into two groups based on the sequence of the second repeat region (see Fig. S4B), either type A-110 (two repeats) or type A-127 (one repeat), although the sequences of the first repeat region (see Fig. S4A, one repeat) were identical in both types. The nucleotide sequences of these repeats are quite conserved (see Fig. S4C).
To amplify as many unknown archaeal species as possible, a newly designed Archaea-specific primer set (340F2 and 932R) also was used for the PCR cloning of 16S rRNA sequences from the sample of Yunohama genomic DNA. The primer set was designed to detect uncultured Archaea based on 307 recently available archaeal 16S rRNA gene sequences obtained from the RDP (August 2010). The PCR amplification of the 16S rRNA genes from the Yunohama genomic DNA again produced two bands, approximately 700 and 1,200 nt in size (see Fig. S1B in the supplemental material). In this case, most of the 700-nt band corresponded to ARMAN, as described above (plus only a single clone of the same size classified in the Marine Group [37]). However, the 1,200-nt band (which included an approximately 680-nt insertion in the rRNA sequence; clone A-9 in Table S1 in the supplemental material) corresponded to the Hot Water Crenarchaeotic Group (HWCG) I (41). Other smaller clones that did not correspond to any major band also were isolated and belonged to the following three taxa: the Ancient Archaeal Group (AAG) (53), the Forest Soil Crenarchaeotic Group (FSCG) (26), and the Thermoplasmatales. These sequences were amplified only by the newly designed primers, suggesting that the new primer set designed in this study can capture additional diverse minor archaeal species.
Among the clones isolated using the originally designed primers, AAG is an interesting taxon, rooted at the deepest position on the archaeal tree, and is found only in deep-sea hydrothermal vent environments (53). Therefore, the discovery of archaeal species related to ARMAN and AAG extends the knowledge of the environments inhabitable by these taxa. Because Yunohama is located on the ocean front, the detection of AAG, the Marine Group, and Thaumarchaeota members (originally found in oceanic environments) is consistent with its geographic location. HWCG I, also known as Caldiarchaeum subterraneum (42), has been isolated from subsurface water in the Hishikari gold mine (Kagoshima, Japan), where the pH (6.19 to 6.80) and temperature conditions (71.5 to 85.0°C) differ from those at Yunohama. Therefore, the archaeal species with rRNA sequences highly similar to those of HWCG I are geographically interesting. Gantner's new archaeal primer set also identified clones corresponding to MCG and SAGMCG, which also were observed at the Hishikari gold mine (41), suggesting that these three taxa are common archaeal groups in subsurface environments in Japan.
The bacterial species in this environment were analyzed using a Bacteria-specific universal primer set (66) to amplify their 16S rRNA gene sequences. In total, 93 sequences derived from a single PCR band (approximately 600 nt in size; see Fig. S1C in the supplemental material) were identified. These were further classified into 15 nonredundant sequences and mapped to the bacterial phylogenetic tree (see Fig. S5). A BLAST search showed that (i) one sequence (B-71) was 100% identical to the 16S rRNA gene sequence of an uncultured bacterium; (ii) one sequence (B-91) was similar (83% sequence similarity) to the 16S rRNA gene sequence of an uncultured bacterium obtained from deep groundwater in a uranium mine in Japan (40); and (iii) the remaining 13 sequences showed 91 to 100% sequence similarity to 16S rRNA gene sequences from uncultured bacteria registered in the National Center for Biotechnology Information (NCBI) database. All of these sequences belong to one of the following six taxa: Betaproteobacteria, Bacteroidetes, Aquificae, Deinococcus/Thermus, Deltaproteobacteria, and Gammaproteobacteria. B-91 remains unclassified, branching outside the domain Bacteria but lacking similarity to the Archaea (Fig. 1B). Three species belonging to the phylum Betaproteobacteria comprised approximately 81% (75/93) of the bacterial clones derived from the Yunohama hot spring environment, and of these, B-35 was rooted at the deepest position in the currently known betaproteobacterial clade (see Fig. S5 in the supplemental material).
Metatranscriptomic analysis of candidate sRNAs from the Yunohama hot spring.
To understand the sRNA population in the Yunohama hot spring and its sequence characteristics, a metatranscriptomic analysis of the sRNA fraction (<200 bp) was conducted using next-generation sequencing technology. Although the sRNA fraction was obtained directly from hot water at 57°C, the extracted sRNA fraction was not degraded and showed a distinct band of approximately 75 nt after electrophoresis on a 6% polyacrylamide gel containing 8 M urea, which is approximately the size of full-length tRNA (data not shown). After the 5′ and 3′ RNA tag sequences were added to the sRNAs for PCR amplification (cDNA library construction), a similar band pattern, including a major band of approximately 120 nt (because of the addition of the tag sequence), was obtained with 10 to 20% polyacrylamide gel electrophoresis (see Fig. S2 in the supplemental material). Pyrosequencing the library then produced 86,236 sequence reads.
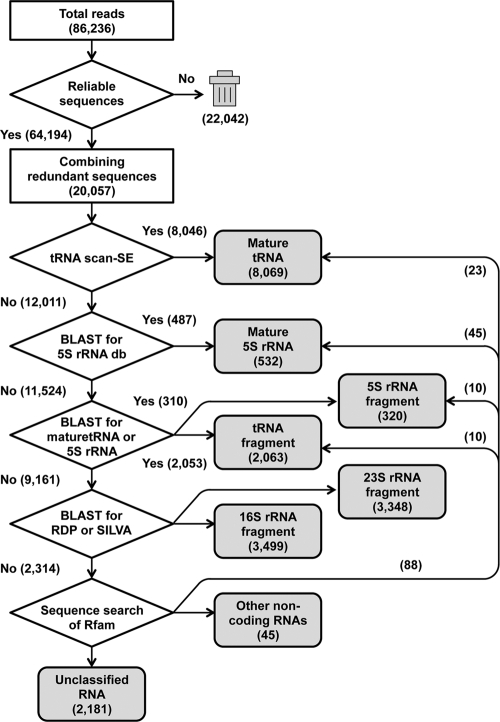
To characterize these reads, a nonredundant data set was prepared by extracting the reliable reads and combining the redundant reads to yield 20,057 nonredundant reads, ranging in length from 11 to 197 nt (average length, 62 nt; Fig. 2). Known noncoding RNAs were extracted with a series of filtering approaches based on homology searches and prediction software (Fig. 2). Of the final reads, 17,831 (88.9%) were either tRNAs, fragmented tRNAs, rRNAs, or fragmented rRNAs. Only 45 reads (0.2%) had certain sequence similarities to the following noncoding RNAs: bacterial signal recognition particle RNA (46), 6S RNA (63), and two known RNA elements, the thiamine pyrophosphate (TPP) riboswitch (67) and the alpha operon ribosome-binding site (48). A recent metatranscriptomic analysis revealed unique microbial sRNAs in the ocean water from the HOT Station ALOHA site in Hawaii (50). Interestingly, the authors identified some candidate sRNAs similar to those identified in the current study, such as signal recognition particle RNA and 6S RNA. Comparative genomics also revealed 104 structured RNAs, including candidate riboswitches, of the Bacteria and Archaea (65). On the contrary, no archaeal-specific transcripts were isolated from the Yunohama hot spring sample. Furthermore, as suggested by the tRNA sequence analysis described below, almost all of the reads are expected to have been derived from the Bacteria. The numbers of these noncoding RNAs are summarized in Table 1.
Fig 2.
Computational scheme for the extraction of known and novel noncoding candidate RNAs. The whole scheme of the computational analysis used in this study is illustrated as a flowchart. The numbers indicate the read numbers at each step. Initially, a data set containing 20,057 nonredundant reads was constructed from a total of 86,236 reads (raw data). These were classified in a stepwise manner as mature tRNA or rRNA (5S rRNA), fragmented tRNA or rRNA (5S, 16S, or 23S rRNAs), or noncoding RNA. The software tRNAscan-SE and BLAST (provided by NCBI) and the databases Ribosomal Database Project (RDP), SILVA, and Rfam were used in our analysis.
The remaining 2,181 reads (10.9%) were categorized as unclassified. These unclassified RNAs may include novel classes of sRNAs. To extract reliable candidate sRNAs, the extremely closely related sequences (with more than 95% sequence similarity) were combined, which further narrowed the sequence set to 1,972 reads, ranging in length from 11 to 176 nt (average length, 55 nt). A BLASTn analysis against the NCBI nucleotide collection (nr/nt) was conducted, which identified 354 reads (18%) with a certain degree of sequence similarity (with E values ranging from 2e−4 to 1e−41) to known nucleotide sequences. Of these 354 reads, 266 (75%) were similar to known genomic sequences (mostly bacterial), whereas 88 (25%) showed similarity to environmental DNA sequences. In particular, 71 reads had a degree of sequence similarity to the following known genes: 5S rRNAs, 23S rRNAs, and protein-coding genes. Although rRNAs were identified in the previous steps (Fig. 2), a few sequences still were retained in the unclassified data set because the numbers and kinds of environmental 5S and 23S rRNA gene sequences were incomplete in the databases.
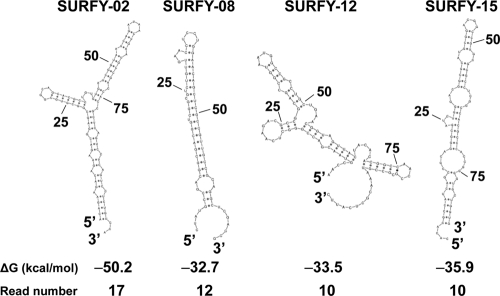
The abundances of the unclassified RNAs showed that 1,574 reads (79.8%) were detected only once, whereas 398 reads (20.2%) were found more than twice (maximum of 19 reads). The 15 unclassified RNAs that appeared 10 or more times then were further characterized as candidate sRNAs, because they were relatively highly abundant in the environment (see Table S2 in the supplemental material). These 15 sequences were designated small unique RNAs from Yunohama (SURFYs). Fourteen of the SURFYs had no sequence similarity to any known nucleotide sequence, whereas SURFY03 shared some degree of similarity (91% identity in 57 nt) to the gene encoding a prokaryotic elongation factor (EF-Tu) in the nitric bacterium Nitrosococcus halophilus. The secondary structures of these candidate sRNAs were predicted with Sfold, and the free energies of the SURFYs (mean, −20 kcal/mol) were calculated (see Table S2). The sequence lengths (average, 60 nt) and GC contents (average, 58%) of the SURFYs also were calculated and are shown in Table S2 in the supplemental material. Because the secondary structures of noncoding RNAs are known to be important for their functions (62), the secondary structures of the four SURFYs with the lowest free energies are shown (Fig. 3). Their high copy numbers and stable RNA structures suggest that these SURFYs are stable in the Yunohama hot spring, probably reflecting their roles in specific biological functions. It has been reported recently that unique sRNAs exist in some environmental samples (50, 65). Depth-dependent variations in putative sRNAs have been reported in a metatranscriptomic analysis of an oceanic water sample, and the authors suggested that these play roles in niche adaptation (50).
Fig 3.
Four examples of abundant candidate sRNAs. The nucleotide sequences, predicted secondary structures, and free energies are shown. The secondary structures and free energies (ΔG) were calculated with Sfold. Detailed information about all candidate RNAs is given in Table S2 in the supplemental material.
Environmental tRNAs and their fragments.
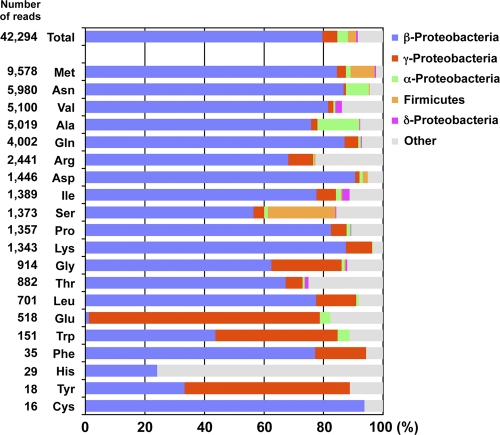
Of the sRNAs (redundant reads) isolated from the microbes in the Yunohama hot spring, 74.1% were either mature tRNA sequences or were their fragments (Table 1). In total, 8,069 tRNAs (43,092 redundant reads) were classified into 20 groups based on their corresponding amino acids. Neither selenocystenyl-tRNA nor pyrrolysyl-tRNA was detected in the sRNA library constructed from the Yunohama microbes. The microbial taxa corresponding to each tRNA group were predicted based on the genomic tRNA database (GtRNAdb), and most tRNAs were shown to derive from bacterial species (Fig. 4). This indicates that bacterial species are more abundant and active than Archaea in the Yunohama hot spring environment. Approximately 80% of the mature tRNAs belonged to the Betaproteobacteria (Fig. 4), which is consistent with the rRNA gene sequence analysis of the bacterial clade (Fig. 1B). However, four tRNA groups (Glu, Trp, His, and Tyr) corresponded to the different microbial taxa, such as Gammaproteobacteria or Alphaproteobacteria, but the reason for this remains unclear.
Fig 4.
Numbers of environmental tRNAs and their corresponding taxa identified in this study. In total, 42,294 redundant tRNA sequences were classified based on the amino acids transferred by them. Suppressor tRNAs and pseudo-tRNAs (tRNAscan-SE could not identify their anticodons) are not included. Colored bars indicate the percentage of each taxon identified with a BLAST search of the genomic tRNA database. The five most abundant taxa are Betaproteobacteria (blue), Gammaproteobacteria (red), Alphaproteobacteria (green), Firmicutes (orange), and Deltaproteobacteria (magenta); other minor taxa are colored gray.
We also found 2,063 tRNA fragments (4,484 redundant reads) with 100% nucleotide sequence identity to one (or more) of the sequenced full-length tRNAs, suggesting that these fragments were produced through either tRNA processing or the nonspecific degradation of the tRNAs. There was no correlation between the abundances of full-length tRNA reads and the abundances of their corresponding tRNA fragment reads (see Fig. S6 in the supplemental material), which indicates that these fragments are not the consequence of nonspecific degradation but are instead generated by specific exonucleolytic/endonucleolytic degradation pathways. To verify this hypothesis, the position of the tRNA cleavage site in each of the 20 tRNA groups was analyzed, and the three most frequently cleaved positions are represented in a piled bar graph (Fig. 5). Because most of the tRNA fragments were very short and parts of the tRNA sequence are common to all tRNAs, many tRNA fragments could be mapped to several independent tRNAs. Therefore, the number of truncated positions was normalized by dividing the number of observed tRNA fragments by the number of corresponding mature tRNAs. Previous studies of tRNA degradation usually are performed in E. coli and have demonstrated the specific degradation of six types of tRNAs: Lys-tRNA is truncated between nucleotides 33 and 34 (33/34) by PrrC; Tyr-tRNA, His-tRNA, Asn-tRNA, and Asp-tRNA are truncated at 34/35 by colicin E5; and Arg-tRNA is truncated at 38/39 by colicin D (36). The current study has shown the truncation positions of all types of tRNA. These truncation positions are located mainly in the anticodon loop and the D-loop, although tRNA cleavage also was observed at several other positions, such as in the D-stem and variable region. Figure 5 shows that the truncation positions of the Lys-tRNAs in the Yunohama hot spring are concentrated in the D-loop. Furthermore, the frequent truncation position at 15/16, commonly seen in tRNA-Ile, tRNA-Lys, tRNA-Asp, tRNA-Asn, and tRNA-Val in this sample, is a novel cleavage site for bacterial tRNA degradation. Nucleotide positions 15 and 16 are known to be exposed in the tertiary tRNA structure, so an as-yet-uncharacterized RNase may have access to this site for its specific cleavage. The most frequently cleaved sites for each tRNA anticodon are listed in Fig. S7 in the supplemental material. These results suggest that there is a nucleotide preference for site-specific degradation at the first base of the anticodon. Specifically, tRNAs that are truncated at 34/35 have a purine nucleotide (A or G) as the first base of their anticodons, whereas tRNAs that are cleaved at positions outside the anticodon region have a pyrimidine nucleotide (C or U) as the first base of their anticodons (see Fig. S7). These results suggest that microbes living in the Yunohama hot spring (possibly Betaproteobacteria) express specific ribonucleases that can digest specific tRNAs by distinguishing the tRNA types.
Fig 5.
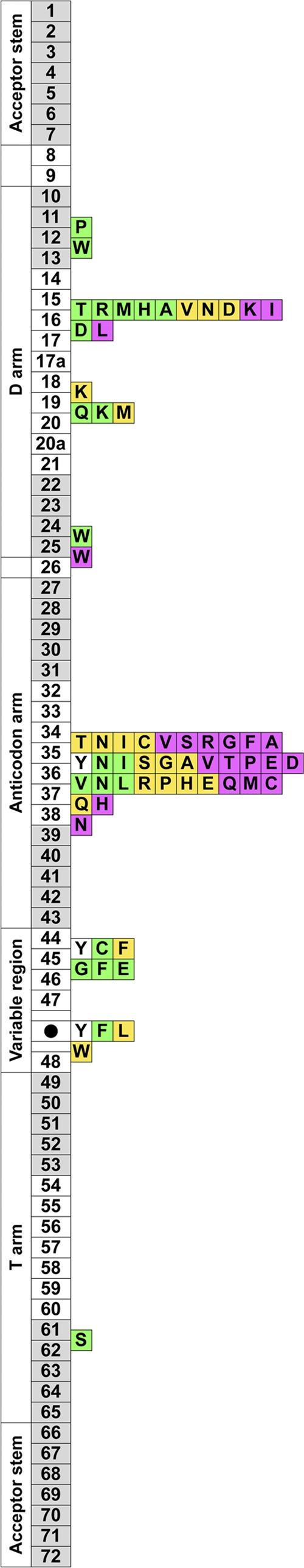
Summary of the fragmented tRNAs isolated from the environment. Numbered boxes (1 to 72) indicate the tRNA nucleotide positions based on a comprehensive report of tRNAs (35). The gray boxes indicate the stem regions, and the white boxes indicate the loop regions. The three most frequently cleaved positions are shown for each tRNA that transfers a specific amino acid (boxes containing letters): most frequent (magenta), second most frequent (yellow), and third most frequent (green). For example, a magenta box with “A” located between nt 34 and 35 means that tRNAs that transfer alanine were most frequently cleaved between nt 34 and 35 in the anticodon loop. In the case of asparagine tRNA, tryptophan tRNA, and phenylalanine tRNA, two positions were observed as the third most frequently cleaved sites, so two green boxes occur at these positions. The variable stem/loop region is compressed into one box, marked with a black circle, between nt 47 and 48, and the cleaved positions observed in the variable region are mapped together on the black circle. Because there were few fragments of tyrosine tRNA, the three positions observed twice (most frequently) in the truncated fragments are mapped as white boxes.
In eukaryotes, tRNA cleavage is known to be a conserved response to some stresses (56), and several studies have reported position-specific tRNA degradation in some types of human cells (11, 28) and in yeast (8, 57). However, the knowledge of bacterial tRNA degradation still is limited. In this study, more than 8,000 putative bacterial tRNAs and more than 2,000 fragments of them in the environment were analyzed, and the results suggest that this tRNA fragmentation was not the consequence of nonspecific degradation. Because certain sRNAs are reported to occur in specific environments (50), many of the fragmented tRNAs isolated in the current study may have been induced by environmental factors, like heat or metal ions. Although the present study may be one of the earliest steps toward defining the unique characteristics of sRNAs in environments, further analyses are required to reveal the possible role(s) of these sRNAs in microbes. To this end, it should be very useful to collect as many sRNA samples as possible from various microbial habitats.
Supplementary Material
ACKNOWLEDGMENTS
We thank Junichi Sugahara and Motomu Matsui (Institute for Advanced Biosciences, Keio University) for their insightful discussions.
This work was supported in part by Grant-in-Aid for Scientific Research (B) 22370066 from the Ministry of Education, Culture, Sports, Science and Technology of Japan, by a research fund at the Institute for Fermentation, and by research funds from the Yamagata Prefectural Government and Tsuruoka City, Japan.
Footnotes
Published ahead of print 9 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Aiba H. 2007. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 10:134–139 [DOI] [PubMed] [Google Scholar]
- 2. Argaman L, et al. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941–950 [DOI] [PubMed] [Google Scholar]
- 3. Arumugam M, et al. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baker BJ, et al. 2010. Enigmatic, ultrasmall, uncultivated Archaea. Proc. Natl. Acad. Sci. U. S. A. 107:8806–8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baker BJ, et al. 2006. Lineages of acidophilic archaea revealed by community genomic analysis. Science 314:1933–1935 [DOI] [PubMed] [Google Scholar]
- 6. Barns SM, Fundyga RE, Jeffries MW, Pace NR. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. U. S. A. 91:1609–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bomar L, Maltz M, Colston S, Graf J. 2011. Directed culturing of microorganisms using metatranscriptomics. mBio 2:e00012–00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buhler M, Spies N, Bartel DP, Moazed D. 2008. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat. Struct. Mol. Biol. 15:1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan PP, Lowe TM. 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37:D93–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen S, et al. 2002. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems 65:157–177 [DOI] [PubMed] [Google Scholar]
- 11. Cole C, et al. 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15:2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding Y, Chan CY, Lawrence CE. 2005. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA 11:1157–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ding Y, Lawrence CE. 2003. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 31:7280–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emara MM, et al. 2010. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 285:10959–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evguenieva-Hackenberg E. 2005. Bacterial ribosomal RNA in pieces. Mol. Microbiol. 57:318–325 [DOI] [PubMed] [Google Scholar]
- 17. Gama-Castro S, et al. 2008. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 36:D120–D124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gantner S, Andersson AF, Alonso-Saez L, Bertilsson S. 2011. Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods 84:12–18 [DOI] [PubMed] [Google Scholar]
- 19. Gardner PP, et al. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res. 37:D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gottesman S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 21:399–404 [DOI] [PubMed] [Google Scholar]
- 21. Grice EA, et al. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. 2008. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 36:732–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hall JR, et al. 2008. Molecular characterization of the diversity and distribution of a thermal spring microbial community by using rRNA and metabolic genes. Appl. Environ. Microbiol. 74:4910–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Inskeep WP, et al. 2010. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One 5:e9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jost D, Nowojewski A, Levine E. 2011. Small RNA biology is systems biology. BMB Rep. 44:11–21 [DOI] [PubMed] [Google Scholar]
- 26. Jurgens G, Saano A. 1999. Diversity of soil Archaea in boreal forest before, and after clear-cutting and prescribed burning. FEMS Microbiol. Ecol. 29:205–213 [Google Scholar]
- 27. Kato M, Chen X, Inukai S, Zhao H, Slack FJ. 2011. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA 17:1804–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawaji H, et al. 2008. Hidden layers of human small RNAs. BMC Genomics 9:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kawano M, Reynolds AA, Miranda-Rios J, Storz G. 2005. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 33:1040–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kvist T, Ahring BK, Westermann P. 2007. Archaeal diversity in Icelandic hot springs. FEMS Microbiol. Ecol. 59:71–80 [DOI] [PubMed] [Google Scholar]
- 31. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 32. Lee YS, Shibata Y, Malhotra A, Dutta A. 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23:2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128 [DOI] [PubMed] [Google Scholar]
- 34. Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marck C, Grosjean H. 2002. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8:1189–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Masaki H, Ogawa T. 2002. The modes of action of colicins E5 and D, and related cytotoxic tRNases. Biochimie 84:433–438 [DOI] [PubMed] [Google Scholar]
- 37. Massana R, Murray AE, Preston CM, DeLong EF. 1997. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63:50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Masse E, Escorcia FE, Gottesman S. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Milbury CA, Lee JC, Cannone JJ, Gaffney PM, Gutell RR. 2010. Fragmentation of the large subunit ribosomal RNA gene in oyster mitochondrial genomes. BMC Genomics 11:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miyoshi T, Iwatsuki T, Naganuma T. 2005. Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters. Appl. Environ. Microbiol. 71:1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nunoura T, et al. 2005. Genetic and functional properties of uncultivated thermophilic crenarchaeotes from a subsurface gold mine as revealed by analysis of genome fragments. Environ. Microbiol. 7:1967–1984 [DOI] [PubMed] [Google Scholar]
- 42. Nunoura T, et al. 2011. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 39:3204–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ogawa T, Inoue S, Yajima S, Hidaka M, Masaki H. 2006. Sequence-specific recognition of colicin E5, a tRNA-targeting ribonuclease. Nucleic Acids Res. 34:6065–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raghavan R, Groisman EA, Ochman H. 2011. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 21:1487–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Regalia M, Rosenblad MA, Samuelsson T. 2002. Prediction of signal recognition particle RNA genes. Nucleic Acids Res. 30:3368–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rivas E, Klein RJ, Jones TA, Eddy SR. 2001. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 11:1369–1373 [DOI] [PubMed] [Google Scholar]
- 48. Schlax PJ, Xavier KA, Gluick TC, Draper DE. 2001. Translational repression of the Escherichia coli alpha operon mRNA: importance of an mRNA conformational switch and a ternary entrapment complex. J. Biol. Chem. 276:38494–38501 [DOI] [PubMed] [Google Scholar]
- 49. Serganov A. 2009. The long and the short of riboswitches. Curr. Opin. Struct. Biol. 19:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shi Y, Tyson GW, DeLong EF. 2009. Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature 459:266–269 [DOI] [PubMed] [Google Scholar]
- 51. Shinhara A, et al. 2011. Deep sequencing reveals as-yet-undiscovered small RNAs in Escherichia coli. BMC Genomics 12:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 2002. 5S Ribosomal RNA Database. Nucleic Acids Res. 30:176–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Takai K, Horikoshi K. 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK. 2001. Archaeal diversity in waters from deep South African gold mines. Appl. Environ. Microbiol. 67:5750–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tang TH, et al. 2002. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 30:921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thompson DM, Lu C, Green PJ, Parker R. 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14:2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thompson DM, Parker R. 2009. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 185:43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tjaden B, et al. 2002. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 30:3732–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Venter JC, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74 [DOI] [PubMed] [Google Scholar]
- 60. Vogel J, et al. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31:6435–6443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Warnecke F, Hess M. 2009. A perspective: metatranscriptomics as a tool for the discovery of novel biocatalysts. J. Biotechnol. 142:91–95 [DOI] [PubMed] [Google Scholar]
- 62. Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF. 2005. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 23:1383–1390 [DOI] [PubMed] [Google Scholar]
- 63. Wassarman KM, Storz G. 2000. 6S RNA regulates E. coli RNA polymerase activity. Cell 101:613–623 [DOI] [PubMed] [Google Scholar]
- 64. Weinberg Z, Perreault J, Meyer MM, Breaker RR. 2009. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature 462:656–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Weinberg Z, et al. 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 11:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wilms R, et al. 2006. Specific bacterial, archaeal, and eukaryotic communities in tidal-flat sediments along a vertical profile of several meters. Appl. Environ. Microbiol. 72:2756–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Winkler W, Nahvi A, Breaker RR. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419:952–956 [DOI] [PubMed] [Google Scholar]
- 68. Xie W, et al. 2011. Comparative metagenomics of microbial communities inhabiting deep-sea hydrothermal vent chimneys with contrasting chemistries. ISME J. 5:414–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yachie N, Numata K, Saito R, Kanai A, Tomita M. 2006. Prediction of non-coding and antisense RNA genes in Escherichia coli with gapped Markov model. Gene 372:171–181 [DOI] [PubMed] [Google Scholar]
- 70. Zhang A, et al. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50:1111–1124 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.