Abstract
The evolution of fungicide resistance within populations of plant pathogens must be monitored to develop management strategies. Such monitoring often is based on microbiological tests, such as microtiter plate assays. Molecular monitoring methods can be considered if the mutations responsible for resistance have been identified. Allele-specific real-time PCR approaches, such as amplification refractory mutation system (ARMS) PCR and mismatch amplification mutation assay (MAMA) PCR, are, despite their moderate efficacy, among the most precise methods for refining SNP quantification. We describe here a new real-time PCR method, the allele-specific probe and primer amplification assay (ASPPAA PCR). This method makes use of mixtures of allele-specific minor groove binder (MGB) TaqMan probes and allele-specific primers for the fine quantification of SNPs from a pool of DNA extracted from a mixture of conidia. It was developed for a single-nucleotide polymorphism (SNP) that is responsible for resistance to the sterol biosynthesis inhibitor fungicide fenhexamid, resulting in the replacement of the phenylalanine residue (encoded by the TTC codon) in position 412 of the enzymatic target (3-ketoreductase) by a serine (TCC), valine (GTC), or isoleucine (ATC) residue. The levels of nonspecific amplification with the ASPPAA PCR were reduced at least four times below the level of currently available allele-specific real-time PCR approaches due to strong allele specificity in amplification cycles, including two allele selectors. This new method can be used to quantify a complex quadriallelic SNP in a DNA pool with a false discovery rate of less than 1%.
INTRODUCTION
Fungicide resistance and its management are of great importance in crop protection. The monitoring of this resistance is a crucial area of research, one on which our knowledge of the distribution, evolution, and effect of fungicide resistance in the field depends. In most cases, the degree of sensitivity of fungal populations to one or more fungicides is assessed by biological methods (17). These bioassays, conducted in vitro or in vivo, have been miniaturized (i.e., microtiter plate methods), but nonetheless they consume considerable resources and time. When the molecular mechanisms of resistance are known (e.g., target mutation, target overexpression, and increased drug efflux), and particularly when the underlying DNA polymorphisms (single-nucleotide polymorphism [SNPs], deletions, or insertions) have been defined, various molecular methods can be used to monitor antimicrobial resistance (8, 14, 15). The principle methods for quantifying resistance are based on real-time PCR technology. Alleles are amplified in a specific manner, either independently or in multiplex systems, with allele-specific probes or primers. Polymorphic alleles then are quantified by the cycle of quantification values and compared to the wild-type values (Cq; i.e., at a given threshold, Cq is the number of PCR cycles at which reporter fluorescence becomes significant or is distinguishable from the background noise). However, one limitation of this method concerns the nonspecific amplification of alleles, which may affect precision. This limitation does not generally hinder the detection of the polymorphism, but it may affect quantification capacity, particularly for mutated alleles with low abundance (reviewed in reference 16).
The hydroxyanilide derivate fenhexamid is a fungicide targeting ergosterol biosynthesis, and it is used on grapevine and other crops to control the gray mold disease caused by Botrytis cinerea. Fenhexamid inhibits the sterol 3-ketoreductase activity of the protein encoded by the erg27 gene and is involved in the C-4 demethylation process in the ergosterol biosynthesis pathway (5). A survey of natural populations of B. cinerea has identified several phenotypes of resistance to this hydroxyanilide fungicide (i.e., HydR1, HydR2, HydR3−, and HydR3+) (13). The strongest resistance was recorded for HydR3+ strains, in which resistance is fully accounted for by a single polymorphic substitution in the target. The underlying DNA polymorphism is the modification of the TTC codon encoding the phenylalanine 412 residue, which is converted into a TCC (serine), GTC (isoleucine), or ATC (valine) codon (7).
The evolution of HydR3+ strains within natural populations of B. cinerea is monitored biologically on annual bulk field samples from infected berries. We developed and investigated a sensitive, real-time PCR method for quantifying the underlying DNA polymorphism responsible for the HydR3+ resistance phenotype to decrease the time required for analysis. Allele-specific real-time PCR assays were conducted, first with allele-specific primers and then with allele-specific probes. These assays independently had low allelic quantification capacities (data not shown) due to nonspecific amplification resulting from the assay having to distinguish between large numbers of alleles and the complexity of the polymorphism. Taking these results into account, we investigated the development of a new technique for quantifying, with high precision, the three different erg27 alleles from the HydR3+ phenotype in pooled DNA from bulk samples of conidia harvested in the field. The best result was obtained with a nonmultiplexed method combining four allele-specific minor groove binder (MGB) TaqMan probes and four mismatched specific primers. This technique was named the allele-specific probe and primer amplification assay (ASPPAA) PCR.
MATERIALS AND METHODS
Fungal strains and culture conditions.
The B. cinerea HydR3+ natural isolates 223a, 440a, and 05-PV Reims, carrying the erg27F412S, erg27F412I, and erg27F412V alleles, respectively, were described in a previous study (7). B. cinerea strain B05.10 (4) is used here as the wild-type reference strain. Its genome has been fully sequenced (2). Strains were grown on MY medium (20 g liter−1 malt extract, 2 g liter−1 yeast extract, 12.5 g liter−1 agar) at 20°C under exposure to continuous white light for 7 to 10 days until conidiation. DNA from Saccharomyces cerevisiae, Plasmopara viticola, and Erysiphe necator was available in our laboratory.
DNA manipulation.
The nuclear DNA used for assessments of assay performance was extracted from a 1-week-old Botrytis cinerea mycelium according to a sarcosyl-based protocol (6). Gel analysis and DNA quantification were carried out according to standard protocols. The DNA used for testing mixtures of conidia was extracted from B. cinerea conidia by grinding twice, for 1 min each in a cetyltrimethylammonium bromide (CTAB) buffer (18) in a Fastprep grinder (MP Biomedicals, Solon, OH). All extracts were treated with RNase H at 0.5 μg/μl for 30 min at 37°C with a subsequent phenol-chloroform extraction. Nuclear DNA was quantified with a UV spectrophotometer (Nanodrop, Wilmington, DE); two independent quantifications were carried out for each DNA preparation before analysis. For assessments of the linearity and efficiency of amplification and for the optimization of reactions, DNA was diluted in nuclease-free water to concentrations of 170, 17, 1.7, and 0.17 ng μl−1. DNA preparations were stored at 4°C or were frozen at −20°C.
Probe and primer design.
The sequence of the erg27 gene of B. cinerea is available in GenBank (AY220532). Four pairs of reverse primers and probes for each DNA strand were designed with Primer Express software, version 3.0 (Applied Biosystems, Foster City, CA), using the default parameters as recommended for allele discrimination (for details, see the 2006 real-time PCR application guide from Bio-Rad). Primers were designed with a melting point (tm) between 58 and 60°C and were purchased from Sigma-Aldrich (Saint Louis). Probes were designed with a tm 7 to 10°C higher than that of the primers (Table 1; polymorphic nucleotides are shown in boldface). A common forward primer was designed (5′-TGTTTCGGAGATCATGCCC-3′) by following the same recommendations as those for allele-specific primers. The terminal 3′ positions of primers and probes were hybridized to the F412 mutation. Deliberate additional mismatches (underlined in Table 1) were introduced to improve hybridization specificity (10, 21). To prevent nonspecific hybridization and amplification, overlaps between probes and primers did not exceed three nucleotides. TaqMan Probes were purchased from Applied Biosystems (Foster City, CA) and were labeled at the 5′ end with 6-carboxyfluorescein (FAM) and at the 3′ end with minor groove binder-nonfluorescent quencher (MGB-NFQ).
Table 1.
Sequences of the four combinations of allele-specific probes and primers used for quantification of the erg27 alleles
| Gene | Codon | Allele-specific reverse primer sequence (5′ to 3′)a | Probe sequence (5′ to 3′) |
|---|---|---|---|
| erg27WT | TTC | CCATCCATCTTACAAGGTCGAAG | FAM-TTATCTACAGATTGATCTTC-MGB-NFQ |
| erg27F412S | TCC | CCATCCATCTTACAAGGTCGG | FAM-TTTATCTACAGATTGATCTCC-MGB-NFQ |
| erg27F412I | ATC | CCATCCATCTTACAAGGTCGATG | FAM-TTATCTACAGATTGATCATC-MGB-NFQ |
| erg27F412V | GTC | CATCCATCTTACAAGGTCGACG | FAM-TTTATCTACAGATTGATCGTC-MGB-NFQ |
Additional mismatches in primers are underlined, and allele-specific nucleotides are shown in boldface.
Real-time PCR assays.
All analyses were conducted on an HT 7900 fast real-time PCR system run with ABI Prism SDS software, version 2.2 (Applied Biosystems, Foster City, CA). Reactions were performed in 96-well plates with optical adhesive films from Applied Biosystems. For measurements of efficiency and linearity, all PCRs were carried out in a reaction volume of 25 μl. Each reaction mixture contained quantitative PCR (qPCR) MasterMix Plus without UNG (Eurogentec, Liege, Belgium). The thermal profile used for PCR was 10 min at 95°C for Hot Gold Start activation and 40 cycles of amplification (95°C for 15 s and 61°C for 60 s). Several annealing temperatures (60, 61, 62, and 63°C) and extension times (60, 75, and 90 s) were tested. During the optimization steps, we assessed primer concentrations of 300, 600, and 900 nM and MGB TaqMan probe concentrations of 100, 200, and 300 nM (Applied Biosystems, Foster City, CA). All reactions were performed in three independent assays with three technical replicates. The threshold for our tests was set at 0.15. We checked that Cq values and sample concentrations were proportional by assessing the linearity of each amplification assay through the calculation of the determination coefficient (R2) of the regression curve obtained by plotting the Cq values for concentrations of 170, 17, 1.7, and 0.17 ng μl−1 against the logarithm of the corresponding amount of DNA. PCR efficiencies (E) were calculated as E = [10(−1/a) − 1], where a is the slope of the regression curve.
Nonspecific amplification rates were estimated by interference limit (IL) calculation, as described by Germer et al. (9), for the determination of the specificity of each amplification. IL values were determined at a DNA concentration of 85 ng μl−1. The four pairs of primers and probes were used independently with the four DNA samples (erg27WT, erg27F412S, erg27F412I, and erg27F412V). Cqs (cycle of specific quantification) and Cqn (cycle of nonspecific quantification) values were obtained for the 12 nonspecific and the four specific possible amplifications (see the y-intercept values in Table 3). These reactions were performed in three independent assays, each carried out in triplicate, and the mean values and corresponding standard deviations (σa for aspecific amplifications and σs for specific amplifications) were calculated. The background threshold cycle for a given amount of DNA (Cqlim) was set at , where n is the number of observations (n = 9) and t0.01 is the tabulated value of the Student's t test for the 1% probability level and n − 1 degrees of freedom. The lowest allele concentration that could be significantly distinguished from the background was calculated as IL = 100/(2Cqs − Cqlim + 1) (9).
Table 3.
Slopes, efficiencies, R2 values, and y-intercepts of the four ASPPAA PCR amplifications for F412 mutations (qPCR threshold at 0.15)
| Gene | Slope | Efficiency (%) | R2 | y-intercepta (σ) |
|---|---|---|---|---|
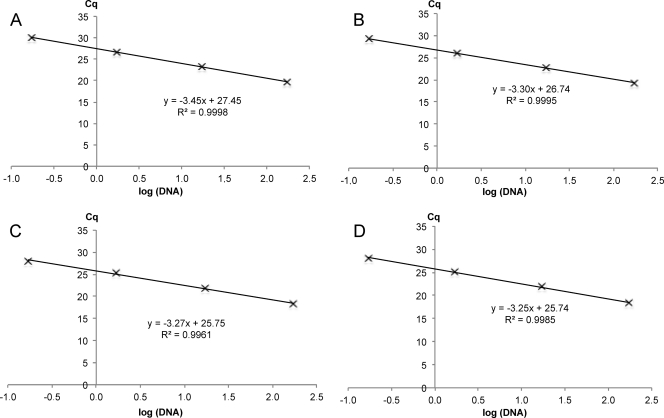
| erg27WT | −3.45 | 95.4 | >0.99 | 27.45 (0.03) |
| erg27F412S | −3.30 | 100.4 | >0.99 | 26.74 (0.01) |
| erg27F412I | −3.27 | 100.9 | >0.99 | 25.75 (0.02) |
| erg27F412V | −3.25 | 101.4 | >0.99 | 25.74 (0.03) |
Results are given as means with standard deviations in parentheses (n = 9 observations).
We determined the limit of detection (LOD), corresponding to the lowest concentration of DNA that could be distinguished from the background, by carrying out the amplification of a range of concentrations of DNA (170, 85, 42.5, 21.25, and 10.625 pg μl−1) for each condition in three independent PCRs with three replicate wells each in the same run.
Assays on artificial mixtures of B. cinerea conidia.
ASPAA PCR was performed on DNA extracted from calibrated mixtures of mutant and wild-type spores. Spores were harvested from the wild-type and mutant strains in sterile water and counted with a hematocytometer. The spore concentrations were adjusted to 107 spores ml−1 prior to mixing the volumes necessary to obtain the mixtures A to D cited in Table 4. Spores (2 × 108) were collected by centrifugation and used for DNA extraction.
Table 4.
Comparison between theoretical and measured concentrations of each allele in DNA pools from four mixtures of conidiaa
| Gene | Result for DNA mix: |
|||||||
|---|---|---|---|---|---|---|---|---|
| A |
B |
C |
D |
|||||
| Theoretical | Measured | Theoretical | Measured | Theoretical | Measured | Theoretical | Measured | |
| erg27WT | 1 | 1.15 (0.05) | 5 | 5.62 (0.07) | 35 | 35.08 (0.05) | 59 | 60.22 (0.07) |
| erg27F412S | 5 | 5.78 (0.08) | 1 | 0.99 (0.05) | 59 | 59.69 (0.07) | 35 | 34.50 (0.04) |
| erg27F412I | 35 | 40.53 (0.04) | 59 | 59.98 (0.05) | 1 | 0.88 (0.06) | 5 | 4.50 (0.1) |
| erg27F412V | 59 | 52.53 (0.07) | 35 | 33.41 (0.07) | 5 | 4.35 (0.05) | 1 | 0.78 (0.09) |
Values are expressed as percent DNA for one allele in the DNA pool. Standard errors are shown in parentheses.
ASPPAA consists of four independent runs with 85 ng of calibrated nuclear DNA, each with one of the four pairs of probes and primers (for erg27WT, erg27F412S, erg27F412I, and erg27F412V). All reactions were performed in three independent assays, each carried out in triplicate to ensure robustness. The resulting Cq values at a threshold of 0.15 obtained for each amplification were averaged and converted into amounts of DNA by the application of the appropriate regression equations.
RESULTS
Principle of the method.
We maximized the precision of quantification by investigating a combination of two principle elements: an allele-specific primer binding to one DNA strand and an allele-specific MGB TaqMan probe binding to the other (Fig. 1). For the discrimination of the complex mixture of the four erg27 alleles described above, we designed four pairs of probes and primers. Each of the pairs is specific to one allele (erg27WT, erg27F412S, erg27F412I, and erg27F412V). The common primer and the combinations of allele-specific probes and primers selectively amplify each allele in independent runs of real-time PCR. The resulting fluorescence, corresponding to the increase in DNA concentration, was monitored. Cq values were obtained and used to calculate the final DNA ratio.
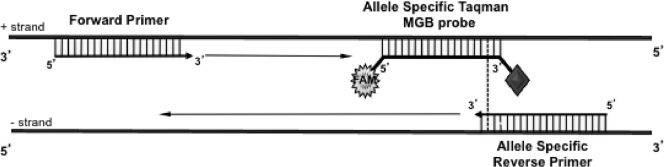
Fig 1.
Schematic diagram of the principle of the ASPPAA PCR. Four allele-specific TaqMan MGB probes, corresponding to additional mismatched allele-specific primers and a common forward primer, were designed to amplify an amplicon that was as short as possible (76 bp for erg27F412). The dotted line indicates the position of the allele-specific nucleotide.
This method depends on rates of nonspecific amplification being low. Allele specificity was increased by designing primers with an additional mismatch to the SNPs at the 5′ end. Given the many parameters used in the design of MGB TaqMan probes, SNPs were placed at the 5′ end in accordance with the manufacturer's recommendations, and a maximum overlap of three nucleotides between a primer and its corresponding probe was tolerated. One or two mismatches, at different positions along the sequence, were analyzed, and more than 100 different mismatched primers were tested (data not shown). Efficiency was highest for a single additional mismatch between nucleotides in positions −3 to −5 with respect to the 3′ end. We tested various combinations of primer and probe concentrations and annealing temperatures, and the best discrimination was obtained with high concentrations of primers (900 nM) and a standard concentration of MGB TaqMan probes (200 nM), with an annealing temperature of 61°C. Probe cleavage was optimized by designing a common forward primer to produce the shortest possible amplicon, 76 bp in our case (Fig. 1 and Table 1).
IL.
The interference limit (IL), expressed as a percentage, reflects the rate of nonspecific amplification for each condition (i.e., the pair consisting of the erg27WT probe and its corresponding primer on erg27F412r nuclear DNA). IL values were calculated for each probe/primer pair with the formula IL = 100/(2Cqs−Cqlim + 1) for each nonspecific amplification on pure DNA calibrated at 85 ng μl−1. IL values ranged from 0.0001 to 0.1448% (Table 2). With a mean value of 0.0201%, the overall IL of the ASPPAA for four-allele quantification was low. This performance of the assay is sufficiently good to ignore the effect of nonspecific amplification on our final DNA ratio calculations (Table 3).
Table 2.
ILa for the 12 possible nonspecific amplifications encountered in ASPPAA for erg27F412 mutations (qPCR threshold at 0.15)
| Gene | IL (%) for: |
|||
|---|---|---|---|---|
| erg27WT | erg27F412S | erg27F412I | erg27F412V | |
| erg27WT | 0.026 | 0.0036 | 0.0006 | |
| erg27F412S | 0.0004 | 0.0005 | 0.0045 | |
| erg27F412I | 0.0481 | 0.0001 | 0.1448 | |
| erg27F412V | 0.0011 | 0.0001 | 0.0111 | |
IL = 100/(2Cqs − Cqlim + 1) according to reference 9 (n = 9 observations); see the text for further explanations.
We also carried out assays on DNA from Saccharomyces cerevisiae, Plasmopara viticola, and Erysiphe necator to assess the potential effect of other microorganisms within natural samples on the performance of the method. However, the amplification obtained was weak or nonsignificant (data not shown).
Amplification efficiency.
The amplification parameters for each reaction are presented in Table 3, and standard curves are presented in Fig. 2. The three assays, each run in triplicate, for each condition displayed good repeatability between runs, as indicated by the slope and y-intercept standard deviations. All R2 values were greater than 0.99, demonstrating the linearity of the method for the four dilution ranges. Amplification efficiency was lowest for the wild-type probe and primer pair (95.4 ± 0.2%). The other three conditions yielded similar but higher values for efficiency. Taken together with the low variation in y-intercepts, these data indicate that this assay is reliable for the purposes of detection and quantification.
Fig 2.
Standard curves of quantification cycles (Cq) against the logarithm of the corresponding amount of DNA for the wild-type (A), erg27F412S (B), erg27F412I (C), and erg27F412V (D) amplifications. y-intercept values correspond to the Cq for a DNA concentration of 1.
The LOD was below 42.5 pg for each condition, which is sufficiently good for the quantification of a rate of 1% in a DNA sample calibrated at 85 ng (850 pg).
Assays on mixtures of Botrytis cinerea conidia.
We mimicked the conditions in field trials by carrying out tests on nuclear DNA extracted from calibrated mixtures of Botrytis cinerea conidia. Four different mixtures with ratios of 1, 5, 35, and 59% for the four alleles (Table 4) were tested in triplicate (three independent assays with three technical replicates each).
It was not possible to include all of the standards in each assay/plate. We therefore relied on the high degree of reproducibility between independent assays observed above (Table 3). Instead of standards, we included in each plate, as calibrator, 85 ng μl−1 of pure DNA for each allele. This calibrator was used to correct variations from the standard curves potentially introduced in each assay.
A close correlation between the theoretical ratios of conidia and their calculated concentrations was obtained (Table 4). The lowest level of DNA tested (1%) was correctly detected and quantified for each allele. The standard errors of the means were low, indicating a high level of reproducibility among the triplicates.
DISCUSSION
Clinical research is a useful source of new diagnostic technologies. Many molecular quantification techniques have been developed for disease diagnosis, including SNP primer extension assays (12), denaturing high-performance liquid chromatography (DHPLC) (1), microarrays (22), pyrosequencing (11), nanoparticle assays (19), and quencher extension assays (20). Allele-specific real-time PCR with allele-specific primers or probes (3, 16) are the best methods currently available for allelic quantification. However, it is difficult to quantify an SNP precisely when its abundance in a DNA pool is less than 5%, principally due to nonspecific amplification caused by false hybridizations of allele-specific probes or primers. We developed a new real-time PCR method that improves the performance of SNP quantification. This method, the allele-specific probe and primer amplification assay (ASPPAA), was developed for a four-allele SNP responsible for strong resistance to fenhexamid in B. cinerea. We demonstrated that fine SNP quantification was possible in this system. The ASPPAA technique combines the mismatch amplification mutation assay (MAMA) method (10), based on the use of mismatched allele-specific primers, with classical quantification using allele-specific probes. We decided to use minor groove-binding (MGB) TaqMan probes because of their properties. Indeed, MGB chemistry increases the tm of the probes, allowing the design of short probes. Shorter probes generally hybridize more specifically than longer probes.
Strong reproducibility between regression curve replicates was observed. However, due to the exponential scale, small variations in Cq values have exponential effects on the DNA concentrations deduced from the regression curves. We found that a correction between Cq and the standard regression curve was required to correct small deviations and to increase precision. This correction was achieved with a standard run calibrated at the same concentration as that of the analyzed sample on the same plate.
It was necessary to establish an optimal balance between acceptable amplification efficiency for all primer/probe pairs (R2 values greater than 0.99, efficiency greater than 95%, and stable y-intercepts) and the lowest possible level of nonspecific amplification. We achieved this by adding one mismatch between nucleotides −3 and −5 with respect to the 3′ end of the primer. The positioning of the mutation-specific nucleotide at position −1 to −2 (position −1 was more efficient for G or C nucleotides) maximized both allele specificity and efficiency. The positioning of a G or C residue at the 3′ end of the primer gave the best result. High primer concentrations also increased specificity. For probes, recommendations for SNP genotyping analysis include designing the shortest probes possible and placing the allele-specific nucleotide in the third part, on the 3′ side, to promote specific hybridization and to ensure the effective cleavage of the TaqMan MGB probes (according to Bio-Rad's 2006 real-time PCR application guide). In ASPPAA PCR, overlap with the allele-specific primer could be reduced by placing the allele-specific nucleotide between positions −2 and −4 with respect to the 3′ end. Annealing optimization showed that an extension time of more than 1 min did not increase allele specificity, but that an increase of 1°C in annealing temperature slightly increased the allele specificity of the primers without decreasing efficiency.
With these technical parameters, strong correlations between the ratios of conidia in mixtures and the percentages calculated by ASPPAA were obtained. Given the deviations introduced during the preparation of mixtures of conidia, a precision for the minimal SNP concentration of 1% was satisfactory. Given the standard errors of the means for each condition, it would be difficult to consider a lower level of quantification. The high degree of precision of the method may be accounted for by the low rates of nonspecific amplification and the strong allele specificity conferred by the combination of allele-specific probes and allele-specific optimized primers. The mean IL for ASPPAA was below 0.02%, whereas a mean IL of about 1% would be expected in the best cases of biallelic quantification with classical allele-specific real-time PCR methods (16). Sensitivity would be expected to be even higher for biallelic SNP quantification.
The high level of performance of the ASPPAA PCR for the quantification of fenhexamid resistance should facilitate more rapid monitoring analysis. In the future, the multiplexing, in the same run, of the analysis of several polymorphisms at different genomic loci with probes picked up in different fluorophore channels is conceivable and would be expected to decrease the time required for monitoring, as well as its cost, significantly. This tool also could be applied to analyze target site resistance to pesticides and biocides in general or in any diagnostics based on SNP variations, including clinical studies, to help improve SNP quantification.
Footnotes
Published ahead of print 9 December 2011
REFERENCES
- 1. Abbas A, Lepelley M, Lechevrel M, Sichel F. 2004. Assessment of DHPLC usefulness in the genotyping of GSTP1 exon 5 SNP: comparison to the PCR-RFLP method. J. Biochem. Biophys. Methods 59:121–126 [DOI] [PubMed] [Google Scholar]
- 2. Amselem J, et al. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Breen G, Harold D, Ralston S, Shaw D, St. Clair D. 2000. Determining SNP allele frequencies in DNA pools. Biotechniques 28:464–466 [DOI] [PubMed] [Google Scholar]
- 4. Büttner P, et al. 1994. Variations in ploidy among isolates of Botrytis cinerea: implications for genetic and molecular analyses. Curr. Genet. 25:445–450 [DOI] [PubMed] [Google Scholar]
- 5. Debieu D, Bach J, Hugon M, Malosse C, Leroux P. 2001. The hydroxyanilide fenhexamid, a new sterol biosynthesis inhibitor fungicide efficient against the plant pathogenic fungus Botryotinia fuckeliana (Botrytis cinerea). Pest Manag. Sci. 57:1060–1067 [DOI] [PubMed] [Google Scholar]
- 6. Dellaporta SL, Wood J, Hicks JB. 1983. A plant DNA minipreparation: version 2. Plant Mol. Biol. Rep. 1:19–21 [Google Scholar]
- 7. Fillinger S, et al. 2008. Genetic analysis of fenhexamide-resistant field isolates of the phytopathogenic fungus Botrytis cinerea. Antimicrob. Agents Chemother. 52:3933–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fraaije BA, Butters JA, Coelho JM, Jones DR, Hollomon DW. 2002. Following the dynamics of strobilurin resistance in Blumeria graminis f. sp. tritici using quantitative allele-specific real-time PCR measurements with the fluorescent dye SYBR green I. Plant Pathol. 51:45–54 [Google Scholar]
- 9. Germer S, Holland MJ, Higuchi R. 2000. High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res. 10:258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glaab WE, Skopek TR. 1999. A novel assay for allelic discrimination that combines the fluorogenic 5′ nuclease polymerase chain reaction (TaqMan) and mismatch amplification mutation assay. Mutat. Res. 430:1–12 [DOI] [PubMed] [Google Scholar]
- 11. Gruber JD, Colligan PB, Wolford JK. 2002. Estimation of single nucleotide polymorphism allele frequency in DNA pools by using pyrosequencing. Hum. Genet. 110:395–401 [DOI] [PubMed] [Google Scholar]
- 12. Kurg A, et al. 2000. Arrayed primer extension: solid-phase four-color DNA resequencing and mutation detection technology. Genet. Testing 4:1–7 [DOI] [PubMed] [Google Scholar]
- 13. Leroux P, et al. 2002. The hydroxyanilide botryticide fenhexamid/mode of action and mechanism of resistance, p. 29–40 In Dehne HW, Gisi U, Kuck KH, Russel PE, Lyr H. (ed.), Modern fungicides and antifungal compounds III. AgroConcept GmbH, Bonn, Germany [Google Scholar]
- 14. Ma Z, Michailides TJ. 2005. Advances in understanding molecular mechanisms of fungicide resistance and molecular detection of resistant genotypes in phytopathogenic fungi. Crop Prot. 24:853–863 [Google Scholar]
- 15. Ma Z, Michailides TJ. 2004. A real-time PCR assay for the detection of azoxystrobin-resistant Alternaria populations from pistachio orchards in California. Crop Prot. 23:1259–1263 [Google Scholar]
- 16. Mattarucchi E, et al. 2005. Different real time PCR approaches for the fine quantification of SNP's alleles in DNA pools: assays development, characterization and pre-validation. J. Biochem. Mol. Biol. 38:555–562 [DOI] [PubMed] [Google Scholar]
- 17. Meletiadis J, Meis JF, Mouton JW, Verweij PE. 2001. Analysis of growth characteristics of filamentous fungi in different nutrient media. J. Clin. Microbiol. 39:478–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moller EM, Bahnweg G, Sandermann H, Geiger HH. 1992. A simple and efficient protocol for isolation of high-molecular-weight DNA from filamentous fungi, fruit bodies, and infected-plant tissues. Nucleic Acids Res. 20:6115–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qin WJ, Yung LY. 2007. Nanoparticle-based detection and quantification of DNA with single nucleotide polymorphism (SNP) discrimination selectivity. Nucleic Acids Res. 35:e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rudi K, Zimonja M, Hannevik SE, Dromtorp SM. 2006. Multiplex real-time single nucleotide polymorphism detection and quantification by quencher extension. Biotechniques 40:323–329 [DOI] [PubMed] [Google Scholar]
- 21. Skopek TR, Glaab WE, Monroe JJ, Kort KL, Schaefer W. 1999. Analysis of sequence alterations in a defined DNA region: comparison of temperature-modulated heteroduplex analysis and denaturing gradient gel electrophoresis. Mutat. Res. 430:13–21 [DOI] [PubMed] [Google Scholar]
- 22. Yin JQ, Zhao RC, Morris KV. 2008. Profiling microRNA expression with microarrays. Trends Biotechnol. 26:70–76 [DOI] [PubMed] [Google Scholar]