Abstract
The ideal host-associated genetic fecal marker would be capable of predicting the presence of specific pathogens of concern. Flowthrough freshwater microcosms containing mixed feces and inocula of the pathogens Campylobacter jejuni, Salmonella enterica serovar Typhimurium, and adenovirus were placed at ambient temperature in the presence and absence of diurnal sunlight. The total Enterococcus DNA increased during the early periods (23 h) under sunlight exposure, even though cultivable Enterococcus and DNA in intact cells, as measured by propidium monoazide (PMA), decreased with first-order kinetics during the entire period. We found a significant difference in the decay of host-associated Bacteroidales cells between sunlight exposure and dark conditions (P value < 0.05), whereas the persistence of host-associated Bacteroidales DNA was comparable. The 2-log reduction times of adenovirus were 72 h for sunlight exposure and 99 h for dark conditions with similar decay rate constants (P value = 0.13). The persistences of fecal Bacteroidales cells and Campylobacter cells exposed to sunlight were similar, and host-associated Bacteroidales DNA and waterborne pathogen DNA were degraded at comparable rates (P values > 0.05). Overall, the ratio of quantitative PCR (qPCR) cycle threshold (CT) values with and without PMA treatment was indicative of the time elapsed since inoculation of the microcosm with (i) fecal material from different animal sources based on host-associated Bacteroidales and (ii) pure cultures of bacterial pathogens. The use of both PMA-qPCR and qPCR may yield more realistic information about recent sources of fecal contamination and result in improved prediction of waterborne pathogens and assessment of health risk.
INTRODUCTION
The threat to human health posed by fecal contamination of surface waters is usually estimated by measuring fecal indicator bacteria (FIB) such as total and fecal coliforms, Escherichia coli, and Enterococcus. However, traditional fecal indicators to monitor recreational water quality are often not associated with health risks in ambient water where nonpoint sources are dominant fecal contributors, and these results suggest a need for alternative indicators of water quality (47). Furthermore, FIB are inadequate to identify the source of fecal pollution because they are observed in both warm- and cold-blooded animal feces (30, 35). Microbial source tracking (MST) can discriminate between human and nonhuman fecal contamination such as cow, dog, and pig in water using microbiological or chemical traits of source identifiers (19, 22). Bacteroidales have been proposed as an alternative fecal indicator as well as source identifier because they are abundant in the gastrointestinal tract and genetic markers based on 16S rRNA have host-associated distributions (7, 13, 14, 17, 18, 25, 30, 32). The persistence of Bacteroidales was investigated by controlling variables such as the presence or absence of sunlight, temperature, and the use of filtered versus unfiltered water (3, 30, 36, 48, 50). According to these previous studies, the DNA of fecal Bacteroidales was detected by quantitative PCR (qPCR) for days or even weeks and the persistence of Bacteroidales genetic markers was influenced by incubation temperature, the effect of artificial sunlight, and the presence of indigenous microorganisms (2, 8, 26). However, little is known regarding the persistence of host-associated Bacteroidales cells and their genetic markers in freshwater. In two studies, Bacteroidales RNA was measured as an equivalent of whole cells using fluorescent in situ hybridization (FISH) (29, 42), but it is difficult to compare FISH results with qPCR results because the number of rRNA gene operons in fecal Bacteroidales is unknown. Furthermore, the relationship of Bacteroidales with FIB or waterborne pathogens needs to be established.
Previous studies have suggested PCR with propidium monoazide (PCR-PMA) to discriminate intact from dead cells as a means of reducing the PCR signal from DNA originating from dead bacterial cells (25). PMA in combination with PCR or qPCR has been successfully applied to identify intact pathogens in a simple matrix (21, 23) and in environmental samples (2, 5, 38, 42). Basically, PMA is a DNA-intercalating dye with an azide group and will penetrate only the membrane of dead cells with compromised cell walls/membrane and then bind their DNA or attach to free DNA. Upon exposure to bright visible light, the photoactive azide group on the dye is converted to a nitrene radical, which is readily attached to a carbon atom of the DNA through a C-H insertion reaction, resulting in an inhibition of amplification of DNA from dead cells or free DNA. Residual unbounded PMA is simultaneously transformed as hydroxylamine, which is no longer capable of covalently binding to DNA. Recently, an optimized PMA-qPCR method successfully discriminated between intact and dead fecally derived Bacteroidales cells (2).
Cultivation-dependent methods to measure FIB require 18- to 96-h incubation periods, during which human exposure to fecal pathogens can occur. A rapid method such as qPCR could improve the protection of public health by reducing the time between exposure measurement and management decisions, potentially providing same-day results (24). In addition, recent epidemiological studies suggested that rapid qPCR methods can predict swimming-associated gastrointestinal illness at freshwater beaches impacted by publicly owned treatment works (POTWs) (44, 45). On the other hand, there is currently no knowledge about the relationship between the DNA of fecal genetic markers and health risks at freshwater beaches that receive non-point source pollutants.
The objectives of this study were, therefore, (i) to evaluate the persistence of FIB measured by both cultivation and molecular methods; (ii) to determine the decay functions of host-associated Bacteroidales cells and their DNA as well as those of the waterborne pathogens Campylobacter jejuni, Salmonella, and adenovirus in freshwater using PMA-qPCR and qPCR; and (iii) to compare the decay profiles and slopes and determine the predictive relationship between the ratio of cycle threshold (CT) values for both qPCR methods and the time elapsed since fecal contamination occurred.
MATERIALS AND METHODS
Fecal sample collection.
Individual fecal samples were collected with a sterile utensil placed in a sterile 50-ml tube and kept on ice until used as inoculum. Fresh human feces was collected within 24 h from six healthy adults, and animal fecal samples were obtained on the day of inoculation from a cattle farm (8 samples; Winters, CA) and a dog park (12 samples; Davis, CA). Each fecal sample was immediately transported on ice to the laboratory. Before mixing fecal samples, each fecal sample was serially diluted and then analyzed by qPCR without DNA extraction (13) to confirm the specificity of each host-associated Bacteroidales assay for the corresponding host fecal samples.
Preparation of inocula.
Salmonella enterica serovar Typhimurium (ATCC 13311) and Campylobacter jejuni (ATCC 43431) were purchased from the American Type Culture Collection (ATCC) and used as spikes in the microcosm study and to generate plasmid quantification standards and optimize qPCR performance. Also, Enterococcus faecalis (ATCC 29212) was purchased from ATCC and grown in brain heart infusion broth for 24 h at 37°C for the preparation of plasmid standards and optimization of qPCR. Adenovirus (VR-680) from ATCC was used as inoculum without further propagation. Salmonella Typhimurium (ATCC 13311) was incubated overnight in LB broth at 37°C, and Campylobacter jejuni (ATCC 43431) was incubated in Trypticase soy broth for 36 h at 37°C under microaerophilic conditions in GasPak anaerobic jars (Becton Dickinson Microbiology Systems, Cockeysville, MD) using CampyPack hydrogen plus CO2 (BD, Franklin Lakes, NJ).
Cell densities of Salmonella Typhimurium and Campylobacter jejuni were measured with the Live/Dead BacLight bacterial viability kit (Molecular Probes Inc., Eugene, OR). The cells were stained by adding 1.5 μl each of the provided fluorescent stains SYTO 9 and propidium iodide. SYTO 9 stains all bacterial cells and fluoresces green, whereas propidium iodide penetrates only bacterial cells with compromised membranes and fluoresces red. Upon mixing and a 10-min incubation in the dark, a 10-μl sample was enumerated three times with a hemacytometer using an Axioskop2 Plus epifluorescence microscope (Zeiss, Thornwood, NY) equipped with two filter sets (fluorescein isothiocyante [FITC] and Texas Red).
Establishment of microcosms.
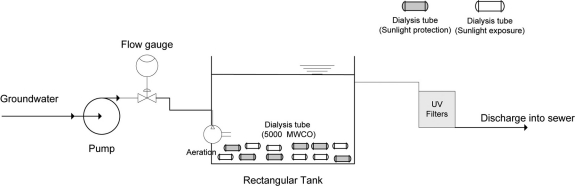
A ready-to-use Spectra/Por 7 membrane dialysis tubing (molecular weight cutoff [MWCO], 50,000; 34-mm flat width; Spectrum Laboratories, Inc., Rancho Dominguez, CA) was cut into segments of 12 cm in length. Four grams of each fresh feces was mixed with 2.5 liters groundwater obtained from the Aqua Toxicology Laboratory, UC Davis. In addition, 2 ml each of approximately 107 cells of Salmonella Typhimurium and Campylobacter jejuni cell suspensions and 1 ml of adenovirus were inoculated. Half of the dialysis tubes were placed inside a 6.4-cm-long polyvinyl chloride (PVC) pipe protected from sunlight. The remainder of the dialysis tubes were exposed to sunlight and immersed by attaching them to a glass ball without the PVC pipe to counteract the buoyancy. Triplicates of the dialysis tubes for both exposure and nonexposure conditions were randomly sampled every day for 3 days and every other day for 6 days.
The microcosm experiments were conducted outdoors in the Aqua Toxicology Laboratory at UC Davis. All microcosms were placed in a rectangular tank with dimensions (length, width, and depth) of 80 cm by 45 cm by 30 cm, providing ambient conditions such as diurnal cycle, temperature fluctuation, the presence of predators, and so forth (Fig. 1). Fresh groundwater was continuously pumped into the tank at a flow rate of 1 gallon per min (5.45 m3 day−1). Temperature, salinity, conductivity, pH, and dissolved oxygen concentration were measured using the probes YSI 63 and YSI 550A (YSI, Yellow Springs, OH), respectively. To confirm the presence or absence of any host-associated Bacteroidales prior to adding feces to the microcosm experiments, DNA was extracted from a 100-ml freshwater sample using the Ultraclean Water DNA isolation kit (Mo Bio Laboratories Inc., Carlsbad, CA).
Fig 1.
Schematic diagram of the experimental setup for microcosm studies. Permeable dialysis tubes were either placed inside PVC pipe for protection from sunlight or exposed to sunlight. Triplicate dialysis tubes from sunlight exposure and nonexposure conditions were randomly sampled for further analysis.
Enumeration of cultivable fecal indicators.
The cultivable fecal indicators E. coli and Enterococcus (cENT) were enumerated using membrane filter techniques (40, 41). Preliminary experiments were conducted to determine the background concentrations of E. coli and Enterococcus in a 100-ml freshwater sample in the Aqua Toxicology Laboratory. For the microcosm study, 10 ml of feces samples from dialysis tubes was diluted with a sterile buffer solution and filtered through a 47-mm-diameter sterile grid-marked membrane with an 0.45-μm pore size (Millipore Corp., Bedford, MA). The filters were subsequently placed on mEI agar and MI agar plates. Triplicate plates from three dialysis tubes were then incubated at 35°C for 24 h and at 41°C for 24 h for E. coli and Enterococcus, respectively.
PMA treatment and DNA extraction.
PMA (Biotium Inc., Hayward, CA) was dissolved in 20% dimethyl sulfoxide (DMSO) to create a stock concentration (1 mg ml−1) and stored at −20°C in the dark. Exposure time and PMA concentration were in accordance with previous optimization experiments for Bacteroidales spp. (2). PMA was added to samples at a final concentration of 100 μM. Following a 5-min incubation in the dark, samples were exposed to light for 10 min with a 600-W halogen light source at a distance of 20 cm from the light source. Microcentrifuge tubes were placed on ice prior to light exposure to avoid excessive heating.
A 400-μl sample was extracted using the FastDNA spin kit for soil (Biomedicals, Solon, OH). Cell lysis was achieved by bead beating using a bead mill Minibead beater (Biospec Products Inc., Bartlesville, OK) at 2,400 rpm for 20 s. Cell debris was removed by centrifugation for 5 min at 14,000 × g before adding 250 μl of PPS solution (provided with the kit). DNA was eluted twice with 80 μl DES buffer (provided in the kit). Otherwise, DNA extraction was performed according to the manufacturer's instructions. Also, viral DNA was extracted from dialysis tubes using a QIAamp MinElute Virus Spin kit (Qiagen, Valencia CA) as specified by the manufacturer.
Quantification standards for real-time PCR.
To create Campylobacter, Salmonella, and Enterococcus qPCR standards, genomic DNA was extracted from a pure culture using a DNeasy Blood & Tissue kit (Qiagen, Valencia CA) and quantified by a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE). Target sequences were amplified by PCR for each organism (see Table S1 in the supplemental material). The size of resultant amplicons was confirmed by gel electrophoresis prior to cloning with TOPO TA cloning vector (Invitrogen, Carlsbad, CA). Plasmids were extracted with a QIAprep Spin MiniPrep kit (Qiagen, Valencia, CA), and the concentration of each plasmid was measured by a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE). Optimal performance of qPCR was achieved by selecting the primer and probe concentrations with the lowest CT and highest ΔRn for a fixed amount of target templates. Assay limits of detection (ALOD) were determined as described previously (13, 27, 31) according to U.S. EPA recommendations for determining method detection limits (40 CFR 136, appendix B). The limit of detection (LOD) was calculated with the following equation: Here, V (ml) represents the volumes of sample that was extracted (Vex), of elute from nucleic acid extraction (Vel), and of nucleic acid template added to the PCR (VT). ALOD indicates assay limit of detection of each target DNA on the assumption of 100% extraction efficiency.
Real-time PCR amplification of Bacteroidales, Salmonella, Campylobacter, and adenovirus.
Amplification and detection were carried out in 96-well plates in an ABI Prism 7700 Sequence Detection system (Applied Biosystems, Foster City, CA). Each 25-μl PCR mixture contained 12.5 μl of qPCR MasterMix Plus (Eurogentec, San Diego, CA), 10 μl of DNA template, and optimal primer-probe concentrations (Table 1). For a negative control, DNA-free water was used instead of templates. The thermal protocol for host-associated Bacteroidales, Enterococcus, and waterborne pathogens consisted of 2 min at 50°C, followed by 10 min at 95°C and 40 cycles of 15 s at 95°C and 60 s at 60°C. Each qPCR experiment was conducted in duplicate on DNA template from three dialysis tubes. A serial dilution approach was employed for each duplicate, and then the CT values were plotted against log of the dilution to estimate the effects of potentially present PCR inhibitors.
Table 1.
Kinetic parameters of host-associated Bacteroidales, Enterococcus, Campylobacter, Salmonella, and adenovirus in freshwater microcosms at 22°Cd
| Condition | Target DNA | Cell |
DNA |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N0 (gc ml−1)a |
k1 (h−1) |
N1 (gc ml−1) |
k2 (h−1) |
t1 (h) | R2 | T99c (h) | N0 (gc ml−1) |
k1 (h−1) |
t1 (h) | R2 | T99 (h) | |||||
| Mean | SEb | Mean | SE | Mean | SE | |||||||||||
| Light | Universal Bacteroidales | 1.63 × 108 | 0.245 | 0.016 | 5.15 × 105 | 0.028 | 0.005 | 0.98 | 22 | 3.79 × 108 | 0.050 | 0.008 | 0.91 | 98 | ||
| Human Bacteroidales | 4.38 × 107 | 0.187 | 0.026 | 2.64 × 106 | 0.052 | 0.016 | 0.95 | 25 | 1.07 × 108 | 0.048 | 0.006 | 0.99 | 103 | |||
| Cow Bacteroidales | 3.47 × 106 | 0.234 | 0.021 | 6.40 × 103 | 0.012 | 0.006 | 0.98 | 20 | 9.84 × 106 | 0.030 | 0.010 | 0.89 | 151 | |||
| Dog Bacteroidales | 3.63 × 106 | 0.228 | 0.032 | 1.77 × 104 | 0.021 | 0.007 | 0.88 | 20 | 8.59 × 106 | 0.049 | 0.003 | 0.98 | 102 | |||
| Cultivable Enterococcus | 3.12 × 106 | 0.081 | 0.006 | 0.94 | 56 | |||||||||||
| Enterococcus | 3.21 × 106 | 0.038 | 0.006 | 0.84 | 118 | 2.65 × 107 | 0.090 | 0.002 | 23 | 0.89 | 98 | |||||
| Campylobacter jejuni | 3.19 × 104 | 0.174 | 0.021 | 3.19 × 103 | 0.045 | 0.01 | 0.81 | 34 | 2.20 × 105 | 0.055 | 0.013 | 46 | 0.90 | 123 | ||
| Salmonella Typhimurium | 4.42 × 105 | 0.052 | 0.01 | 23 | 0.84 | 122 | 4.13 × 106 | 0.047 | 0.002 | 23 | 0.94 | 129 | ||||
| Adenovirus | 5.43 × 104 | 0.056 | 0.010 | 0.76 | 72 | |||||||||||
| Dark | Universal Bacteroidales | 1.54 × 108 | 0.082 | 0.016 | 1.00 × 107 | 0.031 | 0.01 | 0.99 | 57 | 4.01 × 108 | 0.042 | 0.006 | 0.89 | 101 | ||
| Human Bacteroidales | 4.62 × 107 | 0.071 | 0.011 | 2.53 × 105 | 0.025 | 0.017 | 0.95 | 62 | 1.08 × 108 | 0.059 | 0.005 | 0.98 | 95 | |||
| Cow Bacteroidales | 3.40 × 106 | 0.077 | 0.008 | 3.85 × 104 | 0.012 | 0.006 | 0.95 | 59 | 1.09 × 107 | 0.045 | 0.003 | 0.87 | 125 | |||
| Dog Bacteroidales | 3.60 × 106 | 0.098 | 0.011 | 1.77 × 104 | 0.021 | 0.007 | 0.95 | 47 | 9.01 × 106 | 0.050 | 0.003 | 0.97 | 91 | |||
| Cultivable Enterococcus | 7.28 × 106 | 0.087 | 0.006 | 23 | 0.96 | 99 | ||||||||||
| Enterococcus | 2.65 × 107 | 0.105 | 0.001 | 23 | 0.99 | 90 | 5.89 × 107 | 0.093 | 0.008 | 23 | 0.82 | 95 | ||||
| Campylobacter jejuni | 3.65 × 104 | 0.041 | 0.003 | 0.97 | 121 | 2.20 × 105 | 0.064 | 0.017 | 46 | 0.88 | 118 | |||||
| Salmonella Typhimurium | 3.39 × 106 | 0.041 | 0.006 | 23 | 0.96 | 144 | 6.37 × 106 | 0.043 | 0.007 | 23 | 0.97 | 154 | ||||
| Adenovirus | 5.43 × 104 | 0.049 | 0.007 | 0.84 | 99 | |||||||||||
gc ml−1 = number of gene copies per milliliter.
SE, standard error.
Time for 2-log reduction.
The models are as follows: , where N is number of gene copies ml−1 of target DNA at time t, N0 and N1 are the initial concentration of target DNA, and k1 and k2 are the decay rate constants; , where N is number of gene copies or bacteria ml−1 at time t, N0 is the initial concentration, and k1 is the decay rate constant; and , where t1 is time at the end of lag period, t is time at any point after a lag period, and other parameters are as given in the text.
Decay rate calculations and statistical analysis.
We employed two different exponential decay models proposed by previous studies to calculate decay rates as well as T99 (time for 2-log reduction) (3). First, the decay rates were calculated by a simple first-order decay model where N is the number of gene copies or number of bacteria ml−1 at time t, N0 is the initial concentration, and k1 is the decay rate constant. A modification of this model has a lag period before the beginning of logarithmic decay where the population increases or persists to some extent. The logarithmic decay in this situation can be modeled by where t1 is time at end of lag period, t is time at any point after lag period t1, and the other terms are as previously noted. Also, a biphasic model was used to fit the experimental data of host-associated Bacteroidales cells to characterize two subpopulations with different decay rates: where k1 and k2 are the exponential decay rate for N0 and N1, respectively.
Nonlinear regression analysis and fitting of persistence data were performed using Origin Pro 8. A P value was determined by computing the F-test provided in Origin Pro 8 to statistically analyze whether two decay models were significantly different from each other. Furthermore, we compared the slopes (decay rates) by testing the null hypothesis that the slopes were all identical using GraphPad Prism version 5.
Calculation of the ratio of CT values with and without PMA.
The ratio was calculated as described by Bae and Wuertz (2) based on the equation
where CT(PMA) = CT number in PMA treatment, CT(cycle) = total number of cycles of PCR amplification (e.g., 40 cycles), and CT(without PMA) = CT number without PMA treatment. The ratio indicates the age of fecal contamination by measurement of threshold values obtained from qPCR runs.
RESULTS
A microcosm study was conducted in the outdoor Aqua Toxicology Laboratory at UC Davis. The average pH, temperature, and dissolved oxygen values throughout the study were 7.3 mg liter−1, 22.4°C, and 8.7 mg liter−1, respectively. Background concentrations of cultivable E. coli and Enterococcus (cENT) were not detected. In contrast, the universal fecal Bacteroidales gene marker (BacUni-UCD) was detected at concentrations of 3.4 × 102 gene copies (gc)/ml in freshwater prior to inoculation, whereas human-, cow-, and dog-associated Bacteroidales were not found.
Survival of fecal indicator bacteria in freshwater.
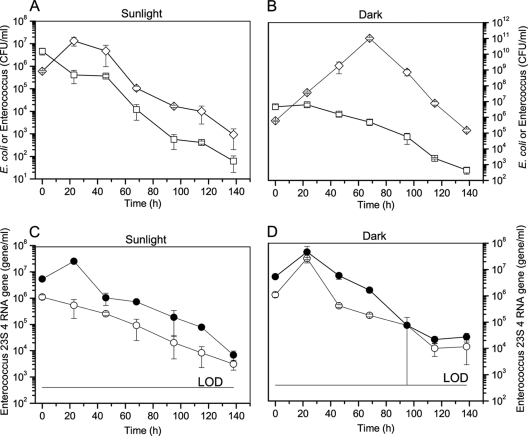
Fecal indicator bacteria such as E. coli and Enterococcus were enumerated by cultivation methods and, for Enterococcus alone, using quantitative PCR methods to determine the persistence of both cells and DNA in freshwater. Regrowth of cultivable E. coli was observed under both sunlight exposure and dark conditions, and all numbers declined after 45 h for diurnal sunlight exposure and after 70 h for dark conditions, respectively (Fig. 2A and B). Cultivable Enterococcus (cENT) experienced a lag period (23 h) and subsequently followed a log-linear inactivation phase under dark conditions (Fig. 2B). Interestingly, the number of Enterococcus 23S rRNA gene copies from total Enterococcus DNA (tENT) exposed to sunlight increased initially even though Enterococcus genes from intact cells (vENT) followed first-order decay as measured by PMA-qPCR during the entire period (Fig. 2C). In contrast, the number of 23S rRNA gene copies increased initially in the dark treatment regardless of whether qPCR or PMA-qPCR was used to amplify DNA (Fig. 2D), indicating that cells were dividing during the first 20 h. Afterwards, tENT declined, with vENT accounting for most of the detected gene marker. Enterococcus gene marker from both intact cells (vENT) and total DNA (tENT) decreased by 4 to 5 logs in both diurnal sunlight and dark conditions but did not approach the detection limit.
Fig 2.
Decay curves for cultivable E. coli and Enterococcus and the 23S rRNA gene of Enterococcus measured by PMA-qPCR and qPCR in freshwater. n = 3; error bar represents standard deviation. (A) Cultivable E. coli or Enterococcus (cENT) under natural sunlight exposure. (B) Cultivable E. coli or Enterococcus (cENT) under dark conditions. White squares and diamonds denote Enterococcus and E. coli, respectively. (C) The number of gene copies of Enterococcus 23S rRNA measured by qPCR under sunlight exposure. (D) The number of gene copies of Enterococcus 23S rRNA measured by qPCR under dark conditions. White circles represent Enterococcus cells (vENT) measured by PMA-qPCR, whereas black circles represent total DNA (tENT) including intact and dead cells and extracellular DNA.
The decay rates calculated by the fitting curve except for the vENT exposed to sunlight, are shown in Table 1. At 107 h and 159 h for diurnal sunlight and dark conditions, respectively, the T99 values of cultivable E. coli suggest that E. coli was more persistent under dark conditions. Similarly, T99 values of cENT were 56 h and 99 h for diurnal sunlight and dark conditions, respectively. However, the T99 of tENT exposed to sunlight was similar to that of vENT exposed to dark conditions: for example, 118 h (vENT) and 98 h (tENT) for sunlight exposure and 90 h (vENT) and 95 h (tENT) h for dark conditions, respectively. We compared the curve fitting parameters of nonlinear regression models for Enterococcus based on either cultivation or quantitative PCR methods. The regression models of tENT for sunlight exposure (Fig. 2C) and dark condition (Fig. 2D) resulted in similar decay rates (P value = 0.73), revealing only trivial effects of sunlight on DNA degradation. The slopes of cENT for sunlight and dark conditions indicated identical decay rates (P value = 0.55). Additionally, the F-test proved that the slopes of tENT and cENT were not significantly different within either condition (P value = 0.25 for sunlight and P value = 0.49 for dark condition, respectively). Note that different decay models for cENT, vENT, and tENT were applied for sunlight exposure and dark conditions because of regrowth of Enterococcus cells in the dark.
Persistence of Bacteroidales in freshwater.
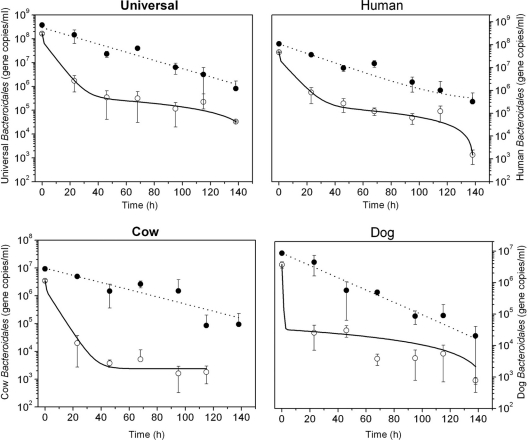
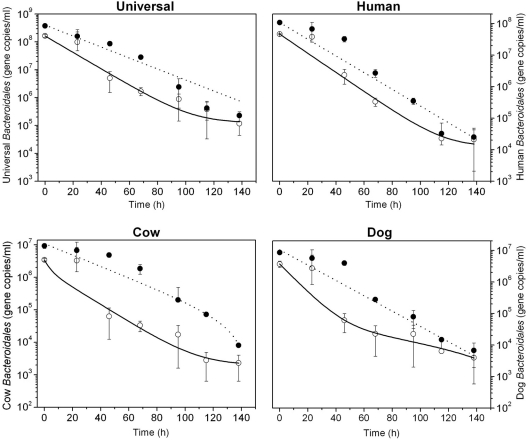
The host-associated Bacteroidales cells and their DNA in the permeable dialysis tubes containing a mixture of fecal samples from humans, cows, and dogs were measured throughout the experiment, except for the cow-associated Bacteroidales cells, which declined more rapidly. The Bacteroidales 16S rRNA gene concentrations from intact cells at the beginning of the experiment were 1.64 × 108, 4.65 × 107, 3.48 × 106, and 3.69 × 106 gene copies (gc) ml−1 for universal and human-, cow-, and dog-associated Bacteroidales, respectively. The concentrations of the host-associated Bacteroidales cells decreased by 3 orders of magnitude under both diurnal sunlight and dark conditions. Cow-associated Bacteroidales cells exposed to sunlight were no longer present at the end of the experiment whereas human- and dog-specific cells were still detectable. The decay of Bacteroidales DNA followed first-order kinetics in both diurnal exposure and dark conditions. The final concentrations in gc ml−1 at the end of the experiment under diurnal sunlight exposure and dark conditions, respectively, were 8.13 × 105 and 2.26 × 105 for universal Bacteroidales DNA; 3.21 × 105 and 2.50 × 104 for human-associated Bacteroidales; 9.40 × 104 and 8.11 × 103 for cow-associated Bacteroidales; and 2.03 × 104 and 6.72 × 103 for dog-associated Bacteroidales.
An exponential model, was used to fit the decay functions of host-associated Bacteroidales DNA markers in both diurnal sunlight exposure and dark conditions, but the biphasic model was the best-fitting curve for intact cells of Bacteroidales, providing higher R2 values (Table 1). Survival times of Bacteroidales cells were significantly shorter compared to their DNA under both diurnal sunlight and dark conditions (P value < 0.05). However, the persistence of Bacteroidales DNA exposed to diurnal sunlight (Fig. 3) and that of host-associated Bacteroidales DNA under dark conditions (Fig. 4) were identical (P value > 0.05). Accordingly, the T99 values of host-associated Bacteroidales DNA were similar under diurnal sunlight and dark conditions; for example, the T99 values of total Bacteroidales DNA were 98 h (universal), 103 h (human), 151 h (cow), and 102 h (dog) for diurnal sunlight exposure and the T99 values under dark conditions were 101 h (universal), 95 h (human), 125 h (cow), and 91 h (dog), respectively. The slopes of decay function of host-associated Bacteroidales DNA in the two conditions were similar, resulting in no statistical differences in the decay rates of each host-associated Bacteroidales marker (average P value > 0.50). In contrast, T99 values of host-associated Bacteroidales cells were shorter under both diurnal sunlight and dark conditions. The T99 values for diurnal sunlight exposure were 22 h (universal), 25 h (human), 20 h (cow), and 20 h (dog). As expected, T99 values under dark conditions were longer than those under diurnal sunlight exposure at 57 h (universal), 62 h (human), 59 h (cow), and 47 h (dog). The F-test showed that the two data sets of Bacteroidales cells between diurnal sunlight and dark condition were statistically different (P values < 0.05).
Fig 3.
The survival of host-associated Bacteroidales cells measured by PMA-PCR and the persistence of DNA including intact/dead cells and extracellular DNA under natural sunlight exposure in freshwater. Open and closed circles represent intact cells and total DNA, respectively. The dotted line of DNA and the dashed line were plotted using a simple first-order decay model and a biphasic model, respectively, as shown in Table 1.
Fig 4.
Survival of host-associated Bacteroidales cells measured by PMA-PCR and the persistence of DNA including intact/dead cells and extracellular DNA under dark conditions in freshwater. Black and white circles represent the DNA and intact cells, respectively. The dotted line of DNA and the dashed line were plotted by the simple first-order decay model and a biphasic model, respectively, as shown in Table 1.
Survival of Salmonella, Campylobacter, and adenovirus in freshwater.
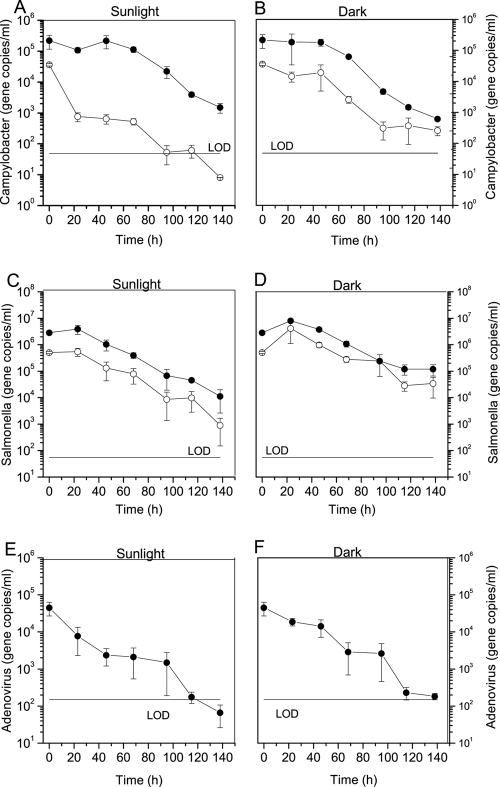
The persistence of waterborne pathogens mixed with feces was examined by PMA-qPCR and qPCR to determine the decay profiles in freshwater. Intact Campylobacter jejuni and Salmonella Typhimurium cells and their total DNA, including dead cells and extracellular DNA, and adenovirus DNA were detected under dark and natural sunlight conditions throughout the experimental period. Campylobacter cells exposed to sunlight decreased exponentially with a rate constant k of 0.127 h−1 whereas the decay rate in the dark condition was lower with k = 0.042 h−1. However, Campylobacter DNA under both sunlight and dark conditions had similar decay rate constants after a lag period in the early phase (Fig. 5A and B). The persistences of intact Salmonella Typhimurium cells and their DNA were similar under sunlight conditions (Fig. 5C). Likewise, the slopes for decay of Salmonella cells and total DNA were not statistically different under dark conditions (P value = 0.55). For adenovirus, the initial concentration under both sunlight and dark conditions was 4.64 × 104 genome copies/ml, which decreased by 3 orders of magnitude, yet only adenovirus exposed to sunlight was below the limit of detection at the end of the experiment (Fig. 5E and F).
Fig 5.
Survival of waterborne pathogens in freshwater measured by PMA-qPCR or qPCR in freshwater. n = 3 for each mean; error bar represents standard deviation. (A to D) The concentration of Campylobacter jejuni under sunlight exposure (A) and dark conditions (B) and Salmonella Typhimurium under sunlight exposure (C) and dark conditions (D). Open and closed circles denote target concentration (gene copies/ml) measured by PMA-qPCR and qPCR, respectively. (E and F) The decay of adenovirus DNA under sunlight exposure (E) and dark conditions (F). LOD, limit of detection.
The T99 values were calculated based on the biphasic model for intact Campylobacter cells, the exponential model including a lag period for dead/intact Campylobacter and extracellular DNA and Salmonella, and the single exponential model for adenovirus (Table 1). The T99 values of intact Campylobacter cells were significantly different for sunlight and dark conditions at 34 h and 121 h, respectively (Fig. 5A and B, P value < 0.05). Campylobacter DNA decayed with T99 values of 123 h and 118 h for sunlight and dark conditions, respectively. The T99 values of Salmonella were 122 h (cells) and 129 h (DNA) for sunlight exposure and 144 h (cells) and 154 h (DNA) for dark conditions. Similarly, the T99 values of adenovirus were calculated as 72 h for sunlight exposure and 99 h for dark conditions. There was no significant difference between sunlight and dark conditions for Campylobacter DNA, Salmonella DNA, and adenovirus DNA (P value > 0.05). Interestingly, the slopes of DNA decay functions for all pathogens in both conditions were the same (P value > 0.05), and furthermore, there was no significant difference in decay rate constants for Salmonella cells and adenovirus genomes for the two conditions (P value = 0.65 for sunlight exposure and P value = 0.64 for dark condition).
CT ratio for qPCR with and without PMA treatment for host-associated Bacteroidales, Enterococcus, and bacterial pathogens.
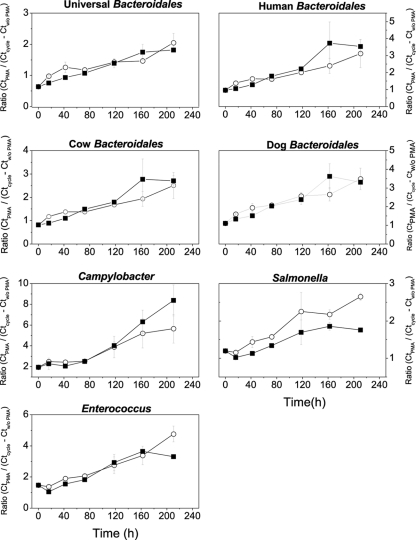
We employed a simple equation to estimate the age of contamination from different fecal contributions based on the ratio of cycle threshold (CT) values obtained from qPCR with and without PMA treatment (3). CT ratios were also determined for Enterococcus and bacterial pathogens. The initial mean ratios for host-associated Bacteroidales, Campylobacter, Salmonella, and Enterococcus at the beginning of the decay experiment were 0.88, 1.85, 1.19, and 1.47, respectively. Over time, the ratios increased linearly and at the end of the microcosm experiment ranged from 3.66 to 10 (Fig. 6). Interestingly, the ratios of Salmonella and Enterococcus decreased initially during the first 24 h due to an increase in gene copy number resulting from lower CT values without PMA treatment. Overall, the ratio of CT values with and without PMA treatment was indicative of the time period passed since inoculation of the microcosm with (i) fecal material from different animal sources based on host-associated Bacteroidales and (ii) pure cultures of bacterial pathogens.
Fig 6.
Ratio of CT values with and without PMA treatment for host-associated Bacteroidales, Campylobacter, Salmonella, and Enterococcus calculated by an equation shown in Materials and Methods. White circles and black squares represent natural sunlight exposure and dark conditions, respectively.
DISCUSSION
This study examined the decay of noncultivable fecal Bacteroidales and of Enterococcus, Campylobacter jejuni, and Salmonella Typhimurium, in freshwater using PMA-qPCR and qPCR, allowing for comparison of intact cells and total target DNA in the same matrix. Adenovirus genomes were measured by qPCR alone. In addition, intact Enterococcus and E. coli cells were enumerated by the membrane filtration method. We employed in situ microcosms in a continuously flowing system under ambient conditions to better simulate environmental conditions and compare the persistence patterns of host-associated Bacteroidales cells and their DNA with those of fecal indicator bacteria and waterborne pathogens.
In general, cultivation-dependent methods to evaluate microbial water qualities require an 18- to 96-h incubation period, during which time the public may continue to be exposed to microbially contaminated water. Rapid detection technologies such as qPCR have been proposed as an alternative tool to monitor water quality (24). Significant concentrations of dissolved DNA are found in marine water, freshwater, and sediments, potentially complicating the interpretation of information from qPCR data due to the relatively long persistence of target DNA (23). The PMA-qPCR method could avoid the issue of persistent DNA and provide a relatively fast, simple, and inexpensive means to discriminate intact/dead cells in mixed populations (2, 22, 42). However, some limitations of PMA-qPCR have been reported, such as the interference of higher particle concentration (2) and highly complex chemical and biological matrices in environmental samples.
Decay of fecal indicator bacteria.
It has been demonstrated that fecal indicators grow in tropical climates under the right conditions (9) and have greater persistence in both sediment and the water column (19). Similar to a previous study (1, 7), regrowth of E. coli but not of cultivable Enterococcus (cENT) was observed, demonstrating the presence of a growing subpopulation. Interestingly, in our study total Enterococcus DNA detected by qPCR (tENT) increased initially (23 h) under sunlight exposure, even though intact cells measured by PMA-qPCR (vENT) decreased with first-order rate kinetics during the entire experiment. It is possible that Enterococcus cells replicated in the microcosm, albeit neither cultivation method nor PMA-qPCR showed any increase in cells. Another possible explanation is the release of extracellular DNA from particulate material during this initial period, which then becomes available for qPCR later. The increase in DNA amplified by both PMA-qPCR and qPCR under dark conditions was not surprising because the availability of dissolved organic carbon can trigger the growth of Enterococcus, which subsequently ceases when the nutrients are depleted, as reported for the growth of Enterococcus in beach sands (51) and of E. coli O157 in natural freshwater at low carbon concentrations (43). The slopes from the cultivation-dependent method and PMA-qPCR were significantly different (P values = 0.001 and 0.02 for sunlight exposure and dark condition, respectively), which was likely due to the presence of intact but noncultivable Enterococcus cells. The higher numbers of tENT compared to cENT in freshwater, estuarine water, and beach sand have previously been reported and attributed to greater persistence of noncultivable organisms and/or free DNA (10, 11, 46).
Decay of Bacteroidales.
Genetic marker DNA targets of Bacteroidales had similar decay rate constants in the freshwater microcosm. In the same vein, Bacteroidales cells possessed similar slopes of exponential decay in each condition (average P value = 0.50 and 0.60 for sunlight exposure and dark condition, respectively). In an earlier study different host-associated genetic markers of Bacteroidales showed similar responses in nonfiltered river water incubated under nonflowing conditions (30). However, our findings contrast with those from a recent study, which reported that the persistence of ruminant Bacteroidales DNA differed from that of human marker DNA (48). This disagreement may be due to a different experimental design such as different qPCR assays (TaqMan versus SYBR green) or type of microcosm (a continuously flowing system versus a batch system). A comparison study between the double-stranded (dsDNA)-binding dye SYBR green I and a specific TaqMan probe proved that the sensitivity of the SYBR green I assay was less than that of the assay using a fluorogenic TaqMan probe due to the accumulation of primer dimers and nonspecific side products bound by the SYBR green dye (12). A batch system could have influenced DNA persistence over the course of the previous study (48) by possibly accumulating nonspecific DNA background and other waste products.
We conclude that the effect of sunlight on decay rates of Bacteroidales DNA was insignificant based on the current study and other microcosm studies (8, 43). However, sunlight significantly affected decay rates of Bacteroidales cells, with a good fit of a complex decay model. These phenomena could be explained by multiple factors (dissolved oxygen, sunlight, and predations) influencing Bacteroidales decay; the subpopulation of Bacteroidales cells could have different decay rates, explaining the biphasic decay pattern observed in this study. It is certain that a simple exponential decay model is inadequate for Bacteroidales cells in surface water, and the single-parameter decay model is simplified to use only the data of a log-linear portion of the curve. In contrast, the biphasic model describes not only the decay rates of Bacteroidales cells but also the persistence of a subpopulation under given conditions.
Decay of intact cells of bacterial pathogens.
Campylobacter jejuni, Salmonella spp., and adenovirus are frequently isolated from ambient waters, which serve as a reservoir and may aid transmission between hosts. We employed PMA-qPCR methods to discriminate intact cells and dead cells or free DNA of C. jejuni and S. Typhimurium. Even though PMA concentration and exposure time used for these experiments were not optimized for the bacterial pathogens, diverse bacterial species have been tested with PMA-qPCR in earlier studies and the signal from dead cells and free DNA was successfully removed (2, 5, 21, 23, 42). The slightly higher Salmonella cell inactivation rates in sunlight exposure compared to those in dark conditions could be explained by the lag period or increase in number of gene copies, which can cause a steep inactivation phase after the lag periods (33). Our results suggest that sunlight inactivation of Campylobacter jejuni was a significant factor. Similarly, the inactivation of host-associated Bacteroidales cells was consistently faster under sunlight exposure than under dark conditions. In agreement with a previous study, the marked difference in inactivation rates between sunlight exposure and dark conditions suggested that rapid inactivation in natural water is due to high susceptibility of Campylobacter to photo-oxidative damage (38). Its sensitivity to short solar radiation (UV-B and UV-C) is likely to be associated with direct DNA (photobiological) damage. However, UV-C wavelengths are removed by the atmosphere and UV-B comprises only a small proportion of solar energy at the earth's surface. UV-B wavelengths are also more rapidly attenuated in natural water than longer and visible wavelengths with the result that photo-oxidative damage becomes more important as a bacterial inactivation mechanism to lead to breakdown of membrane integrity (33, 34) and subsequently allows for the discrimination of intact cells by PMA-qPCR.
Our findings lead to the question as to whether Enterococcus or host-associated Bacteroidales measured by either PMA-qPCR or qPCR displayed a similar persistence to pathogenic bacteria or adenovirus in water. The decay rates of Enterococcus and Salmonella cells during sunlight exposure were similar (P value = 0.307), but Enterococcus cells were more persistent than those of Campylobacter jejuni, indicating that the physiological condition of Campylobacter jejuni at different dissolved oxygen concentrations needs to be considered to understand environmental decay in water. The persistences of fecal Bacteroidales cells (universal Bacteroidales marker, BacUni-UCD) and Campylobacter cells in sunlight exposure were similar as shown by the F-test. Host-associated Bacteroidales DNA and waterborne pathogen DNA were degraded at similar rates from 0.03 to 0.064 h−1 under both dark and sunlight conditions, but decay rates of Enterococcus DNA were significantly different from those of DNA from other pathogens due to regrowth of cells.
Walters and Field further reported that ruminant-associated Bacteroidales cells survived longer than human-associated cells based on rRNA detection used to assess viability of persistent Bacteroidales cells in freshwater microcosms (48). In contrast, ruminant-associated Bacteroidales cells in our study decayed faster than or at a similar rate to human-associated Bacteroidales cells in the presence of sunlight as monitored by PMA-qPCR (Fig. 3). The reason for observed differences in the two studies may be attributed to the stability of rRNA in bacteria after cell death and interference of RNA extraction in cow feces (5). Another recent microcosm study reported higher T99 values of the human-associated DNA marker BacHum (measured as total DNA without PMA) compared to cultivable E. coli (11), whereas BacHum DNA in the present study was more persistent than were cultivable Enterococcus or E. coli cells. Therefore, the question beckons how to interpret and compare data from these different experimental setups to understand decay rates of fecal pollution indicators and source identifiers in impaired water. As Schulz and Childers proposed, a repository database of raw survival and persistence data is needed for a statistical comparison of decay rates from different studies (35). Additionally, comparable kinetic studies are necessary considering inoculum effect, temperature, salinity, quantification methods, and viability of target organism.
The application of qPCR assays with PMA treatment in the same matrix holds promise as an effective tool for identifying sources of fecal pollution and quantifying waterborne pathogens with assessment of health risk. qPCR with PMA should provide greater confidence in fecal pollution assessment and may inform remediation decisions in water. Furthermore, the ratios of CT values in samples with and without PMA treatment are a promising method to estimate recent fecal contamination because ratios of intact cells by PMA-qPCR over dead cells by qPCR reflect different decay rates. For example, a ratio close to unity indicates that fecal contamination occurred recently based on the reasonable assumption that bacterial DNA is more persistent compared with the relatively faster decay of cells in the natural environment. In addition, inhibition of PMA activity after concentration of biomass by filtration and concurrent higher levels of total suspended solids should be considered in field monitoring (42). Even though very little co-occurrence of fecal Bacteroidales detection was observed for E. coli O157, Salmonella, and Campylobacter among samples in a previous study using only (q)PCR (49), PMA-qPCR analysis for both Bacteroidales and waterborne pathogens could be informative to identify the sources of waterborne pathogens due to similar decay rates in the environment.
Several remaining questions have to be investigated. The potential contribution of sediments as secondary habitats of Bacteroidales or waterborne pathogens needs to be established considering that other enteric bacteria and waterborne pathogens can persist by colonizing surfaces and forming biofilms (6, 9, 46). The decay rates should be integrated into models of the fate of enteric bacteria in water to implement a remediation strategy, and long-term and large-scale field studies need to be conducted with the decay model to validate the potential of microbial source tracking. Finally, epidemiological studies are required to determine the correlation between source identifiers, the presence of fecal pathogens, and the occurrence of illness associated with a particular water use (32).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Environmental Division of the California Department of Transportation, contract no. 43A0168, TO 23.
We thank Paul Lutes at the Center for Aquatic Biology and Aquaculture, UC Davis, for providing space and assisting in microcosm studies.
Footnotes
Published ahead of print 2 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alm EW, Burke J, Hagan E. 2006. Persistence and potential growth of the fecal indicator bacteria, Escherichia coli, in shoreline sand at Lake Huron. J. Great Lakes Res. 32:401–405 [Google Scholar]
- 2. Bae S, Wuertz S. 2009. Discrimination of viable and dead fecal Bacteroidales bacteria by quantitative PCR with propidium monoazide. Appl. Environ. Microbiol. 75:2940–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bae S, Wuertz S. 2009. Rapid decay of host-specific fecal Bacteroidales cells in seawater as measured by quantitative PCR with propidium monoazide. Water Res. 43:4850–4859 [DOI] [PubMed] [Google Scholar]
- 4. Bell A, et al. 2009. Factors influencing the persistence of fecal Bacteroides in stream water. J. Environ. Qual. 38:1224–1232 [DOI] [PubMed] [Google Scholar]
- 5. Birch L, Dawson CE, Cornett JH, Keer JT. 2001. A comparison of nucleic acid amplification techniques for the assessment of bacterial viability. Lett. Appl. Microbiol. 33:296–301 [DOI] [PubMed] [Google Scholar]
- 6. Brescia CC, et al. 2009. Cryptosporidium propidium monoazide-PCR, a molecular biology-based technique for genotyping of viable Cryptosporidium oocysts. Appl. Environ. Microbiol. 75:6856–6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bucci V, Vulic M, Ruan XD, Hellweger FL. 2011. Population dynamics of Escherichia coli in surface water. J. Am. Water Resour. Assoc. 47:611–619 [Google Scholar]
- 8. Buswell CM, et al. 1998. Extended survival and persistence of Campylobacter spp. in water and aquatic biofilms and their detection by immunofluorescent-antibody and -rRNA staining. Appl. Environ. Microbiol. 64:733–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desmarais TR, Solo-Gabriele HM, Palmer CJ. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dick LK, et al. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dick LK, Stelzer EA, Bertke EE, Fong DL, Stoeckel DM. 2010. Relative decay of Bacteroidales microbial source tracking markers and cultivated Escherichia coli in freshwater microcosms. Appl. Environ. Microbiol. 76:3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Field KG, Samadpour M. 2007. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res. 41:3517–3538 [DOI] [PubMed] [Google Scholar]
- 13. Fries JS, Characklis GW, Noble RT. 2008. Sediment-water exchange of Vibrio sp. and fecal indicator bacteria: implications for persistence and transport in the Neuse River Estuary, North Carolina, U.S.A. Water Res. 42:941–950 [DOI] [PubMed] [Google Scholar]
- 14. Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559–568 [DOI] [PubMed] [Google Scholar]
- 15. He JW, Jiang S. 2005. Quantification of enterococci and human adenoviruses in environmental samples by real-time PCR. Appl. Environ. Microbiol. 71:2250–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hein I, et al. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67:3122–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kildare BJ, et al. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 41:3701–3715 [DOI] [PubMed] [Google Scholar]
- 18. Layton A, et al. 2006. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 72:4214–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee CM, et al. 2006. Persistence of fecal indicator bacteria in Santa Monica Bay beach sediments. Water Res. 40:2593–2602 [DOI] [PubMed] [Google Scholar]
- 20. Lee YJ, et al. 2008. Temporal assessment of the impact of exposure to cow feces in two watersheds by multiple host-specific PCR assays. Appl. Environ. Microbiol. 74:6839–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malorny B, et al. 2004. Diagnostic real-time PCR for detection of Salmonella in food. Appl. Environ. Microbiol. 70:7046–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mieszkin S, Furet JP, Corthier G, Gourmelon M. 2009. Estimation of pig fecal contamination in a river catchment by real-time PCR using two pig-specific Bacteroidales 16S rRNA genetic markers. Appl. Environ. Microbiol. 75:3045–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D. 2007. Release and persistence of extracellular DNA in the environment. Environ. Biosafety Res. 6:37–53 [DOI] [PubMed] [Google Scholar]
- 24. Noble RT, Weisberg SB. 2005. A review of technologies for rapid detection of bacteria in recreational waters. J. Water Health 3:381–392 [DOI] [PubMed] [Google Scholar]
- 25. Nocker A, Cheung CY, Camper AK. 2006. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67:310–320 [DOI] [PubMed] [Google Scholar]
- 26. Nocker A, Sossa-Fernandez P, Burr MD, Camper AK. 2007. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl. Environ. Microbiol. 73:5111–5117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nocker A, Sossa KE, Camper AK. 2007. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J. Microbiol. Methods 70:252–260 [DOI] [PubMed] [Google Scholar]
- 28. Nogva HK, Bergh A, Holck A, Rudi K. 2000. Application of the 5′-nuclease PCR assay in evaluation and development of methods for quantitative detection of Campylobacter jejuni. Appl. Environ. Microbiol. 66:4029–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okabe S, Okayama N, Savichtcheva O, Ito T. 2007. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 74:890–901 [DOI] [PubMed] [Google Scholar]
- 30. Okabe S, Shimazu Y. 2007. Persistence of host-specific Bacteroides-Prevotella 16S rRNA genetic markers in environmental waters: effects of temperature and salinity. Appl. Microbiol. Biotechnol. 76:935–944 [DOI] [PubMed] [Google Scholar]
- 31. Rajal VB, McSwain BS, Thompson DE, Leutenegger CM, Wuertz S. 2007. Molecular quantitative analysis of human viruses in California stormwater. Water Res. 41:4287–4298 [DOI] [PubMed] [Google Scholar]
- 32. Santo Domingo JW, Bambic DG, Edge TA, Wuertz S. 2007. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Water Res. 41:3539–3552 [DOI] [PubMed] [Google Scholar]
- 33. Saunders AM, Kristiansen A, Lund MB, Revsbech NP, Schramm A. 2009. Detection and persistence of fecal Bacteroidales as water quality indicators in unchlorinated drinking water. Syst. Appl. Microbiol. 32:362–370 [DOI] [PubMed] [Google Scholar]
- 34. Savichtcheva O, Okabe S. 2006. Alternative indicators of fecal pollution: relations with pathogens and conventional indicators, current methodologies for direct pathogen monitoring and future application perspectives. Water Res. 40:2463–2476 [DOI] [PubMed] [Google Scholar]
- 35. Schulz CJ, Childers GW. 2011. Fecal Bacteroidales diversity and decay in response to variations in temperature and salinity. Appl. Environ. Microbiol. 77:2563–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seurinck S, Defoirdt T, Verstraete W, Siciliano SD. 2005. Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ. Microbiol. 7:249–259 [DOI] [PubMed] [Google Scholar]
- 37. Shanks OC, et al. 2006. Basin-wide analysis of the dynamics of fecal contamination and fecal source identification in Tillamook Bay, Oregon. Appl. Environ. Microbiol. 72:5537–5546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sinton L, Hall C, Braithwaite R. 2007. Sunlight inactivation of Campylobacter jejuni and Salmonella enterica, compared with Escherichia coli, in seawater and river water. J. Water Health 5:357–365 [DOI] [PubMed] [Google Scholar]
- 39. Sinton LW. 2006. Biotic and abiotic effects, p 464 In Belkin S, Colwell RR. (ed), Oceans and health: pathogen in the marine environment. Springer Science, New York, NY [Google Scholar]
- 40. US Environmental Protection Agency 2002. Method 1600: enterococci in water by membrane filtration using membrane-Enterococcus indoxyl-β-d-glucoside agar (mEI). US Environmental Protection Agency, Washington, DC [Google Scholar]
- 41. US Environmental Protection Agency 2002. Method 1604: total coliforms and Escherichia coli in water by membrane filtration using simultaneous detection technique (MI medium). US Environmental Protection Agency, Washington, DC [Google Scholar]
- 42. Varma M, et al. 2009. Quantitative real-time PCR analysis of total and propidium monoazide-resistant fecal indicator bacteria in wastewater. Water Res. 43:4790–4801 [DOI] [PubMed] [Google Scholar]
- 43. Vital M, Hammes F, Egli T. 2008. Escherichia coli O157 can grow in natural freshwater at low carbon concentrations. Environ. Microbiol. 10:2387–2396 [DOI] [PubMed] [Google Scholar]
- 44. Wade TJ, et al. 2008. High sensitivity of children to swimming-associated gastrointestinal illness: results using a rapid assay of recreational water quality. Epidemiology 19:375–383 [DOI] [PubMed] [Google Scholar]
- 45. Wade TJ, et al. 2006. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestinal illness. Environ. Health Perspect. 114:24–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wagner AO, Malin C, Knapp BA, Illmer P. 2008. Removal of free extracellular DNA from environmental samples by ethidium monoazide and propidium monoazide. Appl. Environ. Microbiol. 74:2537–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wahman DG, Wulfeck-Kleier KA, Pressman JG. 2009. Monochloramine disinfection kinetics of Nitrosomonas europaea by propidium monoazide quantitative PCR and live/dead BacLight methods. Appl. Environ. Microbiol. 75:5555–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Walters SP, Field KG. 2009. Survival and persistence of human and ruminant-specific faecal Bacteroidales in freshwater microcosms. Environ. Microbiol. 11:1410–1421 [DOI] [PubMed] [Google Scholar]
- 49. Walters SP, Gannon VPJ, Field KG. 2007. Detection of Bacteroidales fecal indicators and the zoonotic pathogens E. coli O157:H7, Salmonella, and Campylobacter in river water. Environ. Sci. Technol. 41:1856–1862 [DOI] [PubMed] [Google Scholar]
- 50. Walters SP, Yamahara KM, Boehm AB. 2009. Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: implications for their use in assessing risk in recreational waters. Water Res. 43:4929–4939 [DOI] [PubMed] [Google Scholar]
- 51. Yamahara KM, Walters SP, Boehm AB. 2009. Growth of enterococci in unaltered, unseeded beach sands subjected to tidal wetting. Appl. Environ. Microbiol. 75:1517–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.