Abstract
Dermacentor andersoni and Dermacentor variabilis from allopatric and sympatric populations near their northern distributional limits were examined for the presence of Francisella species using molecular techniques that targeted 373 bp of the 16S rRNA gene. Although there was no evidence for the presence of Francisella tularensis in any tick, Francisella-like endosymbionts (FLEs) were common in D. andersoni and D. variabilis adults and immatures. A significantly greater proportion of female ticks contained FLEs compared to male ticks. In addition, significantly more D. variabilis adult individuals contained multiple FLE sequence types than did D. andersoni adults. Ten different types of FLEs were identified based on the sequence data, which has implications for diagnostic tests and epidemiological studies of F. tularensis in tick populations in Canada. The three most prevalent types of FLEs have been detected previously in D. andersoni or D. variabilis from other parts of their distributional ranges, whereas the other seven FLE types have not been reported previously. A comparison of the FLEs from both allopatric and sympatric populations of these two tick species provided insight into the relative host-specificity and the modes of transmission of these tick-borne bacteria. In general, each FLE type was specific for one tick species, suggesting vertical transmission of each bacterium. However, there were a few instances of potential cross-transfer of two FLE types to the other tick species at locations where D. andersoni and D. variabilis occurred in sympatry, suggesting that there may be occasional horizontal transmission of some FLEs.
INTRODUCTION
There are four recognized species within the genus Francisella (31), three of which can cause disease in humans (53). Francisella tularensis, the causative agent of tularemia in the Northern Hemisphere (16), varies considerably in its transmission patterns, virulence and disease presentation in different geographical areas (13, 38, 45, 50, 57). In North America, there are two common subspecies of F. tularensis: F. tularensis subsp. tularensis and F. tularensis subsp. holarctica (16). Human infections with F. tularensis subsp. holarctica are mainly acquired through direct contact with infected beavers, muskrats, or lagomorphs, whereas in the United States, F. tularensis subsp. tularensis is often acquired by tick bites (8, 12). The American dog tick, Dermacentor variabilis Say, 1821, and the Rocky Mountain wood tick, Dermacentor andersoni Stiles, 1908, are important for the transmission of F. tularensis in the eastern and western United States, respectively (16, 23). The distributional ranges of D. andersoni and D. variabilis are largely allopatric (i.e., separate), except in some parts of Montana, Wyoming, Colorado, North Dakota, and South Dakota (2, 35, 44, 54, 61) and in Saskatchewan, Canada (S. J. Dergousoff and N. B. Chilton, unpublished data), where they occur in sympatry. These two tick species, as well as Dermacentor albipictus, Dermacentor occidentalis, Dermacentor hunteri, and Dermacentor nitens, are also hosts of a number of bacteria that are closely related to F. tularensis (24, 46, 51). These so-called Francisella-like endosymbionts (FLEs) are generally of undetermined pathogenicity but sometimes assumed to be nonpathogenic (15, 51). However, infection studies with the “Dermacentor andersoni symbiont” (DAS) showed it to be pathogenic for chicken embryos and guinea pigs (6).
Francisella tularensis is also endemic in Canada (62). However, compared to the United States, relatively few human cases of tularemia have been documented (5, 34), some of which have occurred in Saskatchewan and Alberta in western Canada (5, 7, 30, 42, 49). Most human cases of tularemia in western Canada have been associated with contact with infected wildlife (3, 36, 43, 52, 60) or livestock (4, 28). Sporadic occurrences of tularemia have been reported in beavers (40), muskrats (18, 40), jackrabbits (4), snowshoe hares (62), ground squirrels (4), and sheep (28). The most recent outbreak occurred in deer mice (Peromyscus maniculatus), following a population explosion in 2005 near Madison, Saskatchewan. The causative agent was identified as F. tularensis subsp. holarctica, but the source of infection was not determined (63).
Although the transmission cycle of F. tularensis in Canada is not well defined, ticks (Dermacentor spp.) have been implicated as potential vectors in western Canada. For example, adult D. andersoni was important in some of the first recognized cases of human and animal tularemia in southern Alberta (4, 28). Francisella tularensis has been recovered from D. andersoni in British Columbia, southern Alberta, and southern Saskatchewan during surveys conducted between 1938 and 1946 (20, 32, 33). In 1982, F. tularensis was detected in adult D. andersoni from Saskatchewan Landing Provincial Park based on the results of transmission experiments in rabbits (26). Despite these reports, the prevalence of F. tularensis and FLEs in ticks in western Canada is unknown. The aim of the present study was to use PCR-based methods to determine the prevalence of Francisella and FLEs in sympatric and allopatric populations of D. andersoni and D. variabilis from 12 localities near their northern distributional limits, which includes Saskatchewan Landing Provincial Park, a location where F. tularensis in adult ticks has been detected previously. In addition, we examined if the different types of FLEs were specific for either tick species in sympatric populations of D. andersoni and D. variabilis.
MATERIALS AND METHODS
Collection of ticks.
A total of 1,042 adult male and female ticks (425 D. andersoni and 617 D. variabilis) were collected by flagging grass and shrubs or were removed from vertebrate hosts at 12 localities in southwestern Canada (Table 1). Questing ticks obtained by flagging were collected in May and June of 2005 and from April to June in 2006. Some adult ticks were also collected from humans, horses, dogs, skunks, and raccoons between May and June in 2005 and 2007. Immature ticks (nine D. andersoni nymphs, six D. variabilis nymphs and 71 D. variabilis larvae) were collected from 13 deer mice (Peromyscus maniculatus), seven meadow voles (Microtus pennsylvanicus), and eight western jumping mice (Zapus princeps) trapped in Saskatchewan Landing Provincial Park in June and July of 2008, and in April of 2009. An additional seven D. variabilis nymphs and 143 D. variabilis larvae were collected from one deer mouse, one western jumping mouse, five meadow voles and 10 southern red-backed voles (Myodes gapperi) from Blackstrap Provincial Park (Saskatchewan) in May through July of 2009. All adult ticks were identified based on morphological examination. Adults of D. andersoni and D. variabilis are easily distinguished from one another, and from those of D. albipictus (a species that occurs in sympatry with the other two species [61]), based on differences in the morphology of their spiracular plates (27). The species identity of representative individuals was also verified using a PCR-based assay (11). Immature ticks were examined by microscopy to confirm they belonged to the Metastriata. Their species identity was determined using the same PCR assay as for the adults; however, amplicons were subjected to a restriction fragment length polymorphism (RFLP) analysis using AluI (Fermentas) according to the manufacturer's instructions. The RFLP analysis was used to confirm that the internal transcribed spacer 2 (ITS-2) amplicons of D. variabilis individuals were not from D. albipictus, which has an ITS-2 amplicon of the same size. The ITS-2 sequence of D. variabilis lacks the restriction site for AluI present in the ITS-2 sequence of D. albipictus (11).
Table 1.
Localities and coordinates of the collection sites of adult D. andersoni and D. variabilis, and the number of ticks positive for the presence of the Francisella 16S rRNA gene at each locality
| Locality | Coordinates (decimal degrees) |
D. andersoni |
D. variabilis |
|||
|---|---|---|---|---|---|---|
| Latitude (north) | Longitude (west) | No. tested | No. Francisellapositive (%) | No. tested | No. Francisellapositive (%) | |
| Lethbridge, Alberta | 49.73721 | 112.84751 | 100 | 73 (73) | –a | |
| Saskatchewan Landing Provincial Park, Saskatchewan | 50.64528 | 107.96310 | 82 | 79 (96) | 96 | 92 (96) |
| Grasslands National Park, Saskatchewan | 49.21666 | 107.70000 | 17 | 15 (88) | 1 | 1 (100) |
| Buffalo Pound Provincial Park, Saskatchewan | 50.57582 | 105.31356 | 33 | 28 (85) | 79 | 70 (89) |
| Douglas Provincial Park, Saskatchewan | 51.02966 | 106.46590 | 14 | 14 (100) | 40 | 40 (100) |
| Danielson Provincial Park, Saskatchewan | 51.25933 | 106.89580 | 61 | 59 (97) | 99 | 98 (99) |
| Outlook, Saskatchewan | 51.48807 | 107.05817 | 18 | 16 (89) | 12 | 11 (92) |
| Harris, Saskatchewan | 51.73448 | 107.58370 | 100 | 83 (83) | 12 | 9 (75) |
| Blackstrap Provincial Park, Saskatchewan | 51.79760 | 106.45833 | –a | 105 | 95 (91) | |
| Wakaw, Saskatchewan | 52.60297 | 105.85426 | –a | 44 | 44 (100) | |
| Minnedosa, Manitoba | 50.24715 | 99.83870 | –a | 99 | 85 (86) | |
| Kenora, Ontario | 49.90153 | 94.49324 | –a | 30 | 28 (93) | |
| Total | 425 | 367 (86) | 617 | 573 (93) | ||
Tick species does not occur at this locality.
DNA preparation.
Total genomic DNA (gDNA) was extracted and purified from each tick using a modification of the tissue protocol for the DNeasy tissue kit (Qiagen, Valencia, CA). Individual ticks were placed in 1.5-ml micropestle tubes (Kontes), to which 180 μl of buffer ATL (Qiagen) was added. Ticks were homogenized by grinding with micropestles (Kontes) attached to a cordless drill. Proteinase K (20 μl of 15 μg/μl) was added to the homogenate. Samples were incubated for 16 h at 55°C. The gDNA was purified according to the DNeasy tissue kit protocol, except that gDNA was eluted twice from the spin columns using 50 μl of buffer AE. The two elutions derived from the same tick were combined in a single tube and stored at −20°C.
PCR and single-strand conformation polymorphism (SSCP) of 16S rRNA gene.
The presence of Francisella DNA in adult ticks was tested using a PCR targeting 373 bp of the bacterial 16S rRNA gene with the primers NC-Fran16S-F (5′-CAACATTCTGGACCGAT-3′) and NC-Fran16S-R (5′-TGCGGGACTTAACCCAACAT-3′), which were designed to be specific for F. tularensis and FLEs. PCRs were carried out in 25-μl volumes containing 200 μM each deoxynucleoside triphosphate (dNTP) (Fermentas, Burlington, Canada), 3 mM MgCl2, 25 pmol (1 μM) of each primer, 0.5 U of recombinant Taq DNA polymerase (Fermentas), 2.5 μl 10× PCR buffer with (NH4)2SO4 (Fermentas), and 2 μl of template gDNA. A negative control (i.e., without gDNA) and positive control were included in each set of reactions. PCRs were performed in a thermocycler (iCycler; Bio-Rad, Hercules, CA) using the following conditions: 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 5 min. The presence of Francisella DNA in immature ticks was tested using a nested PCR (nPCR) because of the possibility that the relative numbers of bacteria within an individual larva or nymph may be significantly lower than in an adult tick. The first phase of the nPCR targeted 1,141 bp of the bacterial 16S rRNA gene with primers F11 (5′-TACCAGTTGGAAACGACTGT-3′) and F5 (5′-CCTTTTTGAGTTTCGCTCC-3′) (17). Each reaction mixture contained 200 μM each dNTP (Fermentas), 2.5 mM MgCl2, 25 pmol (1 μM) of each primer, 0.5 U of recombinant Taq DNA polymerase (iTaq; Bio-Rad), 2.5 μl 10× PCR buffer (Bio-Rad), and 2 μl of template gDNA. The PCR conditions used were 95°C for 5 min, followed by 30 cycles of 95°C for 60 s, 65°C for 60 s, and 72°C for 60 s and a final extension at 72°C for 5 min. PCR products, including those of the negative controls, were then purified by adding shrimp alkaline phosphatase (0.014 U/μl) (New England BioLabs, Pickering, Canada) and exonuclease I (0.27 U/μl) (Fermentas) and incubating the mixture at 37°C for 15 min and then at 80°C for 15 min. The second phase of the nPCR was conducted with 2 μl of purified PCR products (including the negative-control samples) using primers NC-Fran16S-F and NC-Fran16S-R and the same PCR conditions used for the adult ticks. Additional negative-control samples were also included. Amplicons were subjected to electrophoresis on SYBR safe-stained 1.5% agarose-Tris-borate-EDTA (TBE) gels, and their banding patterns were visualized by UV transillumination. Some of the initial amplicons produced from the adult ticks were subjected to DNA sequencing to verify the specificity of the PCR assay.
SSCP analyses (11) were performed on all samples that were PCR positive. This mutation scanning technique can be used to differentially display DNA sequences that differ by one or more nucleotides (19). In the present study, SSCP was used to prescreen all amplicons for genetic variation before selecting representative samples of each different SSCP profile for DNA sequencing. Samples that were sequenced previously were used as mobility controls in SSCP gels. Where possible, multiple amplicons of each SSCP profile were prepared for DNA sequencing.
DNA sequence analyses.
Amplicons from 76 adult ticks and 21 immature ticks were column purified (MinElute DNA purification kit; Qiagen) and then sequenced using primers NC-Fran16S-F and NC-Fran16S-R in separate reactions. The 16S rRNA gene sequences, excluding primer sites, were manually aligned, and a BLAST search was performed to determine sequence similarity with those of other bacterial 16S rRNA gene sequences deposited in GenBank. Included in the analyses were the 16S rRNA gene sequences of different subspecies of F. tularensis. A minimum spanning network tree depicting the relationships of the different genetic types of FLEs was constructed using the TCS program (Fig. 1) (9).
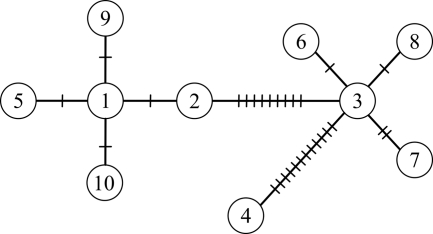
Fig 1.
Minimum spanning network tree depicting the relationships of the 10 FLE sequence types detected in D. andersoni and D. variabilis from western Canada, based on comparisons of the partial sequence of the 16S rRNA gene. FLE types 5, 9, and 10 were found exclusively in D. andersoni, while types 3, 4, 6, 7, and 8 were found only in D. variabilis. Types 1 and 2 were detected in both D. andersoni and D. variabilis. Hatch marks indicate the number of nucleotide differences between sequences of adjacent types.
Nucleotide sequence accession numbers.
The sequences of representative samples obtained in the present study have been deposited in GenBank under accession numbers FR872824 to FR872833.
RESULTS
A total of 1,042 adult and 236 immature (214 larvae and 22 nymphs) ticks were tested individually for the presence of Francisella DNA by PCR. All samples that were positive by PCR produced a single band of the expected size (approximately 370 bp) on an agarose gel. The proportion of adult ticks at each locality that were PCR positive for Francisella DNA ranged from 73% to 100%; however, significantly more D. variabilis (93%) were positive than D. andersoni adults (86%) (χ21 = 12.09, P < 0.05, n = 1,042) (Table 1). There were also significant differences in the proportion of male and female D. andersoni (82% and 90%, respectively) (χ21 = 378.29, P < 0.001, n = 425) and D. variabilis (88% and 97%, respectively) (χ21 = 150.10, P < 0.001, n = 617) that were PCR positive for Francisella DNA. For the immature ticks, 89% of the D. andersoni nymphs, 69% of the D. variabilis nymphs, and 52% of the D. variabilis larvae were PCR positive for Francisella DNA (Table 2). A significantly lower proportion of D. variabilis larvae from Saskatchewan Landing Provincial Park were PCR positive for Francisella DNA than those from Blackstrap Provincial Park (χ21 = 26.75, P < 0.001, n = 214).
Table 2.
Localities of the collection sites of D. andersoni nymphs and D. variabilis larvae and nymphs, and the number of ticks positive for the presence of the Francisella 16S rRNA gene at both localities
| Locality |
D. andersoni nymphs |
D. variabilis larvae |
D. variabilis nymphs |
|||
|---|---|---|---|---|---|---|
| No. tested | No. Francisellapositive (%) | No. tested | No. Francisellapositive (%) | No. tested | No. Francisellapositive (%) | |
| Blackstrap Provincial Park | –a | –a | 143 | 92 (64) | 7 | 7 (100) |
| Saskatchewan Landing Provincial Park | 9 | 8 (89) | 71 | 19 (27) | 6 | 2 (33) |
This species does not occur at this locality.
At least 20 different SSCP banding patterns (i.e., profiles) were detected among the 1,068 PCR products. Many amplicons had SSCP patterns that were comprised of at least two different patterns, suggesting that some ticks contained more than one sequence type that differed from one another by one or more nucleotides. DNA sequencing analyses of representative amplicons of each SSCP banding pattern type revealed that the bacteria present in ticks were not F. tularensis but Francisella-like endosymbionts. A total of 10 different sequence types of FLEs were identified among the tick samples (Tables 3 and 4), some of which were represented by more than one SSCP banding pattern. Multiple FLE sequence types were detected in 24% of the adult ticks, as evident by the presence of two nucleotides at one or more positions in the DNA sequence. The proportion of PCR positive adult ticks that contained more than one sequence type was significantly different between D. andersoni (3%) and D. variabilis (38%) (χ21 = 150.18, P < 0.001, n = 940) (Table 3). None of the 128 Dermacentor immatures contained multiple FLE types (Table 4).
Table 3.
Genetic types of FLEs detected in adult D. andersoni and D. variabilis from each locality
| Localitya | FLE types in D. andersoni |
FLE types in D. variabilis |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 1 and 2 | 1 and 9 | 1 and 10 | 1 | 2 | 3 | 4 | 8 | 2 and 3 | 2, 3, and 4 | 2 and 7 | 3 and 4 | 3 and 7 | 3 and 8 | |
| Allopatric tick populations | ||||||||||||||||
| Lethbridge | 73 | 0 | 0 | 0 | 0 | |||||||||||
| Blackstrap Provincial Park | 0 | 33 | 14 | 0 | 0 | 47 | 0 | 0 | 0 | 1 | 0 | |||||
| Wakaw | 0 | 34 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | |||||
| Minnedosa | 0 | 44 | 4 | 1 | 0 | 34 | 0 | 0 | 0 | 1 | 1 | |||||
| Kenora | 0 | 19 | 0 | 0 | 0 | 8 | 0 | 0 | 1 | 0 | 0 | |||||
| Sympatric tick populations | ||||||||||||||||
| Saskatchewan Landing Provincial Park | 76 | 0 | 2 | 1 | 0 | 0 | 55 | 2 | 0 | 0 | 35 | 0 | 0 | 0 | 0 | 0 |
| Grasslands National Park | 15 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Buffalo Pound Provincial Park | 27 | 1 | 0 | 0 | 0 | 0 | 34 | 8 | 0 | 0 | 24 | 2 | 1 | 1 | 0 | 0 |
| Douglas Provincial Park | 13 | 0 | 0 | 0 | 1 | 0 | 24 | 0 | 0 | 0 | 16 | 0 | 0 | 0 | 0 | 0 |
| Danielson Provincial Park | 59 | 0 | 0 | 0 | 0 | 1 | 64 | 3 | 0 | 1 | 29 | 0 | 0 | 0 | 0 | 0 |
| Outlook | 16 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 |
| Harris | 76 | 0 | 3 | 4 | 0 | 0 | 7 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| Total | 355 | 1 | 5 | 5 | 1 | 1 | 320 | 31 | 1 | 1 | 211 | 2 | 1 | 2 | 2 | 1 |
Coordinates given in Table 1.
Table 4.
Genetic types of FLEs detected in D. andersoni nymphs and D. variabilis larvae and nymphs from each locality
| Locality | FLE types in D. andersoni |
FLE types in D. variabilis |
||||
|---|---|---|---|---|---|---|
| 1 | 5 | 2 | 3 | 6 | 7 | |
| Saskatchewan Landing Provincial Park | 7 | 1 | 93 | 4 | 1 | 1 |
| Blackstrap Provincial Park | 0 | 0 | 20 | 0 | 0 | 1 |
There were 29 variable nucleotide positions in the alignment of the partial 16S rRNA gene sequences (321 to 336 bp) of the 10 FLEs and F. tularensis (Table 5). This included a 12-bp deletion (positions 172 to 183) in the sequence of FLE type 4, which was otherwise identical in sequence to FLE type 3. Pairwise comparisons of all 10 sequences revealed differences at one to 21 nucleotide positions between each type of FLE. A comparison of the partial 16S rRNA gene sequences of the 10 FLEs with sequence data available on GenBank revealed that FLE types 4 to 10 had unique sequences, whereas the sequences of FLE types 1 to 3 were reported previously. For instance, the 336-bp sequence of FLE type 1 was identical to the 16S rRNA gene sequence of FLEs detected in D. andersoni (GenBank accession no. AY375397 and AY375398 [51]). FLE type 2 was identical in sequence to an FLE in D. variabilis (accession no. AY795979 [24] and AY375406 [51]), and FLE type 3 was identical to that of a Francisella-like Dermacentor variabilis endosymbiont (“dermacentor variabilis francisella”) in D. variabilis (accession no. AY795976 to AY795978 [24]). The sequences of all FLE types differed ≥1 bp compared to the 16S rRNA gene sequences of the different subspecies of F. tularensis.
Table 5.
Multiple sequence alignment of the 29 variable nucleotide positions of the 16S rRNA gene fragment (336 bp) obtained from the 10 FLE types found in D. andersoni and D. variabilis adults from 12 localities in western Canada and for the different subspecies of Francisella tularensis
| Taxon | Alignment positiona |
||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 61 | 82 | 172 | 173 | 174 | 175 | 176 | 177 | 178 | 179 | 180 | 181 | 182 | 183 | 188 | 189 | 196 | 198 | 206 | 208 | 215 | 217 | 227 | 260 | 287 | 288 | 289 | 306 | 326 | |
| Type 1 | G | C | G | A | A | T | T | G | A | C | G | G | G | G | G | C | G | T | G | A | T | C | C | T | G | G | G | C | C |
| Type 5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . |
| Type 9 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Type 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . |
| Type 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| Type 4 | A | T | – | – | – | – | – | – | – | – | – | – | – | – | . | . | T | G | C | G | . | T | T | . | A | . | . | . | . |
| Type 3 | A | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | T | G | C | G | . | T | T | . | A | . | . | . | . |
| Type 6 | A | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | T | G | C | G | . | T | T | . | A | . | . | . | G |
| Type 8 | A | T | . | . | . | . | . | . | . | . | A | . | . | . | . | T | T | G | C | G | . | T | T | . | A | . | . | . | . |
| Type 7 | A | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | T | G | C | G | . | T | T | . | A | A | T | . | . |
| F. tularensisb | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | A | . | . | . | . | . |
A dot (.) represents the identical nucleotide, and a dash (-) indicates a deletion with respect to the sequence of the type 1 FLE. The alignment position indicates the nucleotide position relative to the 3′ end of the forward primer.
Most of the 10 FLE types were found in either D. andersoni or D. variabilis. However, two of the FLE sequence types occurred in adult ticks of both species. FLE type 1 was the most prevalent sequence variant in adult D. andersoni, found in 99.7% of the PCR positive ticks; however, it also occurred in a single D. variabilis adult (<1%) (Table 3). This was also the most prevalent FLE type in the D. andersoni nymphs (Table 4). The FLE type 2 sequence variant was the most prevalent type in D. variabilis, occurring in 93% of PCR positive adult ticks and 94% of the PCR positive immature ticks (Tables 3 and 4). This type also occurred in five (1%) D. andersoni females: three from Harris and two from Saskatchewan Landing Provincial Park. All five of these ticks also contained FLE type 1 (Table 3). Infections of FLE type 1 in D. variabilis and FLE type 2 in D. andersoni appeared to be rare and only occurred at localities where these tick species were sympatric. In contrast, seven of the FLE types, which occurred at relatively low prevalences in the present study, were found exclusively in a single tick species. The DNA sequences of the least common FLE types in D. andersoni (types 5, 9, and 10) differed by a single nucleotide compared to the sequence of type 1, the most prevalent type in D. andersoni. Similarly, the FLE types which were relatively rare in D. variabilis (i.e., types 4, 6, 7, and 8) had 16S rRNA gene sequences most similar to that of FLE type 3 (Fig. 1), the second most common type of FLE in D. variabilis (Table 3). FLE type 3 was detected in 44% of the D. variabilis adults found positive by PCR. Most (85%) of D. variabilis adults infected with FLE type 3 also contained FLEs of type 2 (Table 3). FLE type 6 was only detected in a single D. variabilis larva (Table 4).
DISCUSSION
Francisella tularensis is endemic in western Canada (42, 62). Despite this, little is known of the relative prevalence of this pathogen in different geographical areas, or of its natural transmission cycle, in western Canada. In the present study, we did not detect, using PCR-SSCP and DNA sequencing, the presence of F. tularensis in any of the 1,278 ticks collected from 12 localities in western Canada. This includes Saskatchewan Landing Provincial Park, a locality where, in 1982, questing D. andersoni adults were found to be infected with F. tularensis (26). However, this was not unexpected since F. tularensis infections in ticks are often sporadic (25). This appears to be the case for ticks in Saskatchewan Landing Provincial Park because F. tularensis was also not detected in any D. andersoni collected in 1971 and between 1974 and 1981 (26). Given that F. tularensis is often maintained in endemic foci at a low prevalence (21, 25, 29), estimations of the prevalences of F. tularensis in northern populations of D. andersoni and D. variabilis in western Canada will require surveys to be conducted involving a large number of ticks from numerous locations over multiple consecutive years.
Although F. tularensis was not detected in D. andersoni or D. variabilis in our study, we did detect the DNA of FLEs in a large proportion of adult ticks at each locality. This was not surprising given that FLEs have been reported from many genera of ticks (10, 41, 46, 47, 51, 58, 59). The combined results of our SSCP and DNA sequencing analyses revealed the existence of 10 types of FLEs. The partial 16S rRNA gene sequences of seven FLEs (types 4 to 10) represented new sequence types based on comparisons with sequence data available in GenBank, whereas the three most prevalent types of FLEs (types 1, 2, and 3) were identical in sequence to FLEs reported in D. andersoni or D. variabilis from a number of locations in the United States and Canada (24, 39, 51). The partial 16S sequences of FLE types 1 and 2 were also identical in sequence to those previously found in other species of Dermacentor in North America (39, 51). Therefore, a larger fragment of the 16S rRNA gene or a second genetic marker would be needed to distinguish FLE types 1 to 3 in D. andersoni and D. variabilis from those FLEs in the other species of Dermacentor. Nonetheless, we were able to identify different genotypes of FLEs in D. andersoni and D. variabilis based on a relatively small part of the 16S rRNA gene sequence.
A large proportion (>85%) of D. andersoni and D. variabilis at localities near their northern distributional limits contained FLEs, which are similar to rates of infection for FLEs in these two species in more southern parts of their geographical ranges (i.e., 55 to 97%) (24, 46). These bacteria occur in the Malpighian tubules and/or the ovaries of female ticks (6, 46, 47), and transovarial transmission of FLEs has been shown to be very efficient (96 to 100%) in D. andersoni (46) and D. variabilis (24). The infection rate of FLEs in D. variabilis larvae in the present study also suggests that FLEs are vertically transmitted. Transovarial transmission is an important mechanism by which FLEs are maintained in a large proportion of individuals within the tick populations. However, many D. andersoni and D. variabilis males in western Canada were also found to contain FLEs, but it is not known if these individuals would contribute to the maintenance of FLEs in tick populations. Few studies have examined D. andersoni or D. variabilis males for the presence of FLEs. However, the salivary glands and reproductive tissues of D. andersoni males from Bitterroot Mountains (Montana) were found not to contain the DAS FLE found in D. andersoni females from the same locality (46). If vertical transmission is the only means by which FLEs are maintained in a tick population, then Dermacentor males would represent a dead-end host for FLEs.
In the present study, 24% of the ticks containing FLEs were infected with multiple types. This is consistent with the results of previous studies where coinfections of multiple types of FLEs in Dermacentor adults were relatively common (24, 51). However, we found a significant difference between the two tick species in the frequency of multiple infections. Very few D. andersoni (3%) were infected with multiple types of FLEs, whereas significantly more infected D. variabilis (38%) were coinfected with two or three types of FLE.
Previous studies have determined the types of FLEs in D. andersoni and D. variabilis adults from allopatric populations (24, 39, 46, 51). However, the present study differed in that the FLE types present in D. andersoni and D. variabilis adults and immatures were compared from both allopatric and sympatric populations of ticks. Such a comparison provides insights into the modes of transmission (i.e., the occurrence of horizontal and/or vertical transmission) and host specificity of the different FLE types. Our results revealed a significant difference in the types of FLEs present in D. andersoni and D. variabilis, even in sympatric populations of these two species. Most FLE types were specific for a single tick species; however, the two most prevalent types (i.e., types 1 and 2) were found in both species of tick but in different relative frequencies. This infection pattern may be explained by horizontal transmission from one species of tick to the other through a vertebrate host. There is a recent report of the detection of FLEs (e.g., GenBank accession no. EU315913) in wood mice (Apodemus sylvaticus) in Europe (15), which suggests that horizontal transmission of some FLE types from ticks to small mammals can occur. However, horizontal transmission of FLEs has not been demonstrated in experimental infections (1, 6, 46). At localities where D. andersoni and D. variabilis occur in sympatry, immature stages of both species do parasitize the same small mammal hosts (e.g., mice and voles) (Dergousoff and Chilton, unpublished data), thus there is the potential for horizontal transmission of FLEs from one tick species to the other via a vertebrate host. However, at Saskatchewan Landing Provincial Park, there was no evidence of FLEs of type 1 in D. variabilis immatures or of type 2 or 3 in D. andersoni nymphs.
It is not known if there is an epidemiological significance of FLEs in ticks. Studies have not indicated that these organisms are pathogenic to ticks or affect their fecundity (1, 6, 24, 46). Furthermore, multiple FLE types can be cotransmitted transovarially (24), or with Rickettsia spp. (46) or Anaplasma phagocytophilum (1). Thus, the presence of FLEs does not appear to inhibit the vertical transmission of other FLE strains or distantly related organisms. Additional studies are needed to determine if there is a negative correlation between the presence of FLEs and the occurrence of F. tularensis in ticks and if they affect the vectorial capacity of their tick hosts (24).
In conclusion, multiple types of FLEs were found in northern populations of D. andersoni and D. variabilis. However, each FLE type was primarily found in a single tick species, and this host specificity of FLEs was generally maintained at locations where both tick species occurred in sympatry because there were very few examples of potential transfer of FLEs from D. andersoni to D. variabilis and vice versa. Although the three most common FLE types have been found previously in D. andersoni or D. variabilis in other parts of their geographical range, seven of the FLE types detected in the present study represented new sequence types. This finding expands our knowledge on the genetic diversity of FLEs in ticks. The continual discovery of new FLEs in ticks (41, 56) and species of Francisella in fish and mammals (37, 48, 55) and humans (14, 31) shows that the family Francisellaceae is much more diverse than previously realized (38). Furthermore, the presence of multiple FLEs in D. andersoni and D. variabilis that are genetically similar to F. tularensis has important implications for diagnosis and epidemiological studies of tularemia in Canada. These require molecular techniques that can reliably distinguish among the different subspecies and subtypes of Francisella and the different types of FLEs (15, 22, 39). Our study has demonstrated that PCR-SSCP, combined with DNA sequencing, is an effective approach to examine a large number of ticks for the presence of different bacteria within the family Francisellaceae.
ACKNOWLEDGMENTS
This work was approved by the University of Saskatchewan's Animal Research Ethics Board and adhered to the Canadian Council on Animal Care guidelines for humane animal use. Permits to trap rodents were obtained from the Saskatchewan Ministry of Environment.
Funding for this work was provided to N.B.C. from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canadian Foundation for Innovation. S.J.D. received funding through an NSERC postgraduate scholarship.
We thank Andrew Gajadhar and Lorilee Sereda for technical assistance.
Footnotes
Published ahead of print 16 December 2011
REFERENCES
- 1. Baldridge GD, et al. 2009. Transovarial transmission of Francisella-like endosymbionts and Anaplasma phagocytophilum variants in Dermacentor albipictus (Acari: Ixodidae). J. Med. Entomol. 46:625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bishopp FC, Trembley HL. 1945. Distribution and hosts of certain North American ticks. J. Parasitol. 31:1–54 [Google Scholar]
- 3. Black DM, Thomson JA. 1958. Tularaemia in British Columbia. Can. Med. Assoc. J. 78:16–18 [PMC free article] [PubMed] [Google Scholar]
- 4. Bow MR, Brown JH. 1943. Tularemia in the “Seven Persons Coulee,” Alberta. Can. J. Public Health 34:415–418 [Google Scholar]
- 5. Bow MR, Brown JH. 1946. Tularemia. A report on 40 cases in Alberta, Canada, 1931-1944. Am. J. Public Health Nations Health 36:494–500 [PubMed] [Google Scholar]
- 6. Burgdorfer W, Brinton LP, Hughes LE. 1973. Isolation and characterization of symbiotes from Rocky Mountain wood tick, Dermacentor andersoni. J. Invertebr. Pathol. 22:424–434 [DOI] [PubMed] [Google Scholar]
- 7.Canadian Cooperative Wildlife Health Center 1995. Tularemia in muskrats and beaver. Can. Coop. Wildl. Health Centre Newsl. 3-2:1–12 [Google Scholar]
- 8. Choi E. 2002. Tularemia and Q fever. Med. Clin. North Am. 86:393–416 [DOI] [PubMed] [Google Scholar]
- 9. Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9:1657–1659 [DOI] [PubMed] [Google Scholar]
- 10. de Carvalho IL, Santos N, Soares T, Zé-Zé L, Núncio MS. 2011. Francisella-like endosymbiont in Dermacentor reticulatus collected in Portugal. Vector Borne Zoonotic Dis. 11:185–188 [DOI] [PubMed] [Google Scholar]
- 11. Dergousoff SJ, Chilton NB. 2007. Differentiation of three species of ixodid tick, Dermacentor andersoni, D. variabilis and D. albipictus, by PCR-based approaches using markers in ribosomal DNA. Mol. Cell Probes 21:343–348 [DOI] [PubMed] [Google Scholar]
- 12. Eisen L. 2007. A call for renewed research on tick-borne Francisella tularensis in the Arkansas-Missouri primary national focus of tularemia in humans. J. Med. Entomol. 44:389–397 [DOI] [PubMed] [Google Scholar]
- 13. Eisen RJ, et al. 2009. Time course of hematogenous dissemination of Francisella tularensis A1, A2, and type B in laboratory mice. Am. J. Trop. Med. Hyg. 80:259–262 [PubMed] [Google Scholar]
- 14. Escudero R, et al. 2010. A possible novel Francisella genomic species isolated from blood and urine of a patient with severe illness. Clin. Microbiol. Infect. 16:1026–1030 [DOI] [PubMed] [Google Scholar]
- 15. Escudero R, et al. 2008. Molecular method for discrimination between Francisella tularensis and Francisella-like endosymbionts. J. Clin. Microbiol. 46:3139–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Foley JE, Nieto NC. 2010. Tularemia. Vet. Microbiol. 140:332–338 [DOI] [PubMed] [Google Scholar]
- 17. Forsman M, Sandstrom G, Sjostedt A. 1994. Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int. J. Syst. Bacteriol. 44:38–46 [DOI] [PubMed] [Google Scholar]
- 18. Fyvie A, Ross WG, Labzoffsky NA. 1959. Tularemia among muskrats on Walpole Island, Lake St. Clair, Ontario. Can. J. Comp. Med. Vet. Sci. 23:153–156 [PMC free article] [PubMed] [Google Scholar]
- 19. Gasser RB, et al. 2006. Single-strand conformation polymorphism (SSCP) for the analysis of genetic variation. Nat. Protoc. 1:3121–3128 [DOI] [PubMed] [Google Scholar]
- 20. Gibbons RJ. 1939. Survey of Rocky Mountain spotted fever and sylvatic plague in western Canada during 1938. Can. J. Public Health 30:184–187 [Google Scholar]
- 21. Goethert HK, Saviet B, Telford SR., III 2009. Metapopulation structure for perpetuation of Francisella tularensis tularensis. BMC Microbiol. 9:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goethert HK, Shani I, Telford SR., III 2004. Genotypic diversity of Francisella tularensis infecting Dermacentor variabilis ticks on Martha's Vineyard, Massachusetts. J. Clin. Microbiol. 42:4968–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goethert HK, Telford SR., III 2010. Quantum of infection of Francisella tularensis tularensis in host-seeking Dermacentor variabilis. Ticks Tick-borne Dis. 1:66–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goethert HK, Telford SR., III 2005. A new Francisella (Beggiatiales: Francisellaceae) inquiline within Dermacentor variabilis say (Acari: Ixodidae). J. Med. Entomol. 42:502–505 [DOI] [PubMed] [Google Scholar]
- 25. Goethert HK, Telford SR., III 2009. Nonrandom distribution of vector ticks (Dermacentor variabilis) infected by Francisella tularensis. PLoS Pathog. 5:e1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gordon JR, McLaughlin BG, Nitiuthai S. 1983. Tularaemia transmitted by ticks (Dermacentor andersoni) in Saskatchewan. Can. J. Comp. Med. 47:408–411 [PMC free article] [PubMed] [Google Scholar]
- 27. Gregson JD. 1956. The Ixodoidea of Canada. Publication 930. Canada Department of Agriculture, Ottawa, Canada [Google Scholar]
- 28. Gwatkin R, Painter RH, Moynihan IW. 1942. Tularaemia in sheep. Can. J. Comp. Med. Vet. Sci. 6:163–168 [PMC free article] [PubMed] [Google Scholar]
- 29. Gyuranecz M, et al. 2011. Investigation of the ecology of Francisella tularensis during an inter-epizootic period. Vector Borne Zoonotic Dis. 11:1031–1035 [DOI] [PubMed] [Google Scholar]
- 30. Harris TA. 1956. Tularaemia among farmer-trappers in northwestern Saskatchewan. Can. Med. Assoc. J. 74:60–61 [PMC free article] [PubMed] [Google Scholar]
- 31. Huber B, et al. 2010. Description of Francisella hispaniensis sp. nov., isolated from human blood, reclassification of Francisella novicida (Larson et al. 1955) Olsufiev et al. 1959 as Francisella tularensis subsp. novicida comb. nov. and emended description of the genus Francisella.. Int. J. Syst. Evol. Microbiol. 60:1887–1896 [DOI] [PubMed] [Google Scholar]
- 32. Humphreys FA. 1947. Some observations regarding tick and insect-borne infections in western Canada. Can. J. Comp. Med. 11:187–192 [PubMed] [Google Scholar]
- 33. Humphreys FA, Campbell AG. 1947. Plague, Rocky Mountain spotted fever, and tularaemia surveys in Canada. Can. J. Public Health 38:124–130 [PubMed] [Google Scholar]
- 34. Isaac-Renton M, et al. 2010. Tularemia in British Columbia: a case report and review. BCMJ 52:303–307 [Google Scholar]
- 35. James AM, et al. 2006. Distribution, seasonality, and hosts of the Rocky Mountain wood tick in the United States. J. Med. Entomol. 43:17–24 [PubMed] [Google Scholar]
- 36. Jellison WL. 1974. Tularemia in North America 1930–1974. University of Montana Foundation, Missoula, MT [Google Scholar]
- 37. Kamaishi T, et al. 2005. Identification and pathogenicity of intracellular Francisella bacterium in three-line grunt Parapristipoma trilineatum. Fish Pathol. 40:67–71 [Google Scholar]
- 38. Keim P, Johansson A, Wagner DM. 2007. Molecular epidemiology, evolution, and ecology of Francisella. Ann. N. Y. Acad. Sci. 1105:30–66 [DOI] [PubMed] [Google Scholar]
- 39. Kugeler KJ, et al. 2005. Discrimination between Francisella tularensis and Francisella-like endosymbionts when screening ticks by PCR. Appl. Environ. Microbiol. 71:7594–7597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Langford EV. 1954. An outbreak of tularaemia In beaver and muskrat in Waterton Lakes National Park, Alberta. Can. J. Comp. Med. Vet. Sci. 18:28–30 [PMC free article] [PubMed] [Google Scholar]
- 41. Machado-Ferreira E, Piesman J, Zeidner NS, Soares CA. 2009. Francisella-like endosymbiont DNA and Francisella tularensis virulence-related genes in Brazilian ticks (Acari: Ixodidae). J. Med. Entomol. 46:369–374 [DOI] [PubMed] [Google Scholar]
- 42. Martin T, Holmes IH, Wobeser GA, Anthony RF, Greefkes I. 1982. Tularemia in Canada with a focus on Saskatchewan. Can. Med. Assoc. J. 127:279–282 [PMC free article] [PubMed] [Google Scholar]
- 43. McNabb A. 1930. Tularemia: The first case reported in Canada. Can. J. Public Health 21:91–92 [Google Scholar]
- 44. Merten HA, Durden LA. 2000. A state-by-state survey of ticks recorded from humans in the United States. J. Vector Ecol. 25:102–113 [PubMed] [Google Scholar]
- 45. Molins CR, et al. 2010. Virulence differences among Francisella tularensis subsp. tularensis clades in mice. PLoS One 5:e10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Niebylski ML, Peacock MG, Fischer ER, Porcella SF, Schwan TG. 1997. Characterization of an endosymbiont infecting wood ticks, Dermacentor andersoni, as a member of the genus Francisella. Appl. Environ. Microbiol. 63:3933–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Noda H, Munderloh UG, Kurtti TJ. 1997. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl. Environ. Microbiol. 63:3926–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nylund A, Ottem KF, Watanabe K, Karlsbakk E, Krossoy B. 2006. Francisella sp. (Family Francisellaceae) causing mortality in Norwegian cod (Gadus morhua) farming. Arch. Microbiol. 185:383–392 [DOI] [PubMed] [Google Scholar]
- 49.Office of the Medical Health Officer 2007. Human tularemia cases. Public Health Matters MHO Newsl. 13:2 [Google Scholar]
- 50. Reese SM, et al. 2010. Transmission dynamics of Francisella tularensis subspecies and clades by nymphal Dermacentor variabilis (Acari: Ixodidae). Am. J. Trop. Med. Hyg. 83:645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scoles GA. 2004. Phylogenetic analysis of the Francisella-like endosymbionts of Dermacentor ticks. J. Med. Entomol. 41:277–286 [DOI] [PubMed] [Google Scholar]
- 52. Scott JW, Macbeth RAL. 1946. Tularaemia (with a report of nine cases). Can. Med. Assoc. J. 55:564–566 [PMC free article] [PubMed] [Google Scholar]
- 53. Sjöstedt A. 2005. Family III. Francisellaceae fam. nov., p 199–209 In Brenner DJ, Krieg NR, Staley JT. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2. Springer, East Lansing, MI [Google Scholar]
- 54. Sonenshine DE. 1979. Zoogeography of the American dog tick, Dermacentor variabilis, p 123–134 In Rodriguez JG. (ed), Recent advances in acarology, vol 2. Academic Press, New York, NY [Google Scholar]
- 55. Soto E. 2010. In vivo and in vitro pathogenesis of Francisella asiatica in Tilapia nilotica (Oreochromis niloticus). Ph.D. thesis. Louisiana State University, Baton Rouge, LA [Google Scholar]
- 56. Sréter-Lancz Z, Széll Z, Sréter T, Márialigeti K. 2009. Detection of a novel Francisella in Dermacentor reticulatus: a need for careful evaluation of PCR-based identification of Francisella tularensis in Eurasian ticks. Vector Borne Zoonotic Dis. 9:123–126 [DOI] [PubMed] [Google Scholar]
- 57. Staples JE, Kubota KA, Chalcraft LG, Mead PS, Petersen JM. 2006. Epidemiologic and molecular analysis of human tularemia, United States, 1964–2004. Emerg. Infect. Dis. 12:1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Suitor EC, Jr, Weiss E. 1961. Isolation of a Rickettsialike microorganism (Wolbachia persica, n. sp.) from Argas persicus (Oken). J. Infect. Dis. 108:95–106 [Google Scholar]
- 59. Sun LV, Scoles GA, Fish D, O'Neill SL. 2000. Francisella-like endosymbionts of ticks. J. Invertebr. Pathol. 76:301–303 [DOI] [PubMed] [Google Scholar]
- 60. Walker WJ, Moore CA. 1971. Tularemia: experience in the Hamilton area. Can. Med. Assoc. J. 105:390–396 [PMC free article] [PubMed] [Google Scholar]
- 61. Wilkinson PR. 1967. The distribution of Dermacentor ticks in Canada in relation to bioclimatic zones. Can. J. Zool. 45:517–537 [DOI] [PubMed] [Google Scholar]
- 62. Wobeser G, Campbell GD, Dallaire A, McBurney S. 2009. Tularemia, plague, yersiniosis, and Tyzzer's disease in wild rodents and lagomorphs in Canada: a review. Can. Vet. J. 50:1251–1256 [PMC free article] [PubMed] [Google Scholar]
- 63. Wobeser G, Ngeleka M, Appleyard G, Bryden L, Mulvey MR. 2007. Tularemia in deer mice (Peromyscus maniculatus) during a population irruption in Saskatchewan, Canada. J. Wildl. Dis. 43:23–31 [DOI] [PubMed] [Google Scholar]