Abstract
Microscopic localization of endosymbiotic bacteria in three species of mealybug (Pseudococcus longispinus, the long-tailed mealybug; Pseudococcus calceolariae, the citrophilus mealybug; and Pseudococcus viburni, the obscure mealybug) showed these organisms were confined to bacteriocyte cells within a bacteriome centrally located within the hemocoel. Two species of bacteria were present, with the secondary endosymbiont, in all cases, living within the primary endosymbiont. DNA from the dissected bacteriomes of all three species of mealybug was extracted for analysis. Sequence data from selected 16S rRNA genes confirmed identification of the primary endosymbiont as “Candidatus Tremblaya princeps,” a betaproteobacterium, and the secondary endosymbionts as gammaproteobacteria closely related to Sodalis glossinidius. A single 16S rRNA sequence of the primary endosymbiont was found in all individuals of each mealybug species. In contrast, the presence of multiple divergent strains of secondary endosymbionts in each individual mealybug suggests different evolutionary and transmission histories of the two endosymbionts. Mealybugs are known vectors of the plant pathogen Grapevine leafroll-associated virus 3. To examine the possible role of either endosymbiont in virus transmission, an extension of the model for interaction of proteins with bacterial chaperonins, i.e., GroEL protein homologs, based on mobile-loop amino acid sequences of their GroES homologs, was developed and used for analyses of viral coat protein interactions. The data from this model are consistent with a role for the primary endosymbiont in mealybug transmission of Grapevine leafroll-associated virus 3.
INTRODUCTION
Mealybugs, aphids, psyllids, and whiteflies are all plant sap-sucking insects that have cultivated intimate relationships with mutualistic bacteria since their early evolutionary history (14, 15, 22, 35). Mealybugs (Pseudococcoidae, Hemiptera) have an obligate association with prokaryotic primary endosymbionts (P-endosymbionts) of the Betaproteobacteria (23, 33), whose major function appears to be the synthesis of essential amino acids that are lacking in plant sap (16, 29). They are acquired through vertical maternal transmission (6, 7) and are stored within specialized cells called bacteriocytes that form well-defined organs in the mealybug's body cavity (bacteriomes) (5). Each group of these insects has its own coevolved primary endosymbionts, and phylogenetic analyses are consistent with an infection of an ancestor with a precursor of the endosymbiont, followed by a coevolutionary history of vertical transmission of the endosymbiont to progeny (5). Mealybugs are unusual in having betaproteobacterial endosymbionts; the P-endosymbionts of the other, related insects noted above are all gammaproteobacteria. In mealybugs, there can be a further layer of bacterial symbiosis, with the P-endosymbionts themselves harboring secondary endosymbiotic bacteria (S-endosymbionts) of gammaproteobacteria (45). These bacteria are present in most, but not all, mealybugs and form distinct clades, suggesting multiple evolutionary origins, and their transmission mechanism is unknown (24, 41). Gammaproteobacterial endosymbionts are known in many insect species (8), but this arrangement within the P-endosymbiont is thought to be unique to mealybugs. Thus, betaproteobacteria exist as free-living bacteria or P-endosymbionts of eukaryotes, whereas gammaproteobacteria exist as free-living bacteria, P-endosymbionts of eukaryotes, and S-endosymbionts of both prokaryotes and eukaryotes.
Mealybugs are the principal vectors of Grapevine leafroll-associated virus 3 (GLRaV-3), an ampelovirus (26) that causes grapevine leafroll disease (9, 10; J. G. Charles and D. T. Jordan, presented at the New Zealand Grape and Wine Symposium, Auckland, New Zealand, 1993). In New Zealand, three species of mealybugs (Pseudococcus longispinus, the long-tailed mealybug; Pseudococcus calceolariae, the citrophilus mealybug; and Pseudococcus viburni, the obscure mealybug) are known to transmit GLRaV-3 (37). In California, two additional species, the grape mealybug (Pseudococcus maritimus) and the citrus mealybug (Planococcus citri), have also been shown to transmit the virus (20). In France, two additional species of mealybug, Heliococcus bohemicus and Phenacoccus aceris (Pseudococcidae), and the soft scale insect, Parthenolecanium corni (Coccidae), were shown to transmit GLRaV-3 (39). The disease is present in grapevines around the world but is a particular issue in New Zealand (J. G. Charles and D. T. Jordan, presented at the New Zealand Grape and Wine Symposium, Auckland, New Zealand, 1993), where it is usually sufficiently warm during spring and summer for mealybug populations to become very large yet not warm enough in autumn for diseased grapevines to ripen fruit adequately. GLRaV-3 is recognized by the wine industry as the biggest production threat to their economic future (11). Even if vineyards are initially mealybug free, it is impossible to prevent infection from airborne, dispersing young crawlers over time (12).
It has been proposed (1) that GroEL homologs (i.e., proteins homologous to the Escherichia coli GroEL protein) produced by endosymbionts (for mealybugs, the β-endosymbiotic bacteria, which are the primary endosymbiont and always present, were suggested), are involved in virus transmission by insect vectors. GroEL homologs might bind to the virion (in the midgut or as it is being transported into the midgut epithelium) and protect it against degradation in the hemolymph while it is being transported (specifically) to the salivary glands (1). This interaction is partly specific and may exert some control over which viruses a given host can transmit. Thus, GroEL may be essential for circulative transmission of many viruses. Since GLRaV-3 is probably transmitted circularly (13), the GroEL hypothesis is potentially applicable to GLRaV-3. Further, a recent report (18) showed that expression of the relevant endosymbiont GroEL protein in tobacco confers tolerance for a virus with a viral load decreased 1,000-fold, and the plants are essentially asymptomatic.
GroEL is a chaperonin/heat shock-induced protein in E. coli and functions as a complex with another protein, GroES (eukaryotic proteins Hsp60 and Hsp10 are structurally and functionally nearly identical) (25, 27). Because it was found that E. coli GroEL did not bind GLRaV-3 (1), these data have been interpreted as showing that the GroEL mechanism is not relevant for GLRVa3 in mealybugs (13). Stan and coworkers have developed a theoretical model to predict the binding of proteins to E. coli GroEL (40) using the sequence properties of the GroES mobile loop, which also fits the GroEL binding site. We have extended their model to predict potential interactions using the observed ability of various insects to transmit the differing viruses based on the viral coat proteins and the putative binding specificities of the GroEL proteins of their primary endosymbionts (based on the relevant GroES sequences).
In this study, we present data that extend previous observations of multiple evolutionary origins for secondary endosymbiotic bacteria in mealybugs to multiple origins within individual insects of two distinct species, indicating a high degree of mobility for these endosymbionts. Further, we predict interaction between GLRaV-3 coat protein and mealybug betaproteobacterial GroEL (and not with gammaproteobacterial GroEL, which has an E. coli-like GroES) and discuss the evolutionary consequences of these observations.
MATERIALS AND METHODS
Material.
P. longispinus, P. calceolariae, and P. viburni mealybugs were obtained from colonies maintained on potatoes sprouting shoot and root buds at Plant and Food Research in Auckland, New Zealand, or from various plants in orchards around New Zealand.
Microscopy.
Initially, whole immature adult female insects were prepared using two fixation methods: either in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer at pH 7.2 under vacuum for 1 h or in acidified 2,2-dimethoxypropane (DMP) (32). DMP rapidly dehydrates tissue by converting water to ethanol. Fixed whole insects from each species were then dehydrated and embedded in LR White resin (London Resin, Reading, United Kingdom). Serial sections (1 μm thick) were then dried onto slides, stained with 0.05% toluidine blue in benzoate buffer (pH 4.4), dried, and mounted in Shurmount (Triangle Biomedical Sciences, Durham, NC). Sections were examined by light microscopy using an Olympus Vanox AHT3 microscope (Olympus Optical, Tokyo, Japan). Both fixation methods were successful in retaining structural detail.
Once the location of the bacteriome within the insect body was understood, bacteriomes were dissected from mealybugs, isolated, and fixed as isolated organs. The isolated bacteriome, although very small, was fixed and embedded more successfully than when it was part of a whole mealybug. Isolated bacteriomes were also fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer at pH 7.2 under vacuum for 1 h, washed in buffer, postfixed in 1% osmium tetroxide, dehydrated in an ethanol series, and embedded in Spurr's resin. Sections 100 nm thick were collected on Formvar-coated copper grids and stained with 1% (vol/vol) uranyl acetate and lead citrate (38). The sections were viewed in a JEOL (Tokyo, Japan) JEM-1200EX II transmission electron microscope operating at 80 kV.
Isolation of bacteriomes and bacterial DNA.
Bacteriomes were dissected from adult P. longispinus, P. calceolariae, and P. viburni in insect Ringer's solution (10× stock: 1,280 mM NaCl, 15 mM CaCl2, 50 mM KCl, pH 7.4), dried, frozen with liquid nitrogen, and then ground separately in 1.5-ml microcentrifuge tubes prior to the extraction of DNA. Extraction of genomic DNA from ground bacteriomes was carried out following standard protocols for the DNeasy Tissue Kit (Qiagen Inc., Santa Clara, CA), except for the use of 100 μl elution buffer incubated for 5 min prior to the eluate and DNA being centrifuged through the mini-column membrane.
PCR.
All PCRs were carried out using Platinum Taq HiFi (Invitrogen) according to the manufacturer's directions under the following conditions: 40 cycles of melting at 94°C for 20 s, annealing at 55°C for 20 s, and extension at 68°C for 100 s, with a final extension of 5 min. The initial PCR was carried out on DNA isolated from individual bacteriomes using a universal bacterial primer pair designed from the conserved regions of the 16S rRNA gene for E. coli and other species (Table 1). These primers were designed to amplify an ∼1,500-bp 16S sequence from alpha-, beta-, and gammaproteobacteria, i.e., no assumptions as to the nature of the endosymbionts was made. PCR products were of the expected size and were shotgun cloned into the pCR 2.1 TOPO Vector (Invitrogen) according to the manufacturer's instructions and sequenced using M13 standard sequencing primers and conserved internal sequencing primers (Table 1).
Table 1.
PCR primers used in this studya
| Primer name | Organism(s) used | Specificity | Orientation | Sequence (5′–3′) |
|---|---|---|---|---|
| Ec8 | E. coli | Conserved general | Forward | AGAGTTTGATCATGGCTCAGATTG |
| Ec1507 | E. coli | Conserved general | Reverse | TACCTTGTTACGACTTCACCCCAG |
| Bpsf | Betaproteobacteria | Selective | Forward | CACATGCAAGTCGTACGGCAGCAC |
| Gpsf | Gammaproteobacteria | Selective | Forward | CAGRCCTAACACATGCAAGTCGAG |
| BGpsr | Beta- and gammaproteobacteria | Selective | Reverse | TTGTTACGACTTCACCCCAGTCAT |
| BGDf | Beta- and gammaproteobacteria | Conserved sequencing | Forward | CGTGCCAGCAGCCGCGGTAATACG |
| BGDr | Beta- and gammaproteobacteria | Conserved sequencing | Reverse | CGTATTACCGCGGCTGCTGGCACG |
| BGMf | Beta- and gammaproteobacteria | Conserved sequencing | Forward | ACAGGTGCTGCATGGCTGTCGTCA |
| BGMr | Beta- and gammaproteobacteria | Conserved sequencing | Reverse | TGACGACAGCCATGCAGCACCTGT |
All primers were designed against bacterial 16S rRNA genes.
Subsequently, a more detailed study of the endosymbiont populations was carried out on individual bacteria of each species (7 from P. calceolariae, 6 from P. viburni, and 6 from P. longispinus) using selective primer pairs (Table 1). Selective PCR was carried out using primer pairs designed to selectively amplify ∼1,500-bp sequences from beta- and gammaproteobacteria (Table 1). These reactions used a common reverse primer that would select against alphaproteobacteria and forward primers selective for either beta- or gammaproteobacteria. PCR products of the expected size were obtained and were initially sequenced directly using the amplification primers and conserved internal sequencing primers. DNA sequence data were obtained from all individual mealybug bacteriome samples (7 from P. calceolariae, 6 from P. viburni, and 6 from P. longispinus). PCR products from a total of 6 of the gammaproteobacterial reactions were shotgun cloned into pCR 2.1 as described above and sequenced. All reported sequences were fully sequenced in both directions. The search for variant sequences in individuals was not exhaustive, and those sequences found for each species were used to construct the phylogenetic tree (see Fig. 5).
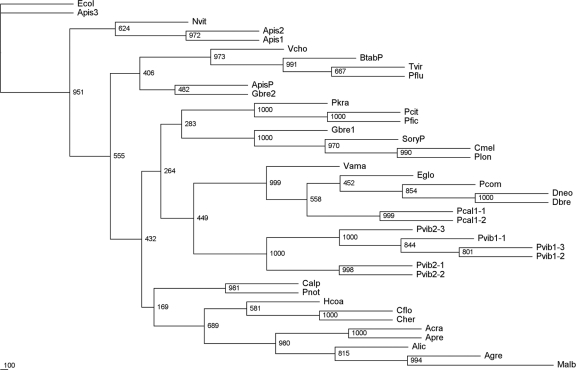
Fig 5.
Secondary (gammaproteobacterial) endosymbiont 16S ribosomal sequence phylogenetic tree. All sequences, except three free-living gammaproteobacterial species, are those of the 16S RNA genes from gammaproteobacterial symbionts of the named insect species. Known primary endosymbionts have “P” suffixed. The 16S ribosomal sequence from the free-living gammaproteobacterium E. coli W3110 (NCBI accession no. AP009048, annotated Ecol) was used as the outgroup.
Phylogenetic analysis.
Alignments were constructed using ClustalX (v.1.83) with the default settings (42). Manual trimming, if required, was carried out using GeneDoc (34). Phylogenetic analysis was carried out using the PHYLIP suite of software (19). The phylogenetic trees (see Fig. 4 and 5) were constructed using the neighbor-joining method. Bootstrap values (percent) were calculated from 1,000 bootstrap replicates. The trees were rooted with a distinct outgroup. TreeView (v.1.6.6) was used to display the resulting trees (36).
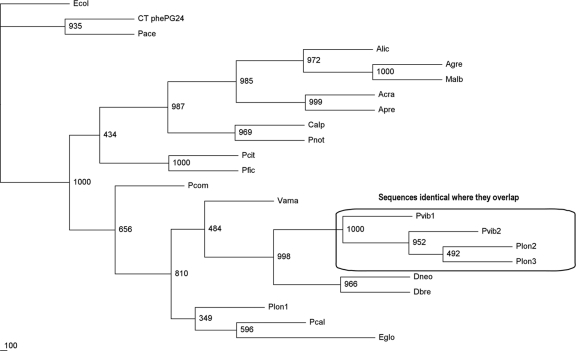
Fig 4.
Primary (betaproteobacterial) endosymbiont 16S ribosomal sequence phylogenetic tree. All sequences are those of the 16S RNA genes isolated from the betaproteobacterial symbionts of the named pseudococcid mealybug species. The sequence for “Candidatus Tremblaya princeps” from P. calceolariae is a first publication and supports a recent mealybug phylogeny study (21). The 16S ribosomal sequence from the free-living gammaproteobacterium Escherichia coli W3110 (NCBI accession no. AP009048, annotated Ecol) was used as the outgroup.
The GroEL binding model.
GroEL is the equivalent of the eukaryotic 60-kDa chaperonin/heat shock protein, and its associated protein, GroES, is the bacterial equivalent of the eukaryotic 10-kDa chaperonin/heat shock protein. GroEL binds proteins by means of a mainly hydrophobic groove and is actively involved in their correct folding.
A model using a sequence-based approach (40) that identifies natural substrates, including viral coat proteins, for these chaperonins has been developed. The authors hypothesized that natural substrate proteins of GroEL contain two or more patterns of residues similar to that of the GroES mobile loop, i.e., G_IVL_G_A (where “_” represents an arbitrary residue).
The GroES pattern was translated into residue chemical types, hydrophobic (H), hydrophilic (P), and positively (+) and negatively (−) charged. The four classes are H (C, F, I, L, W, V, M, Y, and A), P (G, P, N, T, S, Q, and H), + (R and K), and − (D and E). A pattern may contain 4 (P_HHH), 5 (P_HHH_P), or 6 (P_HHH_P_H) GroES-like contacts. The minimum sequence separation between consecutive patterns in each sequence is 23 residues. The authors showed that E. coli GroEL preferentially binds to sequences similar to that of the mobile loop of GroES (P_HHH_P_H). The consensus pattern for the mobile-loop region of GroES in bacteria, including Sodalis spp. (see Fig. 6), generally fits the E. coli model.
Fig 6.
Clustal alignment of HSP10 and GroES sequences from diverse organisms. Light gray, conserved sequence; dark gray, block of similar sequence, as defined in AlignX (Vector NTI, Invitrogen).
However, in this study, we observed that eukaryotic sequences (human, zebrafish, chicken, insect [Tribolium], plant [Arabidopsis]), red alga, green alga, yeast, and fungus) consistently fit a different pattern (P_HHH_−_H) and that the sequence of “Candidatus Tremblaya princeps” GroES (Q8KTR9; AAM75979) is H_HHH_−_H, which nearly fits the eukaryotic pattern. The latter is the pattern searched for in Table 2 under the name of “Ca. Tremblaya princeps.”
Table 2.
Bioinformatic prediction of plant virus coat proteins interacting with E. coli GroEL and “Ca. Tremblaya princeps” GroELa
| Genus | Virus name (acronym), accession no. | Binding of intact virus to E. coli GroEL (1) | Virion shape | Properties of coat protein (Mw, pI, charge, and Arg [%]) | No. of E. coli GroES patternsc |
E. coli GroEL binding predictionb | No. of “Ca. Tremblaya princeps” GroES patternsc |
“Ca. Tremblaya princeps” GroEL binding predictionb | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 6 | 4 | 5 | 6 | |||||||
| Begomovirus | Tomato yellow leaf curl virus (TYLCV), X15656 | Yes | Geminate | 30,285; 10.4; 22.7; 12.9 | 4 | 2 | 2 | Yes | 4 | 3 | 2 | Yes |
| Begomovirus | African cassava mosaic virus (ACMV), AF366902 | Yes | Geminate | 30,129; 10.3; 22.8; 12.5 | 3 | 1 | 0 | Yes | 2 | 2 | 1 | Yes |
| Cucumovirus | Cucumber mosaic virus (CMV), D10538 | Yes | Globular | 24,113; 10.3; 12.2; 13.0 | 2 | 1 | 1 | Yes | 2 | 2 | 1 | Yes |
| Luteovirus | Bean leafroll virus (BLRV), NC 003369 | Yes | Globular | 21,966; 11.2; 22.3; 13 | 3 | 0 | 0 | Yes | 3 | 0 | 0 | Yes |
| Luteovirus | Barley yellow dwarf virus (BYDV), NC 002160 | Yes | Globular | 21,930; 12.1; 23.2; 15.0 | 4 | 2 | 0 | Yes | 1 | 0 | 0 | No |
| Luteovirus | Soybean dwarf virus (SbDV), NC_001747 | Yes | Globular | 22,201; 11.3; 22.3; 13.5 | 3 | 1 | 0 | Yes | 1 | 0 | 0 | No |
| Luteovirus (Enamovirus) | Pea enation mosaic virus (PEMV), NC_003629 | Yes | Globular | 21,104; 11.2; 19.3; 15.6 | 3 | 2 | 1 | Yes | 3 | 1 | 1 | Yes |
| Luteovirus (Polerovirus) | Potato leafroll virus (PLRV), NC_001747 | Yes | Globular | 23,127; 11.6; 24.2; 15.5 | 3 | 2 | 1 | Yes | 1 | 0 | 0 | No |
| Luteovirus (Polerovirus) | Beet western yellows virus (BWYV), NC_003743 | Yes | Globular | 22,459; 11.7; 22.2; 16.0 | 2 | 1 | 0 | Yes | 1 | 1 | 1 | No |
| Potexvirus | Potato virus X (PVX), AF260641 | No | Filamentous | 25,111; 7.0; 0.06; 5.3 | 3 | 2 | 0 | Yes | 3 | 2 | 1 | Yes |
| Potyvirus | Potato virus Y M95491 | No | Filamentous | 29,879; 5.9; 3.5; 7.0 | 1 | 0 | 0 | No | 0 | 0 | 0 | No |
| Tricovirus | Grapevine virus A (GVA), NC_003604 | No | Filamentous | 21,624; 8.4; 1.2; 6.9 | 3 | 2 | 0 | Yes | 3 | 2 | 1 | Yes |
| Ampelovirus (Closteroviridae) | Grapevine leafroll virus 3 (GLRVa 3), NC_004667 | No | Isometric | 4,802.8; 6.78; 0.4; 3.0 | 4 | 2 | 1 | Yes | 4 | 2 | 2 | Yes |
| Llavirus (Bromoviridae) | Prune dwarf virus (PDV), U31310 | Yes | Isometric | 23,922.2; 10.0; 9.0; 6.26 | 4 | 2 | 1 | Yes | 3 | 0 | 0 | Yes |
| Nepovirus (Comoviradae) | Tobacco ringspot virus (TSRV), AF461164 | No | Isometric | 57,177.8; 7.25; 1.78; 6.03 | 6 | 2 | 2 | Yes | 7 | 0 | 0 | Yes |
| Tobamovirus | Tobacco mosaic virus (TMV), NC_001367 | No | Filamentous | 17,620.0; 4.83; 2.04; 9.36 | 2 | 2 | 1 | Yes | 2 | 0 | 0 | Yes |
Adapted from reference 1 with permission of H. Czosnek and Springer-Verlag Wien.
In the yes/no scoring, “yes” means that the number of GroES patterns is at least 2 and that the number of contacts at each site is at least 4 in the (major) coat protein sequence (40).
Pattern match scores are as follows: 4, P_HHH and H_HHH; 5, P_HHH_P and H_HHH_−; or 6, P_HHH_P_H and H_HHH_−_H for E. coli and “Ca. Tremblaya princeps,” respectively (see Materials and Methods for further explanation).
Nucleotide sequence accession numbers.
The accession numbers of the three species' consensus betaproteobacterial sequences are as follows: P. longispinus, JN182336; P. viburni, JN182337; and P. calceolariae, JN182335.
RESULTS
Microscopic localization of endosymbionts.
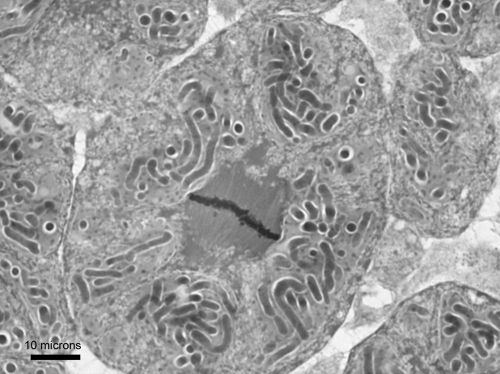
Bacteriomes were identified in a number of individuals from each species of mealybug. The basic structures of the bacteriomes from all three species, as observed using light microscopy (Fig. 1 and 2; P. viburni not shown), were similar. The bacteriome (approximately 300 to 500 by 150 to 200 μm) was located centrally, and each bacteriome contained ∼100 bacteriocytes. Each bacteriocyte was an insect cell containing 8 to 10 globular primary bacteria. Each primary bacterium contained a number (on the order of 10 to 20) of rod-like secondary bacteria.
Fig 1.
Light micrograph of a P. calceolariae bacteriocyte (shown in mitosis) containing six P-endosymbionts, each containing a number of rod-like S-endosymbionts.
Fig 2.
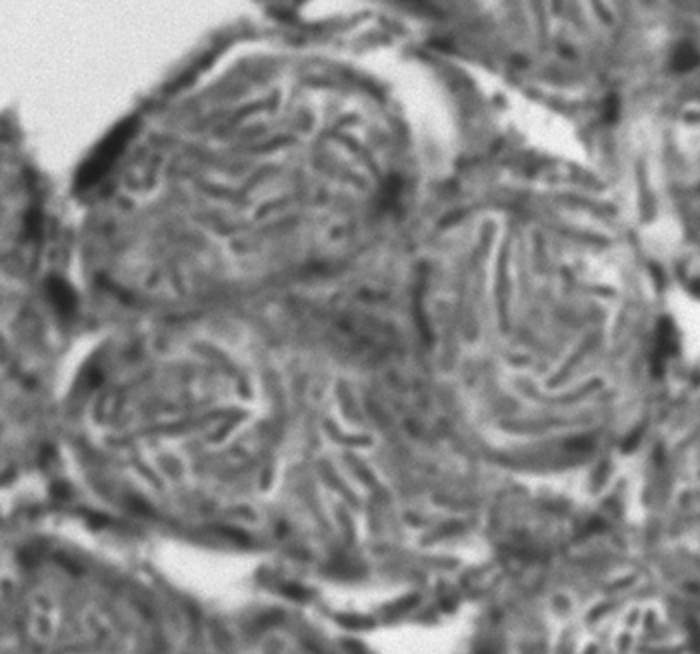
Light micrograph of a P. longispinus bacteriocyte containing four P-endosymbionts, each containing a number of rod-like S-endosymbionts.
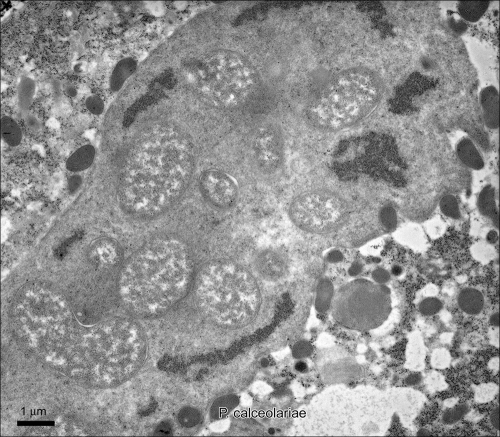
The bacteriomes of two species, P. calceolariae and P. viburni, were examined by electron microscopy (Fig. 3; P. viburni not shown). Secondary bacteria, in particular, have good structure and a clearly visible membrane structure. The structures in the two species were very similar.
Fig 3.
Transmission electron microscopy (TEM) of multiple S-endosymbiont within a P-endosymbiont in a bacteriocyte of P. calceolariae.
Initial PCR-based identification of endosymbionts.
Initial PCR was performed using the general primer pair based on E. coli (Table 1). These primers were designed to amplify any proteobacterium within the bacteriome. PCR products of the expected length (1,500 bp) were obtained from each of two individuals of each species. Twelve random clones from each individual bacterium obtained by shotgun cloning of these products were selected for sequencing.
From these data, we observed that the sequences comprised sequences homologous to those of betaproteobacteria, multiple sequences homologous to those of gammaproteobacteria, and two sequences from a single individual closely homologous to those of alphaproteobacterial soil bacteria and presumed to be contaminants. For the betaproteobacterial type, within any one species, the sequence was invariant, apart from occasional single base changes in the clones consistent with PCR errors and single/double base calls consistent with sequencing compressions (with the exceptions of a C-G and a G-T base change, each in a single clone). The gammaproteobacterial sequences obtained by this method showed variability: P. calceolariae yielded a single sequence and P. viburni a number of closely homologous but nonidentical sequences.
Analysis of the three species' consensus betaproteobacterial sequences using annotated BLAST (Fig. 4) showed that the P-endosymbionts were “Candidatus Tremblaya princeps.” The sequences across the three species were highly homologous (∼98%) to each other and to the already-published sequences for the P-endosymbionts from P. viburni (AF476095) and P. longispinus (M68889 and AF476093). Comparison of our sequences with those published showed that all differences were single base changes or single-base indels. The three published sequences referred to are identical to each other in the region where they overlap, despite being from two different host insect species, and identical to our P. viburni sequence, though different from our sequence from P. longispinus. Possible explanations for these observations include variability between geographically isolated mealybug populations, species misidentification, cross-contamination, or even problems in mealybug taxonomy. In our study, we did not observe any differences in P-endosymbiont sequences among those obtained from endosymbionts isolated from a single individual mealybug that could not be interpreted as PCR errors in individual clones and none in the total PCR product sequences (see below). Our data represent the first sequences for the P-endosymbiont from P. calceolariae.
Based on annotated BLAST, the S-endosymbiont (gammaproteobacterial) sequences are most similar to those of strains of Sodalis glossinidius and are homologous to each other and to other bacteria from this group (Fig. 5). which are P-endosymbionts in some insects and S-endosymbionts in others (46). They are highly homologous to gammaproteobacterial sequences from the mealybug Pseudococcus comstocki (AB374418) (24). The name “Candidatus Moranella endobia” has very recently been proposed for these bacteria (28). We did not find sequences suggesting S-endosymbionts in any individual P. longispinus mealybug by this method, although the search was not exhaustive.
Selective PCR analysis of identified endosymbionts.
The PCR products using the selective primers for betaproteobacteria were sequenced directly, and the sequences were identical to those obtained initially using the universal primer pair (see above). These consensus sequences were used for phylogenetic analysis (Fig. 4) and NCBI submission. The description of sequences used for phylogenetic analysis are shown in Table 3.
Table 3.
| Species used for isolation | 16S Beta accession no. | Tree annotation |
|---|---|---|
| Amonostherium lichtensioides | AF476078 | Alic |
| Antonina crawii | AB030021 | Acra |
| Antonina pretiosa | AF476079 | Apre |
| Australicoccus grevilleae | AF476077 | Agre |
| Cyphonococcus alpines | AF476081 | Calp |
| Dysmicoccus brevipes | AF476082 | Dbre |
| Dysmicoccus neobrevipes | AF476083 | Dneo |
| Erium globosum | AF476084 | Eglo |
| Melanococcus albizziae | AF476087 | Malb |
| Paracoccus nothofagicola | AF476094 | Pnot |
| Phenacoccus aceris | HM449982 | Pace |
| Phenacoccus solani | HM449979 | Psol |
| Planococcus citri | AF322017 | Pcit |
| Planococcus ficus | AF476092 | Pfic |
| Pseudococcus calceolariaeb | JN182335 | Pcal |
| Pseudococcus comstocki | AB374416 | Pcom |
| Pseudococcus longispinusb | JN182336 | Plon1 |
| Pseudococcus longispinus | AF476093 | Plon2 |
| Pseudococcus longispinus | M68889 | Plon3 |
| Pseudococcus viburnib | JN182337 | Pvib1 |
| Pseudococcus viburni | AF476095 | Pvib2 |
| Vryburgia amaryllidis | AF476097 | Vama |
All sequences are those of the 16S RNA gene from the betaproteobacterial symbiont isolated from the named pseudococcid mealybug species. The 16S ribosomal sequence from the free-living gammaproteobacterium E. coli W3110 (NCBI accession no. AP009048, annotated Ecol) was used as the outgroup.
Consensus sequence.
For the gammaproteobacteria, sequencing of the PCR products from the selective primers yielded consistent “mass average” sequences but with clear evidence of sequence variability that was consistent for each of the mealybug species. However, the consensus sequences for the gammaproteobacteria were not identical to those of the relevant clones obtained initially. In order to resolve this problem, PCR products from three individual insects, obtained using the selective primers for gammaproteobacteria, were cloned and sequenced (3 each from two individual P. viburni mealybugs and 2 from a single P. calceolariae mealybug). In all three individual bacteria, the gammaproteobacterial sequences in a given sample bacteriome showed the presence of a population comprising up to three distinct sequences, with only 96% identity between the two most divergent sequences. This represented sequence variation that was greater than that observed between the primary β-endosymbionts from any two of the mealybug species. A phylogenetic analysis of the gammaproteobacteria is shown in Fig. 5, and the sequences used are shown in Table 4.
Table 4.
| Common name | Species used for isolation | 16S Gamma NCBI accession no. | Tree annotationb | Gammaproteobacterium |
|---|---|---|---|---|
| Aphid | Acyrthosiphon pisum | M27039 | ApisP | Buchnera aphidicola |
| Aphid | Acyrthosiphon pisum | AF293616 | Apis1 | “Candidatus Hamiltonella defensa” |
| Aphid | Acyrthosiphon pisum | AF293618 | Apis2 | “Candidatus Regiella insecticola” |
| Aphid | Acyrthosiphon pisum | AB033777 | Apis3 | “Candidatus Serratia” symbiotic |
| Bloodsucking fly | Craterina melbae | EF174495 | Cmel | Sodalis sp. |
| Carpenter ant | Camponotus floridanus | X92549 | Cflo | “Candidatus Blochmannia floridanus” |
| Carpenter ant | Camponotus herculeanus | X92550 | Cher | “Candidatus Blochmannia herculeanus” |
| Free-living bacterium | NC_012660 | Pflu | Pseudomonas fluorescens | |
| Free-living bacterium | AP009048 | Ecol | Escherichia coli W3110 | |
| Free-living bacterium | AJ294747 | Tvir | Thalassomonas viridians | |
| Free-iving bacterium | X74694 | Vcho | Vibrio cholerae | |
| Jewel wasp | Nasonia vitripennis | M90801 | Nvit | Arsenophonus nasoniae |
| Mealybug | Antonina pretiosa | AF476101 | Apre | |
| Mealybug | Amonostherium lichtensioides | AF476100 | Alic | |
| Mealybug | Antonina crawii | AB030020 | Acra | |
| Mealybug | Australicoccus grevilleae | AF476099 | Agre | |
| Mealybug | Cyphonococcus alpines | AF476102 | Calp | |
| Mealybug | Dysmicoccus brevipes | AF476103 | Dbre | |
| Mealybug | Dysmicoccus neobrevipes | AF476104 | Dneo | |
| Mealybug | Erium globosum | AF476105 | Eglo | |
| Mealybug | Melanococcus albizziae | AF476106 | Malb | |
| Mealybug | Paracoccus nothofagicola | AF476109 | Pnot | |
| Mealybug | Planococcus citri | AF322016 | Pcit | |
| Mealybug | Planococcus ficus | AF476108 | Pfic | |
| Mealybug | Planococcus kraunhiae | AB374417 | Pkra | |
| Mealybug | Pseudococcus calceolariae | JN182338 | Pcal1-1 | |
| Mealybug | Pseudococcus calceolariae | JN182339 | Pcal1-2 | |
| Mealybug | Pseudococcus comstocki | AB374418 | Pcom | |
| Mealybug | Pseudococcus longispinus | EU727120 | Plon | |
| Mealybug | Pseudococcus viburni | JN182340 | Pvib1-3 | |
| Mealybug | Pseudococcus viburni | JN182341 | Pvib1-2 | |
| Mealybug | Pseudococcus viburni | JN182342 | Pvib1-1 | |
| Mealybug | Pseudococcus viburni | JN182343 | Pvib2-3 | |
| Mealybug | Pseudococcus viburni | JN182344 | Pvib2-1 | |
| Mealybug | Pseudococcus viburni | JN182345 | Pvib2-2 | |
| Mealybug | Vryburgia amaryllidis | AF476110 | Vama | |
| Sharpshooter | Homalodisca coagulata | AF465793 | Hcoa | “Candidatus Baumannia cicadellinicola” |
| Tsetse fly | Glossina brevipalpis | AP008232 | Gbre1 | Sodalis glossinidius strain Morsitans |
| Tsetse fly | Glossina brevipalpis | BA000021 | Gbre2 | Wigglesworthia glossinidia |
| Weevil | Sitophilus oryzae | AF005235 | SoryP | |
| Whitefly | Bemisia tabaci | Z11925 | BtabP | “Candidatus Portiera aleyrodidarum” |
All sequences except three from free-living gammaproteobacteria are those of the 16S RNA gene from the gammaproteobacterial symbiont isolated from the named insect species. The 16S ribosomal sequence from the free-living gammaproteobacterium E. coli (NCBI W3110) was used as the outgroup.
Known P-endosymbionts are annotated with the suffix “P.” Sequences from individual insects (this study) are annotated as x-y, where x is individual insect and y is independent sequence.
Also detected under these conditions were sequences closely homologous to those of Pseudomonas and Acinetobacter species; both are gammaproteobacteria common in soil and marine samples and not known to include endosymbiont species. They were presumed to be contaminants from the diet, environment, gut, or possibly the hemolymph and are not further discussed. No gammaproteobacteria closely homologous to S. glossinidius were detected in P. longispinus by the above-mentioned molecular methods, but S-endosymbionts were clearly visible within the P-endosymbionts by light microscopy (Fig. 2).
Predicted endosymbiont GroEL-virus interactions.
An alignment of GroES and HSP10 proteins is shown in Fig. 6, and the sequences used are described in Table 5. The bacterial consensus GXIVLXGXA (P_HHH_P_H) is shared by the free-living gammaproteobacterium E. coli and the other gammaproteobacterial symbionts except “Candidatus Carsonella ruddii,” which has GSIFLPFND, which is P_HHH_H_− in terms of hydrophobicity. The free-living betaproteobacterium Neisseria gonorrhoeae also fits the P_HHH_P_H consensus. The protist amoeba is the divergent eukaryote GGIFIPTNK/P_HHH_P_+ (as might be expected), and the Arabidopsis thaliana chloroplast has diverged from its (presumed) ancestral bacterial sequence to an apparently novel sequence, GGVLLPKAA/P_HHH_+_H, with a lysine at the 7th position. Species of the mealybug S-endosymbiont have the typical prokaryotic sequence for the mobile loop of the GroES protein. However, unexpectedly, the GroES mobile loop of “Ca. Tremblaya princeps” (the P-endosymbiont in mealybugs; Q8KTR9; AAM75979) has a binding pattern of CGIVIPDSA/H_HHH_−_H, which closely resembles the eukaryotic consensus sequence GGIVLPEKA/P_HHH_−_P and is quite distinct from the prokaryotic consensus sequence present in other known endosymbionts (Fig. 6). The seventh position is aspartic acid, similar to eukaryotic sequences, which have glutamic acid at this position, and quite distinct from other prokaryotic sequences, which have glycine at the seventh position. Matching the charged residue in the “Ca. Tremblaya princeps” pattern is thought to be the crucial point of difference from the E. coli prediction (40). The E. coli experimental and model results are in fair agreement, considering that the experimental results (Table 2) show binding of GroEL to whole intact viral particles and the model predicts binding to the denatured major viral coat protein. Further, in a personal communication, G. Lorimer (University of Maryland) noted that “your experimental analysis of plant viral coat proteins and their interaction with GroEL, based only on sequence, seems to work quite well with highly charged plant viral coat proteins but fails with proteins that are only slightly charged. Obviously we are missing an electrostatic component. A similar conclusion was reached based on analysis of barnase mutants with GroEL” (4).
Table 5.
HSP10 and GroES sequences from diverse organismsa
| Species | NCBI accession no. | Alignment annotation | Organism | Note |
|---|---|---|---|---|
| Wolbachia strain TRS | YP_198180 | Wolbachia strain TRS GroES | Alphaproteobacterium | Endosymbiont of nematode Brugia malay |
| Wolbachia sp. | NP_966108 | Wolbachia sp. GroES | Alphaproteobacterium | Symbiont of fruitfly Drosophila melanogaster |
| “Candidatus Tremblaya princeps” | Q8KTR9 | “Ca. Tremblaya princeps” GroES | Betaproteobacterium | P-endosymbiont of mealybugs |
| Neisseria gonorrhoeae | YP_002003183 | N. gonorrhoeae GroES | Betaproteobacterium | Free living |
| Buchnera aphidicola | Q9F4E4 | B. aphidicola1 GroES | Gammaproteobacterium | P-symbiont of aphids |
| Buchnera aphidicola | AAC04236 | B. aphidicola2 GroES | Gammaproteobacterium | P-endosymbiont of aphid Myzus persicae |
| “Candidatus Blochmannia pennsylvanicus” | YP_277588 | “Ca. Blochmannia pennsylvanicus” GroES | Gammaproteobacterium | P-symbiont of the carpenter ant |
| “Candidatus Carsonella ruddii” | YP_802449 | “Ca. Carsonella ruddii” GroES | Gammaproteobacterium | P-endosymbiont of psyllids |
| Escherichia coli | NP_418566 | E. coli K12 GroES | Gammaproteobacterium | Free living |
| Unnamed | AAB97669.1 | S. oryzae P-endosymbiont GroES | Gammaproteobacterium | Endosymbiont of aphid Sitophilus oryzae |
| Sodalis glossinidius | YP_453985 | S. glossinidius GroES | Gammaproteobacterium | S-endosymbiont of tsetse fly Glossina morsitans morsitans |
| Wigglesworthia glossinidia | NP_871262 | W. glossinidia GroES | Gammaproteobacterium | P-endosymbiont of tsetse fly Glossina brevipalpis |
| Methanohalophilus portucalensis | ABO16620 | M. portucalensis GroES | Archaeon | |
| Dictyostelium discoideum | XP_636819 | Amoeba HSP10 | Protist | |
| Saccharomyces cerevisiae | NP_014663 | S. cerevisiae HSP10 | Yeast | |
| Ashbya gossypii | NP_984626 | A. gossypii HSP10 | Fungus | |
| Apis mellifera | XP_624910 | Honey bee HSP10 | Bee | |
| Tribolium castaneum | XP_975179 | Tribolium HSP10 | Beetle | |
| Drosophila melanogaster | NP_648622 | Drosophila HSP10 | Fruitfly | |
| Nasonia vitripennis | XP_00159999 | Jewel wasp HSP10 | Wasp | |
| Ostreococcus lucimarinus | XP_001421602 | Green alga HSP10 | Alga | |
| Arabidopsis thaliana | NP_563961 | Arabidopsis HSP10N | Dicot | Nuclear gene |
| Arabidopsis thaliana | NP_566022 | Arabidopsis HSP10C | Dicot | Chloroplast gene |
| Gallus gallus | NP_990398 | Chicken HSP10 | Bird | |
| Danio rerio | NP_571601 | Zebrafish HSP10 | Fish | |
| Homo sapiens | NP_002148.1 | Human HSP10 | Mammal |
These sequences were used in the alignment in Fig. 6.
Using our variation (see Materials and Methods) of the existing binding model (40), GLRaV-3 has two perfect binding sites for a “Ca. Tremblaya princeps” GroEL homolog in its coat protein but only imperfect sites for the prokaryotic GroEL pattern (Table 2). This predicts that the “Ca. Tremblaya princeps” GroEL homolog is able to bind GLRaV-3 coat protein whereas E. coli GroEL cannot, the latter observation having been confirmed (1). The data suggest a role for “Ca. Tremblaya princeps” GroEL in viral transmission in mealybugs. Predictions by computational studies of hydrophobic sites that remain exposed on the surfaces of assembled viral particles and hence available for binding GroEL are possible, given X-ray crystal structures of assembled viral coats or their equivalent, but are beyond this investigation.
DISCUSSION
Previous studies have used 16S RNA sequences of mealybugs to link the sequence variation of P-endosymbionts with the evolutionary histories of their insect hosts and proposed cospeciation (5). Our data are consistent with this hypothesis. In the single case of the divergent sequence we have identified from P. longispinus, it is possible that some sequence drift occurs within these essentially clonally replicating species. However, the alternative explanations (species misidentification and cross-contamination) seem more likely.
The situation with the S-endosymbionts is quite different and complex, showing multiple and divergent sequences within individual mealybugs. The possibility exists that at least some of these divergent ribosomal sequences are chimeric in nature and hence artifactual. Chimera formation in PCR amplification is the most frequent 16S rRNA gene artifact currently displayed by new submissions to the public repositories (3). Chimeras tend also to be the most insidious artifacts, since if undetected they can be responsible for spurious phylogenetic reconstructions, inaccurate taxonomic identifications, and overestimations of microbial diversity. However, the nature of the variations seen in the data (short indels and base changes inconsistent with simple PCR errors) and the close homology with published S. glossinidius sequences over their entire lengths are inconsistent with the six P. viburni sequences being gross cross-species chimeras. While it is still possible for them to represent chimeras between close (S. glossinidius-like) species, this would not invalidate the observation of multiple strains of S-endosymbionts in an individual mealybug, as multiple strains would still need to be present to create the hypothetical chimeras. The two P. calceolariae sequences present insufficient data to draw conclusions in this matter.
Sequence differences in the regions of the PCR primers may explain our failure to detect S-endosymbionts in P. longispinus, as may the P. longispinus S-endosymbionts being from some more divergent group of bacteria. Another explanation is the possibility that the S. glossinidius-like sequences from the other two mealybug species were contaminants, independent endosymbionts, or exosymbionts. This is unlikely, because S. glossinidius-related species have been detected in related mealybugs (5, 24, 28, 41). Furthermore, other bacteria were not seen in the photomicrographs, meaning that, if present, they would be at very low concentrations and hence unlikely to be detected by the initial molecular method employed. Finally, there is a report of a partial sequence for a P. longispinus gammaproteobacterium endosymbiont 16S gene (17) that groups closely with the Sodalis clade and only more loosely with the other mealybug S-endosymbiont clades (Fig. 5).
Thao et al. (41) found S-endosymbionts in many mealybug species, including P. citri, Planococcus ficus, and Paracoccus nothofagus (a New Zealand species), but did not detect S-endosymbionts in Pseudococcus species. The sequences for these mealybug species are closely homologous (Fig. 1, AF476107, AF476108, and AF476109) to the S. glossinidius sequences and our individual sequences from Pseudococcus species. They concluded that the S-endosymbionts had infected their host P-endosymbionts multiple times and coevolved with them, i.e., across a wide range of mealybug species, independent infections occurred, events that then resulted in coevolution. A recent study (24) supports this conclusion, which is in contrast with the results from tsetse flies (2), where no evidence for coevolution of the S-endosymbiont was found. Our data extend previous studies on mealybug S-endosymbiont sequences through the identification of multiple sequences isolated from each individual insect examined where an S-symbiont was detected. This was achieved by sequencing clones from PCR products rather than sequencing PCR products directly, where we had also observed only consensus sequences. It is possible that the sequences previously reported are consensus sequences similar to those we obtained by direct sequencing of PCR products and that multiple sequences also occur in these other mealybug species but were not detected by the methods used in the earlier studies.
The reason for this variability is unknown. The mealybug S-endosymbiont is closely related phylogenetically to S. glossinidius, an S-endosymbiont of tsetse flies that is able to live both inter- and intracellularly in various host tissues, including the midgut and hemolymph. Phylogenetic studies have not indicated a correlation between the evolution of Sodalis spp. and tsetse flies, unlike the tsetse fly P-endosymbiont, Wigglesworthia (2), and the mealybug endosymbiont “Ca. Tremblaya princeps” (5). It seems that S. glossinidius-related species are more parasitic than endosymbiotic and that it is routinely and frequently acquired and may be able to be passed from one individual insect to another. Sodalis species have been cultured in vitro in insect cell lines from species widely diverged from their normal hosts (46), and members of the genus have a genome about 50% the size of the E. coli genome (43), as opposed to that of “Ca. Tremblaya princeps” at 10%. Erosion of the genome is a characteristic of symbionts. S. glossinidius-related species therefore may even have characteristics suitable for maintaining a nonsymbiont life stage in nature, although this has not been observed to our knowledge. It certainly appears to be able to pass from insect to insect with its ability to grow in nonhost cells, whereas normal endosymbionts are unable to survive outside the cells of their usual host. Certainly, individuals of an essentially clonally replicating species carrying multiple strains of an endosymbiotic bacterium strongly suggests multiple acquisitions over time.
The GroES mobile-loop sequence convergence of “Ca. Tremblaya princeps” with the eukaryotic HSP10 mobile loop is a novel observation in this study and provides an explanation for a role for “Ca. Tremblaya princeps” in GLRaV-3 transmission. A role in viral transmission for S-endosymbionts is correspondingly remote. The convergence is, however, unlikely to be a selection to bind the virus coat, since no apparent advantage to the endosymbiont results from the interaction. The convergence might be better explained as a presumed convergence of “Ca. Tremblaya princeps” GroEL with eukaryotic HSP60. However, because the binding site on the GroEL homologs contains contacts from several parts of the GroEL molecule, this can only be determined from the GroES mobile-loop sequence. We hypothesize that “Ca. Tremblaya princeps” has a selective advantage in the ability of its GroEL to recognize, bind, and fold eukaryotic proteins, viz., the proteins of its host mealybug. It seems probable that this ability is a key reason why the endosymbiont has been able to substitute eukaryotic proteins for its own in a folded and hence active form following transport of unfolded protein across the bacterial membrane from the host. Thus, we believe that GroEL convergence to the specificity of the host of this primary endosymbiont is a major factor in allowing the observed genome erosion to occur.
Is convergence of prokaryotic GroEL with eukaryotic GroEL a common feature of endosymbionts and other degraded prokaryotic genomes, such as mitochondria and chloroplasts? If this is likely, we should observe typical prokaryotic GroES mobile loops in free-living bacteria and in the S-endosymbiont of mealybugs that lives within a bacterium rather than a eukaryote but converged GroES mobile loops in P-endosymbionts. Figure 6 shows some tantalizing hints, but further studies are required to explore this hypothesis.
Of particular relevance to the research on the mitigation of grapevine leafroll disease is a study that has implicated the endosymbiotic bacteria of the whitefly Bemisa tabaci in that insect's ability to transmit tomato yellow leaf curl virus (31). At first glance, this may not seem relevant to mealybugs, because the received wisdom is that GLRaV-3 is transmitted via a semipersistent mechanism whereby the virus is restricted to the fore- and/or midgut. If this is the case, then it is difficult to see what effects the endosymbionts, which are restricted to their bacteriocyte on the other side of the gut wall, might have on the ability of a mealybug to transmit a plant virus. However, very recent research suggests that GLRaV-3 may, in fact, be circulatively transmitted in mealybugs (13). This means that the virus is transported from the gut to the salivary glands and hence must cross the gut wall into the hemocoel. This transmission mechanism is considerably more complex and allows much greater opportunities for the endosymbionts to have an impact.
However, the mealybug species that was studied (13) transmits GLRaV-3 much more efficiently than GLRaV-1, and hence, there is a specific and selective step in the process that transports (and protects) the virus from the gut through the hemolymph to the salivary glands. Others (1, 30, 31, 41) have found that (for other insect-transmitted viruses in sucking insects) a specific region of the intact virus (i.e., the assembled viral coat) binds to the endosymbiont GroEL and that mutant viruses do not persist in the insect. For the virus to be specifically transported from the gut to the salivary glands without being degraded there must be some protein specifically binding to the virus. If this protein is the endosymbiont GroEL homolog, which is not identical in sequence to E. coli GroEL, then the fact that the virus does not bind to E. coli GroEL does not conflict with or invalidate the hypothesis.
After initial submission of this paper, details of a novel inversion in the genome of “Ca. Tremblaya princeps” were published, as well as a sequence and proposed name (“Candidatus Moranella endobia”) for the secondary endosymbionts (28). These data raise similar questions about the evolution and maintenance of multiple genetic elements in linked genomes where no compelling case for a selective advantage can readily be made.
ACKNOWLEDGMENTS
We thank G. Lorimer (University of Maryland) for his sage counsel.
The research was funded by the New Zealand Foundation for Science and Technology through Capability Development Fund no. CAP06-56 of the Plant and Food Research Institute.
Footnotes
Published ahead of print 9 December 2011
REFERENCES
- 1. Akad F, Dotan N, Czosnek H. 2004. Trapping of Tomato yellow leaf curl virus (TYLCV) and other plant viruses with a GroEL homologue from the whitefly Bemisia tabaci. Arch. Virol. 149:1481–1497 [DOI] [PubMed] [Google Scholar]
- 2. Aksoy S, Pourhosseini AA, Chow A. 1995. Mycetome endosymbionts of tsetse flies constitute a distinct lineage related to Enterobacteriaceae. Insect Mol. Biol. 4:15–22 [DOI] [PubMed] [Google Scholar]
- 3. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Axe DD, Foster NW, Fersht AR. 1996. Active barnase variants with completely random hydrophobic cores. Proc. Natl. Acad. Sci. U. S. A. 93:5590–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baumann L, Baumann P. 2005. Cospeciation between the primary endosymbionts of mealybugs and their hosts. Curr. Microbiol. 50:84–87 [DOI] [PubMed] [Google Scholar]
- 6. Baumann L, Thao ML, Hess JM, Johnson MW, Baumann P. 2002. The genetic properties of the primary endosymbionts of mealybugs differ from those of other endosymbionts of plant sap-sucking insects. Appl. Environ. Microbiol. 68:3198–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155–189 [DOI] [PubMed] [Google Scholar]
- 8. Baumann P. 2006. Diversity of procaryote-insect associations with the Sternorrhycha (psyllids, whiteflires, aphids, mealybugs), vol 2 CRC Press, Boca Raton, FL [Google Scholar]
- 9. Bovey R, Gartel W, Hewitt WB, Martelli GP, Vuittenez A. 1980. Virus and virus-like diseases of grapevine. Payot Publishing Co., Lausanne, France [Google Scholar]
- 10. Cabaleiro C, Segura A. 1997. Field transmission of grapevine leafroll associated virus 3 (GLRaV-3) by the mealybug Planococcus citri. Plant Dis. 81:283–287 [DOI] [PubMed] [Google Scholar]
- 11. Charles JG, et al. 2006. A review of grapevine leafroll associated virus type 3 (GLRaV-3) for the New Zealand wine industry. HortResearch client report no. 18447 HortResearch, Palmerston North, New Zealand [Google Scholar]
- 12. Charles JG, Froud KJ, van den Brink R, Allan DJ. 2009. Mealybugs and the spread of grapevine leafroll-associated virus 3 (GLRaV-3) in a New Zealand vineyard. Australasian Plant Pathol. 38:576–583 [Google Scholar]
- 13. Cid M, Pereira S, Cabaleiro C, Faoro F, Segura A. 2007. Presence of Grapevine leafroll-associated virus 3 in primary salivary glands of the mealybug vector Planococcus citri suggests a circulative transmission mechanism. Eur. J. Plant Pathol. 118:23–30 [Google Scholar]
- 14. Dale C, Moran NA. 2006. Molecular interactions between bacterial symbionts and their hosts. Cell 126:453–465 [DOI] [PubMed] [Google Scholar]
- 15. Douglas A. 2007. Interactions between insects and their symbiotic microorganisms. Comp. Biochem. Physiol. A 146:S217 doi: 10.1016/j.cbpa.2007.01.4739 [Google Scholar]
- 16. Douglas AE. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17–37 [DOI] [PubMed] [Google Scholar]
- 17. Duron O, et al. 2008. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edelbaum D, Gorovits R, Sasaki S, Ikegami M, Czosnek H. 2009. Expressing a whitefly GroEL protein in Nicotiana benthamiana plants confers tolerance to tomato yellow leaf curl virus and cucumber mosaic virus, but not to grapevine virus A or tobacco mosaic virus. Arch. Virol. 154:399–407 [DOI] [PubMed] [Google Scholar]
- 19. Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. University of Washington, Seattle, WA [Google Scholar]
- 20. Golino DA, Sim ST, Gill R, Rowhani A. 2002. California mealybugs can spread grapevine leafroll disease. California Agric. 56:196–201 [Google Scholar]
- 21. Hardy NB, Gullan PJ, Hodgson CJ. 2008. A subfamily-level classification of mealybugs (Hemiptera: Pseudococcidae) based on integrated molecular and morphological data. Syst. Entomol. 33:51–71 [Google Scholar]
- 22. Jaenike J. 2009. Coupled population dynamics of endosymbionts within and between hosts. Oikos 118:353–362 [Google Scholar]
- 23. Kantheti P, Jayarama KS, Chandra HS. 1996. Developmental analysis of a female-specific 16S rRNA gene from mycetome-associated endosymbionts of a mealybug, Planococcus lilacinus. Insect Biochem. Mol. Biol. 26:997–1009 [DOI] [PubMed] [Google Scholar]
- 24. Kono M, Koga R, Shimada M, Fukatsu T. 2008. Infection dynamics of coexisting beta- and gammaproteobacteria in the nested endosymbiotic system of mealybugs. Appl. Environ. Microbiol. 74:4175–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin Z, Rye HS. 2006. GroEL-mediated protein folding: making the impossible, possible. Crit. Rev. Biochem. Mol. Biol. 41:211–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ling K-S, Zhu H-Y, Gonsalves D. 2004. Complete nucleotide sequence and genome organization of Grapevine leafroll-associated virus 3, type member of the genus Ampelovirus. J. Gen. Virol. 85:2099–2102 [DOI] [PubMed] [Google Scholar]
- 27. Lund PA. 1995. The roles of molecular chaperones in vivo. Essays Biochem. 29:113–123 [PubMed] [Google Scholar]
- 28. McCutcheon JP, von Dohlen CD. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 21:1366–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moran NA, Plague GR, Sandstrom JP, Wilcox JL. 2003. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc. Natl. Acad. Sci. U. S. A. 100:14543–14548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morin S, Ghanim M, Sobol I, Czosnek H. 2000. The GroEL protein of the whitefly Bemisia tabaci interacts with the coat protein of transmissible and nontransmissible begomoviruses in the yeast two-hybrid system. Virology 276:404–416 [DOI] [PubMed] [Google Scholar]
- 31. Morin S, et al. 1999. A GroEL homologue from endosymbiotic bacteria of the whitefly Bemisia tabaci is implicated in the circulative transmission of tomato yellow leaf curl virus. Virology 256:75–84 [DOI] [PubMed] [Google Scholar]
- 32. Muller LL, Jacks TJ. 1975. Rapid chemical dehydration of samples for electron microscopic examinations. J. Histochem. Cytochem. 23:107–110 [DOI] [PubMed] [Google Scholar]
- 33. Munson MA, Baumann P, Moran NA. 1992. Phylogenetic relationships of the endosymbionts of mealybugs (Homoptera: Pseudococcidae) based on 16S rDNA sequences. Mol. Phylogenet. Evol. 1:26–30 [DOI] [PubMed] [Google Scholar]
- 34. Nicholas KB, Nicholas HB, Jr, Deerfield DWI. 1997. GeneDoc: analysis and visualization of genetic variation. Embnew. News 4:14 [Google Scholar]
- 35. O'Fallon B. 2008. Population structure, levels of selection, and the evolution of intracellular symbionts. Evolution 62:361–373 [DOI] [PubMed] [Google Scholar]
- 36. Page RDM. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 37. Petersen CL, Charles JG. 1997. Transmission of grapevine leafroll-associated closteroviruses by Pseudococcus longispinus and P. calceolariae. Plant Pathol. 46:509–515 [Google Scholar]
- 38. Roland JC, Vian B. 1991. General preparation and staining of thin sections, p 1–66 In Hall JL, Hawes C. (ed), Electron microscopy of plant cells. Academic Press, New York, NY [Google Scholar]
- 39. Sforza R, Boudon-Padieu E, Greif C. 2003. New mealybug species vectoring Grapevine leafroll-associated viruses-1 and -3 (GLRaV-1 and -3). Eur. J. Plant Pathol. 109:975–981 [Google Scholar]
- 40. Stan G, Brooks BR, Lorimer GH, Thirumalai D. 2005. Identifying natural substrates for chaperonins using a sequence-based approach. Protein Sci. 14:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thao ML, Gullan PJ, Baumann P. 2002. Secondary (gamma-Proteobacteria) endosymbionts infect the primary (beta-Proteobacteria) endosymbionts of mealybugs multiple times and coevolve with their hosts. Appl. Environ. Microbiol. 68:3190–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Toh H, et al. 2006. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16:149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van den Heuvel J, et al. 1997. The N-terminal region of the luteovirus readthrough domain determines virus binding to Buchnera GroEL and is essential for virus persistence in the aphid. J. Virol. 71:7258–7265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. von Dohlen CD, Kohler S, Alsop ST, McManus WR. 2001. Mealybug beta-proteobacterial endosymbionts contain gamma-proteobacterial symbionts. Nature 412:433–436 [DOI] [PubMed] [Google Scholar]
- 46. Welburn SC, Maudlin I, Ellis DS. 1987. In vitro cultivation of Rickettsia-like-organisms from Glossina spp. Ann. Trop. Med. Parasitol. 81:331–335 [DOI] [PubMed] [Google Scholar]