Abstract
While ingestion of synbiotic yogurts containing Bifidobacterium animalis subsp. lactis and inulin is increasing, their effect on certain microbial groups in the human intestine is unclear. To further investigate this, a large-scale, crossover-design, placebo-controlled study was utilized to evaluate the effect of a synbiotic yogurt containing B. animalis subsp. lactis Bb-12 and inulin on the human intestinal bifidobacteria, clostridia, and enterobacteria. Fecal samples were collected at 14 time points from 46 volunteers who completed the study, and changes in the intestinal bacterial levels were monitored using real-time PCR. Strain Bb-12 could not be detected in feces after 2 weeks of washout. A live/dead PCR procedure indicated that the Bb-12 strain detected in the fecal samples was alive. A significant increase (P < 0.001) in the total bifidobacterial numbers was seen in both groups of subjects during the final washout period compared to the prefeeding period. This increase in total bifidobacteria corresponded with a significant decrease (P < 0.05) in numbers of clostridia but not enterobacteria. No significant differences in numbers of bifidobacteria, clostridia, or enterobacteria were observed between the probiotic and placebo groups during any of the feeding periods. However, subgrouping subjects based on lower initial bifidobacterial numbers or higher initial clostridial numbers did show corresponding significant differences between the synbiotic yogurt and placebo groups. This was not observed for a subgroup with higher initial enterobacterial numbers. While this synbiotic yogurt can increase bifidobacterial numbers and decrease clostridial numbers (but not enterobacterial numbers) in some individuals, it cannot modulate these microbial groups in the majority of individuals.
INTRODUCTION
The human large intestine is host to a wide variety of bacteria, with bifidobacteria being prominent members of this complex ecosystem. These are (often irregularly) rod-shaped Gram-positive, anaerobic, non-spore formers that utilize glucose via a pathway that involves a diagnostic enzyme, fructose-6-phosphate phosphoketolase (F6PPK). As bifidobacteria are generally believed to contribute to good intestinal health, attempts have been made to increase their numbers in the intestine by including them in certain foods as probiotics. They are frequently included in yogurts together with prebiotics, such as inulin or fructooligosaccharides, the combination of which can be referred to as a synbiotic yogurt. Bifidobacteria are implicated in the prevention and treatment of diarrhea, development and maintenance of a healthy microbiota in low-weight preterm infants, and stimulation of certain immune responses (25, 28). It is believed that counts of bifidobacteria begin to decline during old age, coinciding with a proliferation of other bacterial groups, including clostridia and members of the Enterobacteriaceae family (11, 25, 43). Consequently, increasing bifidobacterial counts may be advantageous for controlling the proliferation of certain undesirable bacteria.
Clostridia are toxin producers and have been implicated in many intestinal ailments, such as nosocomial diarrhea, antibiotic-associated diarrhea, necrotizing enterocolitis, and gastrointestinal (GI) infections (5). The genus Clostridium substantially consists of about 140 different species. It has been divided into 19 clusters based on the phylogenetic analysis of the 16S rRNA gene. Cluster I is the largest cluster, consisting of most of the pathogenic species of Clostridium, including C. tetani, C. botulinum, and C. perfringens (41). Gastrointestinal diseases caused by clostridia are mostly a result of an imbalanced gut microbiota, which disrupts the protective effects of the GI flora (21). This imbalance can be precipitated by antibiotic treatment, stress, immunocompromization, and old age (reviewed in references 6 and 10).
Escherichia species such as Escherichia coli belong to the family Enterobacteriaceae and are normal inhabitants of human and animal guts. They are Gram-negative, rod-shaped, facultative anaerobes with excellent survival outside the gut and are frequently utilized as indicators of fecal contamination. While most E. coli strains are nonpathogenic, commensal bacteria, certain strains are harmful and opportunistic due to the presence of virulence factors that support their ability to induce infection (15, 16). The most recognized of these strains is enterohemorrhagic E. coli (EHEC) due to the prevalence of E. coli O157:H7 in food-borne disease outbreaks (16, 27). While it has been proposed that a low level of commensal strains may be beneficial (35), their role is undefined; thus, there is an increasing interest in the role of the intestinal microbiota in maintaining overall and GI health (8, 9).
While there is no legal definition for probiotics in the United States, they are generally referred to as live microbial feed supplements that, when ingested in sufficient amounts, can affect the host beneficially (29). Bifidobacteria are widely used as probiotics in foods. B. animalis is the most common species used in foods, as it has the highest tolerance to oxygen and acids. The continued adaptation of this species to the fermented dairy environment resulted in the evolution of a newly adapted bacterium that was initially classified as a new species, B. lactis (24). However, owing to very close genetic similarities to B. animalis, it was accepted as a subspecies of B. animalis (4). B. animalis subsp. lactis strain Bb-12 is a common commercial probiotic used in foods and is exclusively supplied by Chr. Hansen Inc., a worldwide supplier of fermentation cultures. Studies on Bb-12 ingestion have shown it to have many potential health benefits. These include maintaining the intestinal microbial balance (reviewed in reference 32), diarrhea prevention (37), stimulation of the phagocytic activity in peripheral blood samples (38), and treatment of atopic eczema in infants (14). However, given small subject numbers and limitations of culturing methods, further studies are needed to evaluate the effect of this widely used probiotic culture on the intestinal microbiota. The purpose of this study was to examine the effects of a yogurt supplemented with inulin and strain Bb-12 (∼109 to 1010 CFU/serving) consumed daily by a large number of healthy subjects on intestinal bifidobacterial, clostridial, and enterobacterial numbers using a probe-based real-time PCR approach.
MATERIALS AND METHODS
Bacteria and culture conditions.
Bifidobacterium animalis subsp. lactis Bb-12 (Chr. Hansen Inc., Horsholm, Denmark), B. longum strain DJO10A (13, 19), Clostridium perfringens (Diez laboratory, University of Minnesota), and E. coli (Invitrogen, Carlsbad, CA) were used in this study. E. coli and B. longum strain DJO10A were used as negative controls for strain Bb-12-specific TaqMan real-time PCR. All cultures were streaked on agar plates as follows: strains Bb-12 and DJO10A on BIM-25 (26); C. perfringens on RCM (reinforced clostridial medium) (2); and E. coli on LB. Strains Bb-12 and DJO10A were cultured anaerobically in broth cultures using BliM plus Fe (13) at 37°C for 48 h, C. perfringens was cultured anaerobically in RCM at 37°C for 24 h in anaerobic jars, and E. coli was cultured aerobically in LB medium at 37°C overnight.
Human subjects, study design, and sample collection.
Prior to initiating the study, approval for the study design was obtained from the Institutional Review Board (IRB) of the University of Minnesota. Fifty-two healthy volunteers participated in this study, with 46 of them completing it. The average age, height, and weight for the group of 46 subjects were 31 years, 5 feet 6 in., and 155 lb, respectively. The detailed demographic data are presented in Table 1. The inclusion criteria for this study were that subjects were adults with no known allergies or intolerance to dairy foods and had not consumed foods containing bifidobacteria for at least 2 months prior to the start of the study. To determine the latter criterion, subjects were specifically asked whether they had consumed any dairy products that had the term “bifidus” or “bifidobacteria” or any word beginning with “bifid” on its label. Subjects were asked to maintain their usual diet during the study, with no intake of products containing bifidobacteria with the exception of what was given to them within the study. The subjects were randomly assigned to two groups, A and B, with 26 subjects in each. However, only 46 of the total initial number of subjects completed the study. Consequently, group A finished with 24 subjects and group B with 22 subjects.
Table 1.
Means of ages, weights, and heights for study participants
| Parameter | Group A | Group B | P value |
|---|---|---|---|
| No. of subjects | 22 | 24 | |
| Mean age (yr) | 30.3 ± 9.7 | 30.9 ± 13.8 | 0.87 |
| Mean wt (lb) | 166 ± 46.4 | 145 ± 26 | 0.06 |
| Mean ht (ft) | 5.7 ± 0.33 | 5.49 ± 0.28 | 0.48 |
The study was a double-blind, crossover, placebo-controlled, randomized-feeding trial. It was divided into five consecutive periods: a prefeeding period (1 week), followed by a feeding period (3 weeks), a washout period (4 weeks), a second feeding period (3 weeks), and a final washout period (4 weeks). The prefeeding period was a control period, during which the subjects were not given any yogurt drink. Fecal samples were collected at the start and at the end of the week. During the first feeding period, the subjects consumed daily either 94 g of placebo, which consisted of milk acidified to pH 4.2 with lactic acid, or 94 g of a drinkable yogurt containing 109 to 1010 CFU of strain Bb-12 and 1 g of inulin per serving. The yogurt was prepared with skim milk and a standard yogurt starter blend consisting of Streptococcus thermophilus and Lactobacillus bulgaricus, together with the B. animalis subsp. lactis Bb-12 culture, and had a final pH of 4.2. Both products were flavored with sucrose and strawberry. The shelf life of the yogurt was set as 50 days at 4°C, corresponding to the lower level (109 CFU/serving) of strain Bb-12. Fecal samples were collected at the end of each of the 3 weeks. The subjects consumed neither the yogurt nor the placebo during the subsequent washout period. Single fecal samples were collected at the end of every 2 weeks. Participants were given 50-ml Falcon tubes containing 10 ml of sterile phosphate-buffered saline (PBS) buffer and a sterile plastic spoon and were asked to fill the tube to the 40-ml mark with feces from the midstream defecation period. The tubes were mixed thoroughly and centrifuged to collect a fixed amount of pelleted stool. During the second feeding period, there was a crossover of the feeding design. Again, fecal samples were collected at the end of each of the 3 weeks. The final washout period lasted for 4 weeks. Neither placebo nor probiotic yogurt was consumed during that period. Fecal samples were collected at the end of each week. In all, 14 fecal samples were collected from each subject. For every collection, ∼5 g of feces was collected in 10 ml of phosphate buffer (pH 7) and immediately frozen at −20°C. The study design and sample collection strategy are depicted in Fig. 1.
Fig 1.
Representation of the study design and fecal collection. The study was divided into 5 periods: a prefeeding period, a feeding period, a washout period, a crossover feeding period, and a final washout period. Fecal samples are represented by the sterile tubes containing buffer and collection spoon given to subjects.
DNA extraction.
DNA was extracted from pure cultures by the use of a Qiagen MiniPrep DNA extraction kit protocol with slight modifications. A 1-ml aliquot of culture was centrifuged (1,350 rpm for 10 min in a Hermle Labnet Z 233 MK centrifuge with rotor model C-0230-2A) to reduce cells to pellets. The cells were resuspended in 340 μl of ASL buffer and subjected to 95°C for 5 min. Proteinase K and buffer AL were subsequently added directly to the heat-treated cells in ASL buffer. The rest of the Qiagen protocol was followed according to the manufacturer's instructions. DNA was extracted from feces by the use of a Qiagen MiniPrep DNA extraction kit according to the manufacturer's instructions. The DNA was then quantified using a Nanodrop spectrophotometer and stored at −20°C.
Quantification of B. animalis subsp. lactis Bb-12 and total bifidobacteria in feces by using TaqMan real-time PCR.
Specific primers and TaqMan probes used for amplification in TaqMan real-time PCR are listed in Table 2. Probes were labeled with the fluorescent dye 6-carboxyfluorescein (FAM) and the quencher 6-carboxytetramethylrhodamine (TAMRA). Seven 10-fold dilutions with known cell numbers (ranging from 102 to 109 CFU/ml) of strain Bb-12 were used to spike fecal samples that were free of strain Bb-12, and the DNA extracted was used in real-time PCR to generate a standard curve for quantification of strain Bb-12. DNA from a pure culture of bifidobacteria diluted in seven 10-fold dilutions with known cell numbers (ranging from 102 to 109 CFU/ml) was also used as the template in TaqMan real-time PCR to generate standard curves for quantification of total bifidobacteria.
Table 2.
Primers and probes used for quantitative real-time PCR
| Target | Primer or probe | Sequence (5′→3′) | Concn (nM) | Reference or source |
|---|---|---|---|---|
| Bb-12a | F-Bal 23 | GGT GGT CTG GTA GAG TAT ACC G | 900 | Chr. Hansen Inc. |
| R-Bal 23 | GGC GAC TTG CGT CTT G | 900 | ||
| P-Bal 23 | CGC CCA CGA CCC GCA AG | 200 | ||
| Enterobacteria | F-En | CATGCCGCGTGTATGAAGAA | 900 | 12 |
| R-En | CGGGTAACGTCAATGAGCAAA | 900 | ||
| P-Bal 23 | CGC CCA CGA CCC GCA AG | 200 | ||
| Bifidobacterium genus | F-TAQ | GCG TCC GCT GTG GGC | 300 | 34 |
| R-TAQ | CTT CTC CGG CAT GGT GTT G | 300 | ||
| P-TAQ | TCC ACC GGC ACC AAG AAC GC | 200 | ||
| Clostridium cluster I | F-CI | TAC CHR AGG AGG AAG CCA C | 300 | 40 |
| R-CI | GTT CTT CCT AAT CTC TAC GCA T | 300 | ||
| P-CI | GTG CCA GCA GCC GCG GTA ATA CG | 200 |
These primers are specific for B. animalis subsp. lactis.
Quantification of clostridia and enterobacteria in feces by the use of TaqMan real-time PCR.
Specific primers and probes for Clostridium cluster I and enterobacteria used for amplification in real-time PCR are given in Table 2. Probes were labeled with the fluorescent dye FAM and black hole quencher (BHQ). Seven 10-fold dilutions with known cell numbers (ranging from 102 to 109 CFU/ml) of C. perfringens and E. coli were used to extract DNA. These DNAs were then used as templates in real-time PCR to generate standard curves for quantification of Clostridium cluster I and enterobacteria.
Real-time PCR.
Real-time PCR amplifications were performed on an ABI 7500 real-time PCR machine. Each reaction was carried out in duplicate in a volume of 25 μl using 96-well optical-grade ABI plates. The temperature settings for the different bacteria were as follows: one cycle of 95°C (10 min) and 40 cycles of 95°C (15 s) and 60°C (1 min) for strain Bb-12; one cycle of 95°C (10 min) and 40 cycles of 95°C (15 s) and 58°C (1 min) for total bifidobacteria; one cycle of 95°C (10 min) and 45 cycles of 95°C (15 s), 63°C (30 s), and 72°C (45 s) for clostridia; and one cycle of 95°C (10 min) and 45 cycles of 95°C (15 s) and 60°C (1 min) for enterobacteria. The CT (cycle at which the signal crosses a threshold) values were plotted as a linear function of the base 10 logarithm of the number of respective bacterial cells in the culture as determined by plate counts. These standard curves were then used to quantify the fecal samples with unknown cell concentrations collected during the study. Three negative controls, consisting of a no-template control and two controls for nontarget bacteria, were used to validate the specificity of each real-time PCR procedure.
Determination of numbers of live B. animalis subsp. lactis Bb-12 cells in feces.
Ethidium monoazide bromide (EMA) binds covalently to DNA upon exposure to light and has been validated to differentiate between live and dead cells by the use of PCR (36, 39). To validate this procedure for strain Bb-12 in feces, a 1-ml aliquot of a fully grown culture (8.3 × 108 CFU/ml) of strain Bb-12 was subjected to heat killing at 100°C for 10 min and incubated overnight at 4°C. Two 250-mg aliquots of feces with no detectable B. animalis subsp. lactis were spiked with 1 ml of either dead or live cells. EMA (100 μg/ml) was added to these samples, and the mixture was incubated in the dark at 4°C for 1 h. After incubation, the fecal samples were placed on ice and exposed for 1 h to 600 W of halogen light located at a distance of 20 cm. The samples were then used to extract DNA for real-time PCR. To differentiate between live and dead cells in fecal samples collected during the study, 100 μg of EMA/ml was added to them and the fecal DNA isolation procedure outlined above was repeated.
Statistical analysis.
For statistical analysis, SPSS software was used. Independent-sample t tests were used for between-subject analysis. Paired-sample t tests were used for within-subject analysis.
RESULTS
Enumeration of Bifidobacterium animalis subsp. lactis Bb-12, total bifidobacteria, clostridia, and enterobacteria in feces.
Standard curves were generated by plotting real-time PCR CT values obtained for each bacterial group against the initial number of cells in the culture (Fig. 2). The curves were found to be linear, with R2 values > 0.98, over the ranges of 105 to 109 CFU/ml for total bifidobacteria (105 was therefore taken as the limit of detection); 104 to 109 CFU/ml for strain Bb-12 (104 was taken as the limit of detection); 102 to 107 CFU/ml for clostridia (102 was taken as the limit of detection); and 103 to 108 CFU/ml for enterobacteria (103 was taken as the limit of detection). These curves were then applied to quantify the totals of intestinal bifidobacteria, Clostridium cluster I, enterobacteria, and strain Bb-12 in the fecal samples collected from subjects during the study. This revealed the variations in cell numbers of strain Bb-12, total bifidobacteria, clostridia, and enterobacteria in each subject over the study period.
Fig 2.
CT standard curves for real-time PCR quantitative analysis of B. animalis subsp. lactis Bb-12, total bifidobacteria, clostridia, and enterobacteria.
Detection and viability of strain Bb-12 in feces.
The Bb-12 primers did not detect any B. animalis subsp. lactis in fecal samples collected during the prefeeding period except for the first sample of one subject. As the second fecal sample was not positive, the subject was allowed to continue in the study. Strain Bb-12 was detected in the feces of 70% of subjects during the period in which they were ingesting the supplemented yogurt but in only 2 out of 22 subjects (9%) after 1 week of washout and none after 2 weeks. To evaluate viability using a live/dead PCR approach, a control experiment with fecal samples spiked with live or dead cells of strain Bb-12 was used to validate the procedure. This procedure was applied to 10 fecal samples that had tested positive for strain Bb-12 from 10 different subjects and revealed that strain Bb-12 was alive in 8 out of the 10 samples, substantiating its ability to survive gastric transit (Table 3).
Table 3.
Quantitative real-time PCR data for strain Bb-12 in the feces of 10 subjects before and after EMA treatment
| Sample | CT before EMA | Amt of Bb-12 (log 10 CFU/g) before EMA | CT after EMA | Amt of Bb-12 (log 10 CFU/g) after EMA | % viable Bb-12 in feces |
|---|---|---|---|---|---|
| 1 | 31.93 | 5.65 | 32.50 | 5.50 | 97.35 |
| 2 | 32.38 | 5.53 | 33.63 | 5.20 | 94.03 |
| 3 | 31.86 | 5.67 | 53.21 | 0.00 | 00.00 |
| 4 | 33.25 | 5.30 | 34.00 | 5.10 | 96.23 |
| 5 | 31.44 | 5.78 | 32.61 | 5.47 | 94.64 |
| 6 | 33.48 | 5.24 | 34.38 | 5.00 | 95.42 |
| 7 | 32.42 | 5.52 | 53.21 | 0.00 | 00.00 |
| 8 | 33.85 | 5.14 | 35.81 | 4.62 | 89.88 |
| 9 | 32.08 | 5.61 | 32.53 | 5.49 | 97.86 |
| 10 | 32.31 | 5.55 | 32.42 | 5.52 | 99.46 |
Total bifidobacterial counts in fecal samples.
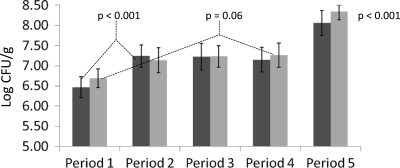
Prior to the feeding trial, the total bifidobacterial counts (as estimated by real-time PCR) in individual stool samples ranged from the detection limit of 5.0 log10 CFU/g to 9.0 log10 CFU/g, the mean bifidobacterial count being 7.2 log10 CFU/g. During the two feeding periods, no statistically significant difference was seen between the placebo and the probiotic groups. However, there was a fluctuating increase in the bifidobacterial numbers within the two groups (Fig. 3). The final washout period was marked with a significant increase in the bifidobacterial population in both groups compared to the prefeeding period (P < 0.001). However, a subgroup of subjects who had bifidobacterial counts < 108 CFU/g in their feces during the prefeeding period harbored a significant increase (P < 0.001 for group A and P = 0.06) when ingesting the synbiotic yogurt (Fig. 4).
Fig 3.
Mean numbers of total bifidobacteria for all group A subjects who consumed probiotic first and all group B subjects who consumed placebo first at different time points during the study.
Fig 4.
Fecal bifidobacterial levels in subjects subgrouped on the basis of bacterial numbers < 108 CFU/g in the prefeeding period. Period 1, prefeeding period; period 2, first supplemented yogurt feeding period; period 3, first washout period; period 4, crossover feeding period; period 5, final washout period. Group A is represented by the darker shading. P values represent differences between the yogurt feeding and final washout periods compared to the prefeeding period.
Clostridial counts in fecal samples.
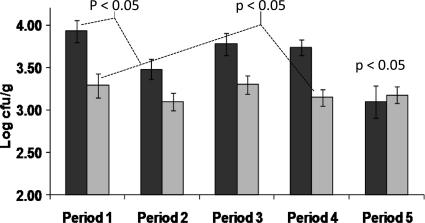
The initial clostridial counts in fecal samples were found to be 3.36 log10 CFU/g (the values ranged from the limit of detection, 2.0 log10 CFU/g, to 5.0 log10 CFU/g). The only statistically significant difference in clostridial numbers between the placebo and yogurt groups occurred during the second feeding period (P < 0.05) (Fig. 5). However, there was no reduction in numbers within each group in comparison to the prefeeding periods. There was a decrease in the clostridial numbers in the final washout period compared with the prefeeding levels in both groups. This reduction was not statistically significant for the whole washout period but was significant for the final fecal sample (P < 0.001 for group A and P < 0.05 for group B). This reduction was significant (P < 0.05) for the whole final washout period when subjects were subgrouped on the basis of high clostridial counts (>3.5 log10 CFU/g) in the prefeeding period (Fig. 6). This subgroup also exhibited a significant decrease in numbers of clostridia (P < 0.05) during the period in which the members of the subgroup were ingesting the supplemented yogurt compared to their prefeeding levels, which was consistent with the concomitant increase in total bifidobacterial numbers.
Fig 5.
Mean numbers of Clostridium cluster I for all group A subjects who consumed probiotic first and all group B subjects who consumed placebo first at different time points during the study. Values below the level of detection (102 CFU/g) have been equated to the value of the limit of detection of 102 CFU/g for mean calculations. The bars represent standard errors.
Fig 6.
Fecal Clostridium (cluster I) levels in subjects subgrouped on the basis of bacterial numbers > 3.5 log10 CFU/g in the prefeeding period. Period 1, prefeeding period; period 2, first supplemented yogurt feeding period; period 3, first washout period; period 4, crossover feeding period; period 5, final washout period. Group A is represented by the darker shading. P values represent differences between the yogurt feeding and final washout periods compared to the prefeeding period.
Enterobacterial counts in fecal samples.
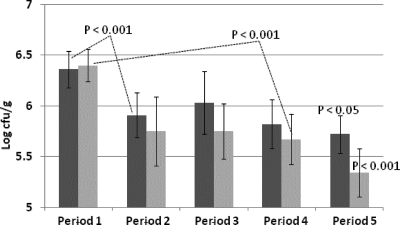
The initial mean Enterobacterium count in fecal samples was found to be 5.68 log10 CFU/g (the values ranged from 3.5 log10 CFU/g to 8.2 log10 CFU/g). There was no statistically significant decrease in Enterobacterium numbers between the synbiotic yogurt and placebo groups or between any group period and the corresponding prefeeding period (Fig. 7). However, subjects with initial Enterobacterium counts > 5.5 log10 CFU/g feces showed a decrease in mean Enterobacterium counts during the feeding study and this decrease was statistically significant (P < 0.001) for both groups (Fig. 8). However, there were no significant differences between the results seen with the synbiotic yogurt and placebo feeding periods even for this subgrouping.
Fig 7.
Mean numbers of enterobacteria for all group A subjects who consumed probiotic first and all group B subjects who consumed placebo first at different time points during the study. Values below the level of detection (103 CFU/g) have been equated to the value of the limit of detection of 103 CFU/g for mean calculations. The bars represent standard errors.
Fig 8.
Fecal enterobacterial levels in subjects subgrouped on the basis of bacterial numbers > 5.5 log10 CFU/g in the prefeeding period. Period 1, prefeeding period; period 2, first supplemented yogurt feeding period; period 3, first washout period; period 4, crossover feeding period; period 5, final washout period. Group A is represented by the darker shading. P values represent differences between the yogurt feeding and final washout periods compared to the prefeeding period.
DISCUSSION
The impact of the intestinal microbial community on human health is currently an expanding field of research. There is currently insufficient evidence to evaluate whether commonly used bifidobacteria, such as strains of B. animalis subsp. lactis, have a modulatory impact on the intestinal microbiota. In this study, we evaluated the effect of B. animalis subsp. lactis Bb-12 and inulin administered in a drinkable yogurt on the numbers of intestinal bifidobacteria, clostridia, and enterobacteria in 46 human volunteers.
As subjects were selected on the basis of not having consumed yogurt-type foods for 2 months, their initial fecal samples were negative for any B. animalis subsp. lactis. One subject's initial fecal sample tested positive, suggesting that that subject had unknowingly ingested it, as has also been found to have occurred in other studies (23, 30) and is plausible given the vast variety of available foods harboring this culture. The percentage of subjects who tested positive for strain Bb-12 during probiotic yogurt consumption in this study was consistent with studies that used comparable levels of this strain (7) and with the correlation between dosage and subsequent recovery of this probiotic from fecal samples (18). The rapid disappearance of Bb-12 from fecal samples following cessation of feeding is consistent with other published clinical studies (7, 22). Some studies have seen persistence in one or two subjects for a week or two longer (42). However, this may have been due to variations in subject reliability or to inadvertent consumption of other foods containing bifidobacteria rather than persistence of the species.
While the use of real-time PCR to monitor the intestinal flora facilitates studies using a larger number of subjects, it is usually limited by lack of data on the viability of the ingested culture. Some studies have included viable plate counts with real-time PCR (3). To address this limitation without including viable plate counts, which would limit the number of subjects that could be used, as fresh feces would be needed, we utilized a live/dead cell differentiation procedure to show that strain Bb-12 cells detected in feces retained their viability. As this procedure was demonstrated in this study to differentiate between live and dead cells of strain Bb-12 directly in feces, it indicated that the strain Bb-12 cells were viable in the majority of subjects at the time of freezing the samples.
The average number of bifidobacteria in human feces in adults has been reported to be ∼7.5 log10 CFU/g (23, 38). This was consistent with the total bifidobacterial counts in this study. Bifidobacterial numbers have been found to vary with age, with infants normally harboring larger numbers (∼1010 CFU/g of feces) (17, 31). Elderly people and those suffering from bowel diseases such as cancer have been reported to have lower numbers of bifidobacteria in their feces (33). In this study, older subjects (age > 50 years) were found to have lower numbers of bifidobacteria in their prefeeding fecal samples, and while the sample size was small, it is consistent with this trend. Some studies have reported an increase in bifidobacterial numbers during probiotic intake (23, 38). In our study, no significant differences between the synbiotic and the placebo groups during yogurt consumption were observed. This differs from another study using Bb-12, which did find a difference (1). However, that study had a subject group of just 10 compared to the 46 subjects in our study, indicating the probability of greater interindividual variability between subjects in this study. When subjects were subdivided on the basis of initial lower bifidobacterial numbers, an increase in total numbers of bifidobacteria during periods of probiotic yogurt consumption became evident (Fig. 4). An increase in the total numbers of bifidobacteria in subjects with low starting numbers of bifidobacteria has been observed previously (20, 22). Given that the total numbers of fecal microbes were not estimated in any of these studies, the possibility that such a result is linked to the fluctuations observed cannot be ruled out.
A significant increase in numbers of bifidobacteria for all groups in the final washout period was very evident and has not been seen in other studies. This significant increase in the final washout period was not due to irregularities with the real-time PCR quantification, as subject participation was staggered and quantification of samples was conducted randomly rather than sequentially. This may suggest that in a long probiotic feeding study, such as this 15-week study, some subjects may change their overall eating habits, given that they are constantly cognizant of the association of fermented foods and a favorable intestinal environment. This increase in bifidobacterial numbers corresponded with a significant decrease in numbers of clostridia but not in numbers of enterobacteria. An inverse correlation between bifidobacterial and clostridial numbers has been observed previously in bifidobacterium feeding studies (23, 33).
Several overall conclusions can be drawn from the study. Strain Bb-12 survived passage through the gastrointestinal tract but did not persist in the gut. Total numbers of bifidobacteria increased and clostridia (but not enterobacteria) decreased at the end of the study for all subjects irrespective of when they consumed the synbiotic yogurt or placebo (acifidied milk). This novel finding may reflect a general change of eating habits of subjects over the course of a long probiotic feeding study. It was also evident that subjects that had lower starting bifidobacterial levels or higher starting clostridial levels benefited the most (by increasing total numbers of bifidobacteria or decreasing numbers of clostridia) from ingesting the supplemented yogurt. However, there were no significant differences in enterobacterial numbers between the synbiotic yogurt and placebo feeding periods even in the subgroup of subjects that had higher initial levels of enterobacteria. These data would therefore support the idea that ingestion of this supplemented yogurt does not statistically modulate enterobacterial numbers but may modulate total bifidobacterial and clostridial numbers in people with either below-average levels of bifidobacteria or above-average levels of clostridia.
ACKNOWLEDGMENTS
This study was funded by General Mills Inc.
We thank Ravi Menon and Maeve Murphy for suggestions throughout the study.
Footnotes
Published ahead of print 18 November 2011
REFERENCES
- 1. Alander M, et al. 2001. Effect of galacto-oligosaccharide supplementation on human faecal microflora and on survival and persistence of Bifidobacterium lactis Bb-12 in the gastrointestinal tract. Int. Dairy J. 11:817–825 [Google Scholar]
- 2. Barnes EM, Ingram M. 1956. The effect of redox potential on the grown Clostridium welchii strain isolated from horse muscle. J. Appl. Bacteriol. 19:177–178 [Google Scholar]
- 3. Bartosch S, Woodmansey EJ, Paterson JCM, McMurdo MET, Macfarlane GT. 2005. Microbiological effects of consuming a synbiotic containing Bifidobacterium bifidum, Bifidobacterium lactis, and oligofructose in elderly persons, determined by real time polymerase chain reaction and counting of viable bacteria. Clin. Infect. Dis. 40:28–37 [DOI] [PubMed] [Google Scholar]
- 4. Cai YM, Matsumoto M, Benno Y. 2000. Bifidobacterium lactis Meile et al. 1997 is a subjective synonym of Bifidobacterium animalis (Mitsuoka 1969) Scardovi and Trovatelli 1974. Microbiol. Immunol. 44:815–820 [DOI] [PubMed] [Google Scholar]
- 5. Fallani M, et al. 2006. Clostridium difficile and Clostridium perfringens species detected in infant faecal microbiota using 16s rRNA targeted probes. J. Microbiol. Methods 67:150–161 [DOI] [PubMed] [Google Scholar]
- 6. Fooks LJ, Gibson GR. 2002. Probiotics as modulators of the gut flora. Br. J. Nutr. 88:S39–S49 [DOI] [PubMed] [Google Scholar]
- 7. Fukushima Y, Kawata Y, Hara H, Terada A, Mitsuoka T. 1998. Effect of a probiotic formula on intestinal immunoglobulin A production in healthy children. Int. J. Food Microbiol. 42:239–244 [DOI] [PubMed] [Google Scholar]
- 8. Gibson GR, Probert HM, Loo JV, Rastall RA, Roberfroid MB. 2004. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr. Res. Rev. 17:259–275 [DOI] [PubMed] [Google Scholar]
- 9. Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401–1412 [DOI] [PubMed] [Google Scholar]
- 10. Hébuterne X. 2003. Gut changes attributed to ageing: effects on intestinal microflora. Curr. Opin. Clin. Nutr. Metab. Care 6:49–54 [DOI] [PubMed] [Google Scholar]
- 11. Hopkins MJ, MacFarlane GT. 2002. Changes in predominant bacterial populations in human feces with age and with Clostridium difficile infection. J. Med. Microbiol. 51:448–454 [DOI] [PubMed] [Google Scholar]
- 12. Huijsdens XW, et al. 2002. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J. Clin. Microbiol. 40:4423–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Islam A. 2005. Iron reversible inhibition by bifidobacteria and microbial diversity of the human intestine, p 40–42 M.S. thesis. University of Minnesota, St. Paul, MN [Google Scholar]
- 14. Isolauri E, Arvola T, Sutas Y, Moilanen E, Salminen S. 2000. Probiotics in the management of atopic eczema. Clin. Exp. Allergy 30:1604–1610 [DOI] [PubMed] [Google Scholar]
- 15. Kaper JB, Karmali MA. 2008. The continuing evolution of a bacterial pathogen. Proc. Natl. Acad. Sci. U. S. A. 105:4535–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 17. Kirjavainen PV, Arvola T, Salminen SJ, Isolauri E. 2002. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut 51:51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Larsen CN, et al. 2006. Dose-response study of probiotic bacteria Bifidobacterium animalis subsp. lactis BB-12 and Lactobacillus paracasei subsp. paracasei CRL-341 in healthy young adults. Eur. J. Clin. Nutr. 60:1284–1293 [DOI] [PubMed] [Google Scholar]
- 19. Lee JH, et al. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Link-Amster H, Rochat F, Saudan KY, Mignot O, Aeschlimann JM. 1994. Modulation of a specific humoral immune response and changes in intestinal flora mediated through fermented milk intake. FEMS Immunol. Med. Microbiol. 10:55–63 [DOI] [PubMed] [Google Scholar]
- 21. Lyerly DM, Krivan HC, Wilkins TD. 1988. Clostridium difficile: its disease and toxins. Clin. Microbiol. Rev. 1:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malinen E, et al. 2002. PCR-ELISA. II: analysis of Bifidobacterium populations in human faecal samples from a consumption trial with Bifidobacterium lactis Bb-12 and a galacto-oligosaccharide preparation. Syst. Appl. Microbiol. 25:249–258 [DOI] [PubMed] [Google Scholar]
- 23. Matto J, et al. 2006. Intestinal survival and persistence of probiotic Lactobacillus and Bifidobacterium strains administered in triple-strain yoghurt. Int. Dairy J. 16:1174–1180 [Google Scholar]
- 24. Meile L, et al. 1997. Bifidobacterium lactis. sp. nov., a moderately oxygen tolerant species isolated from fermented milk. Syst. Appl. Microbiol. 20:57–64 [Google Scholar]
- 25. Mitsuoka T. 1990. Bifidobacteria and their role in human health. J. Ind. Microbiol. 6:263–267 [Google Scholar]
- 26. Muñoa FJ, Pares R. 1988. Selective medium for isolation and enumeration of Bifidobacterium spp. Appl. Environ. Microbiol. 54:1715–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Olsvik O, Wasteson Y, Lund A, Hornes E. 1991. Pathogenic Escherichia coli found in food. Int. J. Food Microbiol. 12:103–113 [DOI] [PubMed] [Google Scholar]
- 28. O'Sullivan DJ. 2001. Screening of intestinal microflora for effective probiotic bacteria. J. Agric. Food Chem. 49:1751–1760 [DOI] [PubMed] [Google Scholar]
- 29. O'Sullivan DJ. 2005. Primary sources of probiotic cultures, p 89–105 In Gopteke I, Juneja VK, Ahmedna M. (ed), Probiotics in food safety and human health. CRC Press, Taylor & Francis Group, Boca Raton, FL [Google Scholar]
- 30. Ouwehand AC, Kurvinen T, Rissanen P. 2004. Use of a probiotic Bifidobacterium in a dry food matrix, an in vivo study. Int. J. Food Microbiol. 95:103–106 [DOI] [PubMed] [Google Scholar]
- 31. Penders J, et al. 2005. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol. Lett. 243:141–147 [DOI] [PubMed] [Google Scholar]
- 32. Playne MJ. 2002. The health benefits of probiotics. Food Aust. 54:71–74 [Google Scholar]
- 33. Rafter J, et al. 2007. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am. J. Clin. Nutr. 85:488–496 [DOI] [PubMed] [Google Scholar]
- 34. Requena T, et al. 2002. Identification, detection, and enumeration of human Bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 68:2420–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roberfroid MB. 2005. Introducing inulin-type fructans. Br. J. Nutr. 93(Suppl. 1):S13–S25 [DOI] [PubMed] [Google Scholar]
- 36. Rudi K, Moen B, Drømtorp SM, Holck AL. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71:1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saavedra JM, et al. 1994. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet 344:1046–1049 [DOI] [PubMed] [Google Scholar]
- 38. Schiffrin EJ, Rochat F, Link-Amster H, Aeschlimannp JM, Donnet-Hughes A. 1995. lmmunomodulation of human blood cells following the ingestion of lactic acid bacteria. J. Dairy Sci. 78:491–497 [DOI] [PubMed] [Google Scholar]
- 39. Soejima T, et al. 2007. Photoactivated ethidium monoazide directly cleaves bacterial DNA and is applied to PCR for discrimination of live and dead bacteria. Microbiol. Immunol. 51:763–775 [DOI] [PubMed] [Google Scholar]
- 40. Song Y, Liu C, Finegold SM. 2004. Real-time PCR quantitation of clostridia in feces of autistic children. Appl. Environ. Microbiol. 70:6459–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stackebrandt E, Kramer I, Swiderski J, Hippe H. 1999. Phylogenetic basis for a taxonomic dissection of the genus Clostridium. FEMS Immunol. Med. Microbiol. 24:253–258 [DOI] [PubMed] [Google Scholar]
- 42. Su P, et al. 2005. Detection and quantification of Bifidobacterium lactis LAFTI B94 in human faecal samples from a consumption trial. FEMS Microbiol. Lett. 244:99–103 [DOI] [PubMed] [Google Scholar]
- 43. Topping DL, Clifton PM. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81:1031–1064 [DOI] [PubMed] [Google Scholar]