Abstract
The study of the microbiotas of 19 Italian sourdoughs used for the manufacture of traditional/typical breads allowed the identification, through a culture-dependent approach, of 20 and 4 species of lactic acid bacteria (LAB) and yeasts, respectively. Numerically, the most frequent LAB isolates were Lactobacillus sanfranciscensis (ca. 28% of the total LAB isolates), Lactobacillus plantarum (ca. 16%), and Lactobacillus paralimentarius (ca. 14%). Saccharomyces cerevisiae was identified in 16 sourdoughs. Candida humilis, Kazachstania barnettii, and Kazachstania exigua were also identified. As shown by principal component analysis (PCA), a correlation was found between the ingredients, especially the type of flour, the microbial community, and the biochemical features of sourdoughs. Triticum durum flours were characterized by the high level of maltose, glucose, fructose, and free amino acids (FAA) correlated with the sole or main presence of obligately heterofermentative LAB, the lowest number of facultatively heterofermentative strains, and the low cell density of yeasts in the mature sourdoughs. This study highlighted, through a comprehensive and comparative approach, the dominant microbiotas of 19 Italian sourdoughs, which determined some of the peculiarities of the resulting traditional/typical Italian breads.
INTRODUCTION
During the last decades, European and worldwide consumers have come ever more to appreciate traditional and typical foods. Traditional is the definition used for foods that historically are part of the cultural heritage of people living in a certain geographical area (29a). Typical is the attribute used for a food produced using one or more ingredients having characteristics strictly depending on the geographical area it comes from (9). Mainly due to the long history of regional political division, about 200 different types of bread are manufactured throughout Italy with large differences of recipes and traditions (24). Some breads have already received the Protected Designation of Origin (PDO) (Pane di Altamura and Pagnotta del Dittaino) or the Protected Geographical Indication (PGI) (Pane di Matera, Pane Casareccio di Genzano, and Coppia Ferrarese). In spite of the differences, almost all traditional/typical Italian breads use sourdough as the natural starter. Sourdough represents a very complex biological ecosystem (19) where yeasts and, especially, lactic acid bacteria (LAB) largely determine the sensory, technology, nutritional, and functional features of the resulting baked goods (20). In mature sourdoughs, LAB occur in numbers >108 CFU g−1, whereas the number of yeasts is at least one order of magnitude lower (19). The microbial composition of the sourdough was subjected to numerous studies which have revealed a large species diversity (for reviews, see references 15, 16, 22, and 43). Overall, Lactobacillus brevis, Lactobacillus paralimentarius, Lactobacillus plantarum, Lactobacillus rossiae, and Lactobacillus sanfranciscensis dominate sourdough processes that are characterized by low incubation temperatures and continuous back slopping (traditional type I sourdoughs). Industrialized, type II sourdoughs are characterized by higher temperature, longer time of fermentation, and higher water content than type I sourdoughs. They harbor thermophilic and acid-tolerant lactobacilli, such as Lactobacillus amylovorus, Lactobacillus fermentum, Lactobacillus pontis, and Lactobacillus reuteri. Species belonging to the genera Enterococcus, Lactococcus, Leuconostoc, Pediococcus, Streptococcus, and Weissella are less frequently isolated (8, 41, 49). Other studies (29, 38) showed that the composition of the sourdough LAB microbiotas is subjected to fluctuations depending on the robustness of strains and ecological factors. Candida krusei, Candida milleri, Kazachstania exigua, Pichia norvegensis, Saccharomyces cerevisiae, Torulaspora delbrueckii, and Wickerhamomyces anomalus (formerly Pichia anomala) are the yeasts most commonly found in sourdoughs (23, 27, 31, 39). Comparative studies on numerous sourdoughs which are used for the manufacture of various traditional/typical breads may (i) highlight the ingredients and technology factors contributing to the stability of the microbiotas and competitiveness between LAB and yeasts and (ii) establish the effects of the microbiotas on the biochemical characteristics of the sourdough (44).
This study aimed at the identification of the LAB and yeast microbiotas of 19 Italian sourdoughs used for the manufacture of traditional/typical Italian breads. The dominating LAB and yeasts were monitored by culture-dependent methods. Multivariate statistical analyses were performed in order to find the correlation between ingredients and the composition of the sourdough microbiotas, as well as the effects of the latter on the biochemical characteristics of sourdoughs.
MATERIALS AND METHODS
Sourdoughs.
Nineteen sourdoughs used for the manufacture of traditional/typical Italian breads were studied (Table 1). The sourdough breads were representative for several Italian regions (see Fig. S1 in the supplemental material). Table 1 summarizes the ingredients and technology parameters of the sourdoughs studied. Three batches of each sourdoughs were collected on three consecutive days at local bakeries. All samples were taken immediately at the end of the final back slopping and were stored at 4°C for a few hours before analyses. All the analyses were carried out at least in duplicate for each batch of sourdough (total of six analyses for each type of sourdough).
Table 1.
Ingredients and technology parameters of sourdoughs used for the manufacture of traditional/typical Italian breads
| Sourdougha | Type of flour | % NaCl added per back slopping | % Baker's yeast | % Sourdough used in back slopping | No. of back sloppings | Time (h) and temp (°C) of back sloppingb | Sourdough as leavening agent (LA) or baking improver (BI) |
|---|---|---|---|---|---|---|---|
| Pane di Altamura PDO | T. durum | 2.0 | 0 | 20 | 3 | 10, 25 | LA |
| Pane di Laterza | T. durum | 2.0 | 0 | 28 | 1 | 6, 25 | LA |
| Pane di Matera PGI | T. durum | 2.5–3.0 | 0 | 10 | 2 | 6, 25 | LA |
| Pane di Montecalvo Irpino | T. aestivum/T. durum (20/80%) | 2.0 | 0 | 20 | 2 | 6, 25 | BI |
| Pane Casereccio di Reggio Calabria | T. aestivum | 1.5–2.0 | 0 | 50 | 2–3 | 6, 22 | BI |
| Pane Casereccio del Molise | T. aestivum | 2.0 | 0 | 30 | 2–3 | 10, 22 | BI |
| Pane Casareccio di Genzano PGI | T. aestivum | 2.0 | 0.4 | 1.5 | 1 | 6, 30 | BI |
| Bozza Pratese | T. aestivum | 0 | 0 | 20 | 1 | 15, 11 | BI |
| Pane di Altopascio Tradizionale | T. aestivum | 0 | 0 | 40 | 1 | 24, 9 | LA |
| Pane di Terni | T. aestivum | 0 | 0 | 14 | 1 | 12, 16 | BI |
| Bastone di Padova | T. aestivum | 2.0 | 0.6 | 45 | 1 | 18, 27 | BI |
| Coppia Ferrarese PGI | T. aestivum | 2.0 | 0 | 10 | 1 | 12, 22 | BI |
| Pane Casereccio Marchigiano | T. aestivum | 2.0 | 0 | 33 | 1 | 6, 23 | BI |
| Pane di Cappelli | T. durum | 1.5 | 0 | 25 | 2 | 4, 26 | LA |
| Moddizzosu | T. durum | 1.8–2.0 | 0 | 30 | 1 | 11, 22 | LA |
| Pane Carasau | T. durum | 1.0–2.0 | 0 | 30 | 1 | 11, 27 | BI |
| Pane nero di Castelvetrano | T. durum | 1.5 | 0 | 1 | 1–2 | 10, 26 | BI |
| Pane di Lentini | T. durum | 2.0 | 0 | 20 | 2 | 11, 25 | LA |
| Pagnotta del Dittaino PDO | T. durum | 2.0 | 0 | 16.5 | 1 | 5, 22 | LA |
Sourdoughs are indicated with the names of the related breads.
The first number indicates the length of back slopping, and the second number indicates the temperature of incubation. Mature sourdough, after the last back slopping, was subjected to analyses.
Determination of pH, carbohydrates, organic acids, ethanol, and free amino acids.
The pH values were determined by using a pH meter. Ten grams of unfermented dough (flour and water, dough yield [DY; (dough weight/flour weight) × 100] of 160) or mature sourdough were homogenized with 90 ml of Tris-HCl 50 mM (pH 8.8) buffer and treated for 3 min in a BagMixer 400P (Interscience, St. Nom, France) blender. After incubation (at 25°C for 30 min with stirring), the suspension was centrifuged (12,857 × g, 10 min, 4°C). The supernatant (water-soluble extract) was analyzed for the concentration of carbohydrates (unfermented dough) and organic acids (mature sourdough). They were determined by fast protein liquid chromatography (FPLC) using an ÄKTA purifier system (GE Healthcare Bio-Sciences, Uppsala, Sweden) equipped with a refractive index detector (Perkin Elmer Corp., Waltham, MA). The quotient of fermentation (QF) was determined as the molar ratio between d,l-lactic and acetic acids. The concentration of ethanol was determined by an enzymatic method (EnzyPlus; BioControl Systems, Inc., United States). The concentrations of free amino acids (FAA) in the water-soluble extracts of unfermented dough and mature sourdough were determined using a Biochrom 30 amino acid analyzer (Biochrom LTD, Cambridge Science Park, England) (11). A mixture of amino acids at known concentration (Sigma Chemical Co., Milan, Italy) was added with cysteic acid, methionine sulfoxide, methionine sulfone, tryptophan, and ornithine and used as the external standard (11).
LAB and yeast enumeration and isolation.
Ten grams of sourdough were homogenized with 90 ml of sterile peptone water (1% [wt/vol] peptone and 0.9% [wt/vol] NaCl) solution. LAB were counted and isolated by using modified MRS (mMRS) (containing 1% [wt/vol] maltose, 5% [vol/vol] fresh yeast extract, pH 5.6), sourdough bacteria (SDB), MRS5 (28), and glucose-M17 (containing 0.5% [wt/vol] glucose instead of lactose) agar media (Oxoid, Basingstoke, Hampshire, United Kingdom). All these media were supplemented with cycloheximide (0.1 g liter−1). Plates were incubated under anaerobiosis (AnaeroGen and AnaeroJar, Oxoid) at 30°C for 48 h. For each medium, at least five colonies were randomly selected from the plates containing the two highest sample dilutions. Gram-positive, catalase-negative, nonmotile rods and cocci isolates were cultivated in mMRS, SDB, MRS5, or glucose-M17 broth at 30°C for 24 h and restreaked onto the same agar media. For those sourdoughs lacking colonies in certain media (e.g., Pane Casereccio del Molise, Bozza Pratese, Pane di Altopascio Tradizionale, Bastone di Padova, Pane Casereccio Marchigiano, and Pagnotta del Dittaino PDO for glucose-M17 agar), more colonies were picked up from the media where colonies had grown, in order to start isolation from 20 colonies for each sourdough. All isolates considered for further analyses were able to acidify the culture medium. Stock cultures were stored at −20°C in 10% (vol/vol) glycerol. The number of yeasts was estimated on malt extract agar (MEA) and Sabouraud dextrose agar (SDA) (Oxoid) media supplemented with chloramphenicol (0.1 g l−1) at 30°C for 48 h. Randomly selected colonies (10 for each medium, except for Pane di Genzano PGI and Bozza Pratese sourdoughs, which had shown no colonies in MEA, and therefore 20 colonies were picked up from SDA plates) of yeasts from the highest plate dilutions were subcultured in malt extract or Sabouraud dextrose broth and restreaked onto the same agar media. Yeast isolates were subjected to the following phenotypical assays: capacity for fermenting galactose, sucrose, maltose, raffinose, and trehalose and capacity for growing at 37°C (3).
Genotypic characterization by RAPD-PCR analysis.
Genomic DNA of LAB was extracted according to the method of de Los Reyes-Gavilán et al. (14). Three oligonucleotides, P1 (5′-GCGGCGTCGCTAATACATGC-3′), P4 (5′-CCGCAGCGTT-3′), and M13 (5′-GAGGGTGGCGGTTCT-3′), with arbitrarily chosen sequences, were used for biotyping LAB isolates. The reaction mixture and PCR conditions for primers P1 and P4 were those described by De Angelis et al. (10). The PCR conditions for primer M13 were according to Siragusa et al. (38). Randomly amplified polymorphic DNA (RAPD)-PCR profiles were acquired with the Gel Doc 2000 documentation system and compared using Fingerprinting II Informatix Software (Bio-Rad Laboratories). The similarities of the electrophoretic profiles were evaluated by determining the Dice coefficients of similarity and using the unweighted-pair group method using average linkages (UPGMA) algorithm. Since RAPD profiles of the isolates from one batch of each type of sourdough were confirmed by analyzing isolates from two other batches, strains isolated from a single batch were further analyzed.
Genomic DNA from yeast isolates was extracted as described by Cardinali et al. (5). RAPD-PCR analysis with inter simple sequence repeat (iSSR) primer (GACA)4 was used to discriminate yeasts at the strain level. The PCR protocol was previously described by Andrade et al. (2), and PCR was performed with EuroTaq enzyme (EuroClone, Milan, Italy) in a One-Gradient thermal cycler apparatus (EuroClone). The similarities of the electrophoretic profiles were evaluated by determining the Dice coefficients of similarity and using the UPGMA algorithm. Since RAPD profiles of the isolates from one batch of each type of sourdough were confirmed by analyzing isolates from two other batches, strains isolated from a single batch were further analyzed.
Genotypic identification of LAB and yeasts.
Identification of strains was performed using two primer pairs (Invitrogen Life Technologies, Milan, Italy), LacbF/LacbR and LpCoF/LpCoR, to amplify the 16S rRNA gene of LAB (13). Primers designed for the recA gene were also used to distinguish L. plantarum, Lactobacillus pentosus, and Lactobacillus paraplantarum species using multiplex PCR (40). PheS primers were used for identification at the species level within the genera Enterococcus, Weissella, and Leuconostoc (30). The primers casei/para were used to discriminate among the species Lactobacillus casei, Lactobacillus paracasei, and Lactobacillus rhamnosus (48).
Genomic DNA from yeasts was extracted as described above. The D1/D2 domain of the 26S rRNA gene was amplified and sequenced according to the procedure described by Kurtzman and Robnett (26). Electrophoresis was carried out on agarose gel at 1.5% (wt/vol) (Gellyphor; EuroClone), and amplicons were purified with GFX PCR DNA and a gel band purification kit (GE Healthcare). Sequencing electropherogram data were processed with Geneious. rRNA gene sequence alignments were carried out using the multiple sequence alignment method (17), and identification queries were fulfilled by a BLAST search (1) in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/).
Statistical analysis.
Data (at least three biological replicates) for maltose, glucose, fructose, FAA, pH, organic acids, ethanol, and cell density were subjected to one-way analysis of variance (ANOVA), and pair comparison of treatment means was achieved by Tukey's procedure at P < 0.05, using the statistical software Statistica for Windows (Statistica 6.0 for Windows 98). Principal component analysis (PCA) using a correlation matrix was carried out to visualize the effects of ingredients and technology parameters on the sourdough microbiotas and the effects of the microbiotas on the biochemical features of the sourdoughs. For each sourdough, a microbial community profile was composed which reflected the qualitative (number of species) and quantitative (number of strains per species) diversity of LAB and yeasts (36). The chemical composition of the flours (concentrations of maltose, glucose, fructose, and total and individual FAA) or other ingredients and microbial community (cell density of LAB and yeasts, number of species, number of strains, and percentages of obligately homofermentative and obligately and facultatively heterofermentative LAB) data were used as variables for PCA analyses. In addition, microbial community data were also analyzed together with the biochemical characteristics (pH, organic acids, quotient of fermentation, ethanol, and FAA) of sourdoughs (36). All data were standardized before PCA analysis using the statistical software Statistica for Windows. Cluster analysis was also performed using data regarding ingredients and microbial community or data regarding microbial community and biochemical characteristics. For cluster analysis, the unweighted pair-group method using arithmetic averages (UPGMA) was applied, and similarities were expressed using the Pearson's r correlation coefficient. Cluster analysis was also used to pick clusters of sourdoughs and overlay confidence ellipses on the PCA score plots. In order to validate the clusters formed in PCA, ANOVA on component scores of PCA was performed.
RESULTS
Biochemical characteristics of sourdoughs.
Compared to T. aestivum, T. durum flours were characterized by higher concentrations (median values) of maltose (0.61 versus 0.21%), glucose (0.47 versus 0.25%), fructose (0.43 versus 0.26%), and total FAA (0.95 versus 0.58 g kg−1) (see Table S1 in the supplemental material). In particular, the concentrations of Ser, Ala, Val, Leu, His, and Lys and those of Thr, Glu, Gly, Cys, Tyr, Trp, and Pro were higher (ca. 1.5-fold and 2-fold, respectively) in T. durum flours than in T. aestivum flours (see Table S1 in the supplemental material).
Mature sourdoughs showed pH values ranging from 3.70 ± 0.02 (mean standard ± deviation) (Pagnotta del Dittaino PDO) to 4.28 ± 0.02 (Pane Casereccio di Reggio Calabria) (Table 2). Thirteen of the 19 sourdoughs had pH values of less than 4.0 (median value of 3.97). The concentrations of d,l-lactic acid were in agreement with the pH values and ranged from ca. 63.7 to 94.0 mM (median value of 80.0 mM). The median value for the concentration of acetic acid was 18.0 mM, with variations from ca. 6.0 to 20.6 mM. Except for Pane di Altopascio Tradizionale, all the other sourdoughs showed low and similar quotients of fermentation ranging from 3.3 ± 0.30 to 5.6 ± 0.14. The median value for the concentration of ethanol was 0.24 M. The 5th and 95th percentiles of the compiled sourdough data were 0.05 M (Pane di Matera PGI) and 0.50 M (Pane Casereccio Marchigiano), respectively. The water-soluble extracts of the 19 Italian sourdoughs contained various concentrations of total FAA. The range was from 339.6 ± 16.9 to 1,090.5 ± 54.5 mg kg−1 (Table 2). The median value was 590.9 mg kg−1. Pane di Laterza, Pane Casereccio di Reggio Calabria, and Pane Casereccio Marchigiano sourdoughs were distributed between the 75th and 95th percentile (625.2 and 763.5 mg kg−1, respectively). Outliers were represented by Pane nero di Castelvetrano (1,090.5 ± 54.5 mg kg−1), Pane di Cappelli (361.7 ± 18.0 mg kg−1), and Bastone di Padova (339.6 ± 16.9 mg kg−1) sourdoughs. Compared to the initial concentrations, FAA varied individually and in total (data not shown). Among sourdoughs, Pane di Matera and Pane di Altopascio Tradizionale were characterized by the highest concentrations of γ-amino butyric acid (GABA) (ca. 68 and 20 mg kg−1, respectively).
Table 2.
pH values, organic acid concentrations, fermentation quotient, concentration of ethanol, and total free amino acids of sourdoughs used for the manufacture of traditional/typical Italian breadsa
| Sourdoughb | pH | Lactic acid (mM) | Acetic acid (mM) | Fermentation quotient | Ethanol (M) | Free amino acids (mg kg−1) |
|---|---|---|---|---|---|---|
| Pane di Altamura PDO | 4.03 ± 0.02 d | 82.0 ± 3.6 cde | 20.0 ± 0.8 ab | 3.9 ± 0.91 e | 0.25 ± 0.01 fg | 612.2 ± 30.6 de |
| Pane di Laterza | 4.05 ± 0.01 d | 79.0 ± 3.9 def | 18.0 ± 0.7 cde | 4.4 ± 0.92 de | 0.24 ± 0.04 fg | 669.1 ± 33.0 c |
| Pane di Matera PGI | 4.21 ± 0.01 b | 68.5 ± 2.8 h | 20.6 ± 0.6 a | 3.3 ± 0.30 f | 0.05 ± 0.02 j | 555.6 ± 27.0 h |
| Pane di Montecalvo Irpino | 4.17 ± 0.03 bc | 84.0 ± 4.2 cd | 20.6 ± 0.9 a | 4.0 ± 0.93 e | 0.25 ± 0.01 fg | 562.2 ± 28.1 gh |
| Pane Casereccio di Reggio Calabria | 4.28 ± 0.02 a | 75.3 ± 3.7 f | 14.5 ± 0.7 gh | 5.2 ± 0.93 bc | 0.19 ± 0.01 h | 664.0 ± 33.2 c |
| Pane Casereccio del Molise | 3.95 ± 0.07 ef | 94.0 ± 4.7 a | 17.0 ± 0.8 ef | 5.5 ± 0.90 b | 0.35 ± 0.02 cd | 412.6 ± 20.6 k |
| Pane Casareccio di Genzano PGI | 4.14 ± 0.02 c | 63.7 ± 3.1 i | 12.1 ± 0.6 i | 5.3 ± 0.91 bc | 0.31 ± 0.02 de | 595.4 ± 29.7 ef |
| Bozza Pratese | 3.89 ± 0.01 fg | 90.0 ± 4.5 ab | 18.8 ± 0.9 bc | 4.7 ± 0.90 cd | 0.15 ± 0.01 i | 570.0 ± 28.5 fgh |
| Pane di Altopascio Tradizionale | 3.97 ± 0.01 e | 87.3 ± 2.1 bc | 6.0 ± 0.3 j | 14.5 ± 0.95 a | 0.48 ± 0.02 ab | 590.9 ± 29.5 efg |
| Pane di Terni | 3.90 ± 0.01 fg | 85.0 ± 3.7 c | 16.4 ± 0.8 f | 5.1 ± 0.88 bc | 0.25 ± 0.05 fg | 613.5 ± 30.6 de |
| Bastone di Padova | 3.90 ± 0.01 fg | 75.1 ± 2.6 f | 14.0 ± 0.7 h | 5.4 ± 0.91 b | 0.24 ± 0.01 fg | 339.6 ± 16.9 m |
| Coppia Ferrarese PGI | 3.89 ± 0.05 fg | 78.5 ± 3.9 ef | 18.3 ± 0.9 cd | 4.3 ± 0.91 de | 0.20 ± 0.01 h | 487.1 ± 24.3 j |
| Pane Casereccio Marchigiano | 3.97 ± 0.08 e | 87.1 ± 4.3 bc | 15.6 ± 1 g | 5.6 ± 0.14 b | 0.50 ± 0.01 a | 763.5 ± 38.1 b |
| Pane di Cappelli | 3.97 ± 0.02 e | 75.0 ± 3.7 fg | 18.0 ± 0.7 cde | 4.1 ± 0.90 e | 0.28 ± 0.01 ef | 361.7 ± 18.0 l |
| Moddizzosu | 3.87 ± 0.11 g | 84.2 ± 1.9 cd | 18.6 ± 0.5 c | 4.5 ± 0.91 de | 0.38 ± 0.01 c | 624.2 ± 18.4 d |
| Pane Carasau | 3.97 ± 0.08 e | 80.5 ± 3.1 de | 17.4 ± 0.4 def | 4.6 ± 0.91 d | 0.18 ± 0.01 hi | 608.7 ± 11.5 ef |
| Pane nero di Castelvetrano | 3.74 ± 0.06 h | 78.0 ± 3.9 efg | 20.0 ± 0.8 ab | 3.9 ± 0.93 e | 0.20 ± 0.01 h | 1090.5 ± 54.5 a |
| Pane di Lentini | 3.89 ± 0.02 fg | 73.4 ± 3.6 g | 17.5 ± 0.7 def | 4.1 ± 0.89 e | 0.45 ± 0.02 b | 520.2 ± 26.0 i |
| Pagnotta del Dittaino PDO | 3.70 ± 0.02 i | 83.2 ± 4.1 cd | 18.2 ± 0.7 cd | 4.5 ± 0.90 de | 0.21 ± 0.01 gh | 553.4 ± 27.6 h |
Values are means ± standard deviation (n = 6). Values within a column with different letters are significantly different (P < 0.05).
Sourdoughs are indicated with the names of the related breads.
Enumeration of LAB and yeasts.
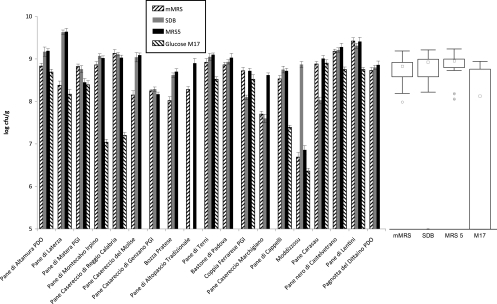
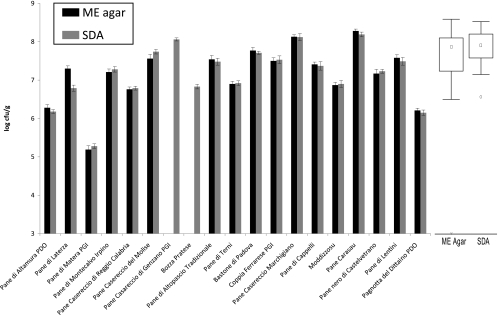
Plate counts using four culture media showed different values of presumptive LAB (Fig. 1). Apart from the culture medium and considering the highest value for each sourdough, the median plate count was 9.01 log CFU g−1. The 25th and 75th percentiles of the compiled data were 8.75 and 9.13 log CFU g−1, respectively. This difference could be attributed to qualitative and quantitative differences among media in terms of nutrients and to the different metabolic capabilities among strains harbored from each sourdough. Almost similar values of yeast cell density were found between MEA and SDA media (Fig. 2). Regardless of the culture medium and considering the highest value for each sourdough, the median value was 7.30 log CFU g−1. The ratio between LAB and yeasts of the 19 Italian sourdoughs ranged from 100:1 to 10:1, the only two exceptions being Pane di Matera PGI (ca. 1,000:1) and Pane Casareccio di Genzano PGI (ca. 1:1) sourdoughs.
Fig 1.
Histogram and box plot representations of cell density (log CFU g−1) of presumptive lactic acid bacteria in sourdoughs used for the manufacture of traditional/typical Italian breads. The center line of each box represents the median (□), and the top and bottom of the box represent the 75th and 25th percentiles of the data, respectively. The top and bottom of the error bars represent the 5th and 95th percentiles of the data, respectively. A circle in a box plot shows an outlier in the data (○), and very extreme points are represented as individual data points (*). Plate counts were carried out on different agar media: modified MRS (mMRS), SDB, MRS5, and glucose-M17. Data are the means of three independent experiments ± standard deviations, performed in duplicate (n = 6). Sourdoughs are indicated with the names of the related breads.
Fig 2.
Histogram and box plot representations of cell density (log CFU g−1) of yeasts in sourdoughs used for the manufacture of traditional/typical Italian breads. The center line of each box represents the median (□), and the top and bottom of the box represent the 75th and 25th percentiles of the data, respectively. The top and bottom of the error bars represent the 5th and 95th percentiles of the data, respectively. A circle in a box plot shows an outlier in the data (○), and a very extreme point is represented as an individual data point (*). Plate counts were carried out on different agar media: malt extract (ME) and Sabouraud dextrose (SDA). Data are the means of three independent experiments ± standard deviations, performed in duplicate (n = 6). Sourdoughs are indicated with the names of the related breads.
Typing and identification of LAB.
Gram-positive, catalase-negative, nonmotile cocci and rods able to acidify mMRS, SDB, MRS5, or glucose-M17 broth (411 isolates) were subjected to RAPD-PCR analysis (Table 3). The reproducibility of the RAPD fingerprints was assessed by comparing the PCR products obtained with primers P1, P4, and M13 and DNA extracted from three separate cultures of the same strain. For this purpose, 10 strains were studied, and the patterns for the same strain were similar at a level of ca. 90% (data not shown), as estimated by UPGMA analysis. The combined RAPD profiles were subjected to cluster analysis by UPGMA (data not shown). On the basis of the reproducibility of the RAPD-PCR analysis, isolates showing significantly (P < 0.10) different profiles were subjected to further analyses. Strains were identified by sequence analysis of at least 700 bp of the 16S rRNA gene (Table 3). Cluster analysis of RAPD profiles revealed that diversity among isolates ranged from ca. 2 to 33%. Strains showing RAPD profiles with a maximum level of diversity of 10% were grouped in the same cluster. Thus, it was possible to group ca. 50% of strains in 23 clusters (numbered I to XXIII). Overall, the clustering of the strains was not related to the sourdoughs they were isolated from. Sourdoughs harbored the following species: Weissella cibaria, Weissella confusa, Leuconostoc citreum, L. sanfranciscensis, L. plantarum, Leuconostoc mesenteroides subsp. mesenteroides, Lactobacillus sakei, Pediococcus pentosaceus, L. paralimentarius, Lactobacillus gallinarum, Lactococcus lactis subsp. lactis, L. brevis, Pediococcus inopinatus, L. casei, Pediococcus argentinicus, L. rossiae, Weissella paramesenteroides, Lactobacillus spicheri, Lactobacillus namurensis, and Enterococcus durans (Table 3; also see Table S2 in the supplemental material).
Table 3.
Species of lactic acid bacteria identified from sourdoughs used for the manufacture of traditional/typical Italian breadsa
| Sourdoughsb | Isolatesc | Clusterd | Closest relative (% identity)e | GenBank accession no. |
|---|---|---|---|---|
| Pane di Altamura PDO | AC13 (AD11p) | UC | Weissella cibaria (99) | JN845527 |
| AC1 (AD5) | XXIII | W. cibaria (99) | JN851736 | |
| AA25 (AB1) | UC | W. cibaria (99) | JN851737 | |
| AA22 | UC | W. cibaria (99) | JN851738 | |
| AA3 | UC | W. cibaria (99) | JN851739 | |
| AB15 | UC | W. cibaria (99) | JN851740 | |
| AB24 (AD16, AB7, AD8, AC9) | UC | W. cibaria (99) | JN851741 | |
| AA1 (AB3) | UC | W. cibaria (99) | JN851742 | |
| AA6 | UC | W. cibaria (99) | JN863639 | |
| AA12 (AD6) | UC | W. confusa (100) | JN851743 | |
| AC26 | UC | W. cibaria (99) | JN851745 | |
| AA15 | UC | W. cibaria (99) | JN863637 | |
| AA10 | UC | W. cibaria (99) | JN851746 | |
| AC22p | UC | W. cibaria (99) | JN851744 | |
| Pane di Laterza | BA1 (BA2) | UC | Leuconostoc citreum (99) | JN851747 |
| BC18 | II | Ln. citreum (99) | JN851748 | |
| BB23 (BB22, BB21) | I | Lactobacillus sanfranciscensis (99) | JN851749 | |
| BB28 (BB6) | UC | L. sanfranciscensis (99) | JN851750 | |
| BC9 | I | L. sanfranciscensis (99) | JN851751 | |
| BB1 (BA13, BA17g, BA14, BA10) | I | L. sanfranciscensis (99) | JN863640 | |
| BC1 (BC2, BC3) | I | L. sanfranciscensis (99) | JN863641 | |
| BD10 (BD9, BD8) | UC | Ln. citreum (99) | JN863643 | |
| BD4 (BD3) | UC | Ln. citreum (99) | JN863642 | |
| Pane di Matera PGI | CA1 (CA8) | XXIII | Ln. citreum (99) | JN851752 |
| CA10 (CA9, CB3) | XXIII | Ln. citreum (99) | JN851753 | |
| CA25 (CB8, CB11, CB20, CB23, CC21, CC25, CD2, CD12) | XVIII | Ln. citreum (99) | JN863680 | |
| CD5 (CD1) | XVII | Ln. citreum (99) | JN863644 | |
| CD14 (CA14, CC6, CC12) | XVIII | Lactobacillus plantarum (100) | JN863645 | |
| DA13 (DA23, DB1, DC6, DB8, DC17, DC3, DB3, DC24, DC9, DC2) | XVIII | L. sanfranciscensis (99) | JN851754 | |
| DA2 | XVII | L. sanfranciscensis (99) | JN851755 | |
| DB2 (DB17) | XVIII | L. sanfranciscensis (99) | JN851756 | |
| DA4 | XXII | L. sanfranciscensis (99) | JN863646 | |
| DB6 | UC | L. sanfranciscensis (99) | JN851757 | |
| DC5 | UC | L. sanfranciscensis (99) | JN851758 | |
| DC7 | IV | L. sanfranciscensis (99) | JN851759 | |
| DA6 | XVIII | L. sanfranciscensis (99) | JN863647 | |
| Pane Casereccio di Reggio Calabria | EA15 | XVIII | L. sanfranciscensis (99) | JN851760 |
| EA8 | UC | Leuconostoc mesenteroides subsp. mesenteroides (99) | JN851761 | |
| EA1 | UC | L. sanfranciscensis (99) | JN851769 | |
| EB3 (EC4) | III | Lactobacillus sakei (99) | JN851762 | |
| EC7 (EC8) | III | L. sakei (99) | JN851763 | |
| EB7 | IV | Ln. mesenteroides subsp. mesenteroides (99) | JN851764 | |
| EB10 | XIV | Ln. mesenteroides subsp. mesenteroides (99) | JN851765 | |
| EC19 (ED3, ED12, EB9, EA19) | XVI | Ln. mesenteroides subsp. mesenteroides (99) | JN851766 | |
| EA6 (EA5, EC1) | UC | L. sakei (99) | JN863648 | |
| EB27 | XIV | Ln. mesenteroides subsp. mesenteroides (99) | JN863649 | |
| ED4 (ED1, ED2) | UC | Ln. mesenteroides subsp. mesenteroides (99) | JN863681 | |
| Pane Casereccio del Molise | FA22 | UC | L. sanfranciscensis (99) | JN851767 |
| FA30 (FB13) | XVII | L. sanfranciscensis (99) | JN851768 | |
| FA13 (FB18, FA19) | VII | L. sanfranciscensis (99) | JN851770 | |
| FA26 | VII | L. sanfranciscensis (99) | JN851771 | |
| FA2 (FC7, FC9, FC5, FC8, FC12) | UC | L. sanfranciscensis (99) | JN882276 | |
| FB5 (FB10, FB11, FB23, FC15) | XVIII | L. sanfranciscensis (99) | JN851772 | |
| FB4 | XV | L. sanfranciscensis (99) | JN851773 | |
| FA4 | XII | L. sanfranciscensis (99) | JN863638 | |
| Pane Casareccio di Genzano PGI | GA6 | UC | L. plantarum (100) | JN851774 |
| GA12 | UC | L. plantarum (100) | JN851775 | |
| GA7 | UC | L. plantarum (100) | JN851776 | |
| GA4 | UC | L. plantarum (100) | JN851777 | |
| GB14 | UC | L. plantarum (100) | JN863682 | |
| GA1 (GC11, GC6, GB2, GB11, GC4) | XVIII | Pediococcus pentosaceus (99) | JN851778 | |
| GB5 (GB1, GA10) | VI | P. pentosaceus (99) | JN851779 | |
| GB3 | UC | P. pentosaceus (99) | JN851780 | |
| GC16 | UC | P. pentosaceus (99) | JN851781 | |
| GC3 | VI | P. pentosaceus (99) | JN851782 | |
| GB22 (GB10, GA2, GB19) | VII | L. plantarum (100) | JN863650 | |
| GC1 | VII | P. pentosaceus (99) | JN863651 | |
| Bozza Pratese | HA18 (HB7, HB11, HB19, HB23, HB4, HB12) | XVIII | L. sanfranciscensis (99) | JN851783 |
| HA3 (HA19, HC27, HC30, HC5, HC9, HC11, HA23, HB1) | XII | L. sanfranciscensis (99) | JN851784 | |
| HA1 (HA4, HA2, HA17) | XII | L. sanfranciscensis (99) | JN851785 | |
| HC23 (HC25) | XI | L. sanfranciscensis (99) | JN863683 | |
| HC26 | XX | Lactobacillus paralimentarius (99) | JN863684 | |
| HB25 | XVI | L. paralimentarius (99) | JN863652 | |
| Pane di Altopascio Tradizionale | IA6 (IA13, IC3) | XXI | Lactobacillus gallinarum (99) | JN851786 |
| IA23 | XXII | L. gallinarum (99) | JN851787 | |
| IA29 (IA2, IA3a, IC8, IC28) | XVII | L. gallinarum (99) | JN851788 | |
| IA1 (IA7, IA3b, IA11b, IC14, IC12, IC6) | XVIII | L. gallinarum (99) | JN851789 | |
| IC13 (IC27) | XIII | L. gallinarum (99) | JN851790 | |
| IC19a | XIII | L. gallinarum (99) | JN851791 | |
| IC7 | XXII | L. gallinarum (99) | JN851792 | |
| IC11 | UC | L sanfranciscensis (99) | JN863685 | |
| Pane di Terni | LC6 | UC | L. sanfranciscensis (99) | JN863686 |
| LC9 | UC | L. plantarum (100) | JN863687 | |
| LC12 (LA4, LC25, LA1, LA6, LA3, LA2, LB1, LB2, LB3, LB4, LB18) | XXI | L. plantarum (100) | JN851793 | |
| LB19 | UC | L. plantarum (100) | JN851794 | |
| LC17 | UC | Lactococcus lactis subsp. lactis (99) | JN851795 | |
| LD4 | XXII | Lc. lactis subsp. lactis (99) | JN851796 | |
| LD26 | UC | Lc. lactis subsp. lactis (99) | JN851797 | |
| LD14 | XXI | Lactobacillus brevis (99) | JN863688 | |
| LD11 | UC | Lc. lactis subsp. lactis (99) | JN863653 | |
| Bastone di Padova | MB3 | UC | Ln. mesenteroides subsp. mesenteroides (99) | JN851798 |
| MC2 | XX | L. plantarum (100) | JN851799 | |
| MA1 | XXII | L. paralimentarius (99) | JN851800 | |
| MC1 | XVIII | L. paralimentarius (99) | JN851801 | |
| MB1 (MA4, MA6, MA7, MA27, MA30, MB4, MB6, MB7, MB8, MB11, MB15, MC7, MC16, MC21, MC22, MC30, MB14) | XXII | L. paralimentarius (99) | JN851802 | |
| MC13 | XVIII | L. plantarum (100) | JN863654 | |
| MC5 | XVII | L. paralimentarius (99) | JN863655 | |
| MC9 | XX | L. paralimentarius (99) | JN863656 | |
| Coppia Ferrarese PGI | NA3 (NA4, NB1, NB3, NA8) | XIX | L. plantarum (100) | JN851803 |
| NA2 (NC2, ND3, ND7) | UC | L. plantarum (100) | JN851804 | |
| NB2 (NB4, NB24) | UC | L. plantarum (100) | JN851805 | |
| NC4 | UC | L. plantarum (100) | JN851806 | |
| ND25 (NC3, NC6) | XVII | L. paralimentarius (99) | JN851807 | |
| ND2 (NA1, NC1, NC7, NC8, ND1, ND4) | VIII | L. paralimentarius (99) | JN851808 | |
| NA28 | XIX | L. plantarum (100) | JN863657 | |
| Pane Casereccio Marchigiano | OA28 (OA2, OB4, OB3) | UC | L. plantarum (100) | JN851809 |
| OC1 | XXII | L. plantarum (100) | JN851810 | |
| OA1 | XVII | Pediococcus inopinatus (99) | JN851811 | |
| OA19 (OA5, OB7, OB8, OC10, OC17, OA18, OA13) | XXI | P. inopinatus (99) | JN851812 | |
| OC6 | XXII | Lactobacillus casei (99) | JN851813 | |
| OA6 (OB1, OB2) | XVIII | P. inopinatus (99) | JN863658 | |
| OA22 (OC14) | UC | L. sanfranciscensis (99) | JN863659 | |
| Pane di Cappelli | PA22 (PA14, PD4) | UC | L. sanfranciscensis (99) | JN863689 |
| PB24 (PA24, PD2) | UC | L. sanfranciscensis (99) | JN863595 | |
| PC30 | UC | L. sanfranciscensis (99) | JN863596 | |
| PB23 (PA23, PD9) | UC | L. plantarum (100) | JN863597 | |
| PA15 (PC4, PD1) | XVII | L. sanfranciscensis (99) | JN863660 | |
| PA16 (PB6) | UC | L. sanfranciscensis (99) | JN863661 | |
| PA21 (PD7) | UC | L. plantarum (100) | JN863662 | |
| PC28 (PB1) | UC | L. sanfranciscensis (99) | JN863663 | |
| PC27 (PB3) | II | L. sanfranciscensis (99) | JN863664 | |
| PC23 | UC | L. sanfranciscensis (99) | JN863666 | |
| Moddizzosu | QB4 | UC | L. sanfranciscensis (99) | JN863598 |
| QB9 (QB12) | UC | L. sanfranciscensis (99) | JN863599 | |
| QC1 | UC | L. plantarum (100) | JN863600 | |
| QA22 | UC | L. brevis (99) | JN863601 | |
| QA23 | UC | L. brevis (99) | JN863602 | |
| QA1 | UC | L. brevis (99) | JN863603 | |
| QD2 (QD3, QD5, QD11) | UC | L. brevis (99) | JN863604 | |
| QD1 | XVII | L. brevis (99) | JN863690 | |
| QA16 | UC | Pediococcus argentinicus (99) | JN863605 | |
| QB24 (QB2) | UC | Lactobacillus rossiae (99) | JN863606 | |
| QB5 | UC | L. rossiae (99) | JN863607 | |
| QA13 | UC | L. brevis (99) | JN863665 | |
| QC8 | UC | L. brevis (99) | JN863667 | |
| QC15 | UC | L. rossiae (99) | JN863668 | |
| QC6 | XI | L. sanfranciscensis (99) | JN863669 | |
| QC2 | UC | L. brevis (99) | JN863608 | |
| QC27 | UC | L. rossiae (99) | JN863670 | |
| Pane Carasau | RD24 | UC | Ln. mesenteroides subsp. mesenteroides (99) | JN863609 |
| RA1 | UC | L. plantarum (100) | JN863610 | |
| RA11 (RC7, RB2, RB13, RC23, RB7, RB5, RB8) | V | L. plantarum (100) | JN863611 | |
| RA2 (RA25) | XVII | L. plantarum (100) | JN863612 | |
| RC5 | V | P. pentosaceus (99) | JN863614 | |
| RC24 | XVII | Lc. lactis subsp. lactis (99) | JN863615 | |
| RA6 (RC15) | UC | L. brevis (99) | JN863616 | |
| RC2 | UC | W. paramesenteroides (99) | JN863617 | |
| RD9 | UC | Ln. mesenteroides subsp. mesenteroides (99) | JN863618 | |
| RD1 | UCe | L. brevis (99) | JN863619 | |
| RD12 | UC | L. plantarum (100) | JN863613 | |
| RD2 | UC | L. plantarum (100) | JN863671 | |
| RD23 | UC | L. plantarum (100) | JN863672 | |
| Pane nero di Castelvetrano | SA3b (SA7b, SA14, SB1) | IX | L. paralimentarius (99) | JN863620 |
| SA22b (SA19, SB5) | XV | L. paralimentarius (99) | JN863621 | |
| SC19 (SB2) | XV | L. paralimentarius (99) | JN863691 | |
| SC5 (SD13, SB7, SC11a) | IX | L. paralimentarius (99) | JN863622 | |
| SC10 (SD7, SB10, SD11b) | XV | Lactobacillus spicheri (99) | JN863623 | |
| SC6 (SC17) | XVII | L. spicheri (99) | JN863624 | |
| SD1 (SD2) | VIII | L. spicheri (99) | JN863625 | |
| SC4 | IX | L. spicheri (99) | JN863673 | |
| SC20 | XXI | L. spicheri (99) | JN863674 | |
| Pane di Lentini | TA22 (TB20, TD2) | UC | L. sanfranciscensis (99) | JN863626 |
| TA1 (TA23, TB6, TB17, TA8, TB1, TA6, TD7) | XVIII | L. paralimentarius (99) | JN863627 | |
| TC13 (TD1) | UC | L. paralimentarius (99) | JN863628 | |
| TC18 | UC | L. paralimentarius (99) | JN863629 | |
| TC2 (TD5, TD6) | UC | Lactobacillus namurensis (99) | JN863630 | |
| TC7 | XVIII | L. plantarum (99) | JN863631 | |
| TB18 | UC | L. namurensis (99) | JN863675 | |
| TC4 | UC | L. paralimentarius (99) | JN863676 | |
| TC25 | UC | L. sanfranciscensis (99) | JN863677 | |
| Pagnotta del Dittaino PDO | UA21 (UB20, UB22) | X | L. sanfranciscensis (99) | JN863632 |
| UC15 | UC | L. sanfranciscensis (99) | JN863633 | |
| UB17 (UB1, UB2, UB4, UA17, UC6) | UC | L. sanfranciscensis (99) | JN863635 | |
| UA27 | X | Enterococcus durans (99) | JN863634 | |
| UC12 (UA12, UC10, UC16, UC23) | UC | E. durans (99) | JN863678 | |
| UA16 (UA23, UA1, UA5) | UC | E. durans (99) | JN863679 | |
| UA3 | XVII | L. sanfranciscensis (99) | JN863636 |
Identification was carried out by 16S rRNA, recA, pheS, or casei/para gene sequencing.
Sourdoughs are indicated with the names of the related breads.
Where more than one isolate per row is present, the first isolate is the representative one for that strain. The second letter of the isolate name indicates the medium used for the isolation (A, mMRS; B, SDB; C, MRS5; D, glucose-M17).
Randomly amplified polymorphic DNA (RAPD)-PCR analysis was carried out to exclude clonal relatedness. RAPD-PCR cluster. Clusters are numbered with Roman numerals from I to XXIII; UC, unclustered.
Species showing the highest identity (%) to the strain isolated from sourdough. The percentage of identity was that shown by performing multiple sequence alignments in BLAST. Ln., Leuconostoc; Lc., Lactococcus.
Typing and identification of yeasts.
Preliminarily, 382 yeast isolates (ca. 20 for each sourdough) were assayed for the ability to ferment galactose, sucrose, maltose, raffinose, and trehalose, as well as for the capacity for growing at 37°C. At least 6 isolates for each sourdough, including all isolates showing different fermentation profiles (data not shown), were genetically characterized and identified. Cluster analysis of iSSR-PCR profiles revealed a diversity level among isolates ranging from 3 to 54% (data not shown). Isolates showing iSSR-PCR profiles with a maximum level of diversity of 10% were grouped in the same cluster. Thus, it was possible to group 127 isolates in 13 clusters (I to XIII), whereas 11 were unclustered. Except for Pane di Matera PGI, Coppia Ferrarese PGI, and Pane di Cappelli, all sourdoughs harbored isolates distributed over two or more clusters. Strains were identified by partial sequence analysis of the 26S rRNA gene (data not shown). S. cerevisiae was identified in all sourdoughs except for Pane di Cappelli, Pane nero di Castelvetrano, and Pagnotta del Dittaino PDO. C. humilis was identified in Pane di Terni, Pane di Cappelli, and Pagnotta del Dittaino PDO sourdoughs. K. barnettii and K. exigua were identified in Pane nero di Calstelvetrano sourdough.
Statistical correlations between ingredients, technology parameters, microbial species diversity, and sourdough biochemical characteristics.
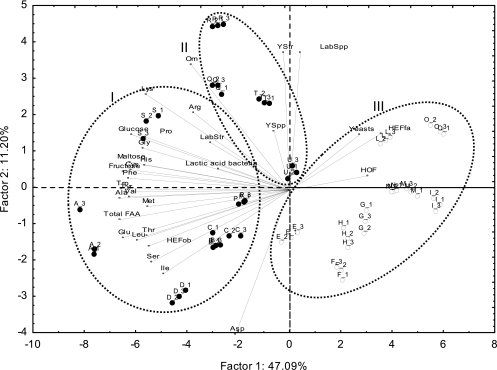
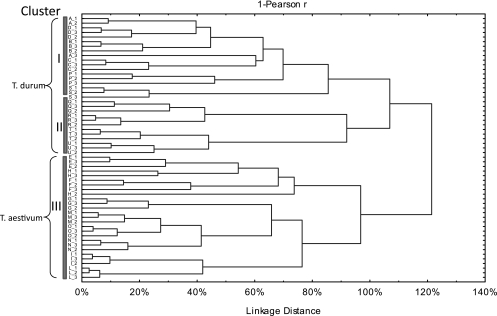
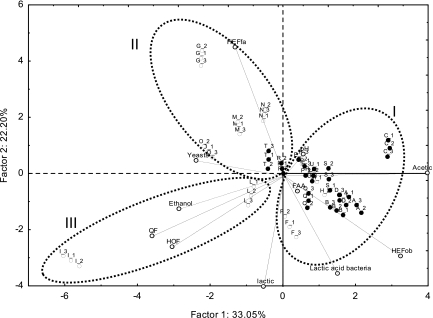
PCA analysis showed that the type of flour was correlated with the sourdough microbiota (Fig. 3). It showed only two significant PCs that explained 47.09% (PC1) and 11.20% (PC2) of the total variance of the data. Cluster analysis between ingredients and microbial community data was also performed (Fig. 4). Cluster analysis was also used to pick clusters of sourdoughs on the PCA score plots (Fig. 3 and 4). As determined by ANOVA, each ellipse on the PCA score plot was statistically (P < 0.001) different from the others (data not shown). All T. durum sourdoughs were grouped at a distance linkage of 107%, whereas T. aestivum sourdoughs were clustered at 97%. At a distance linkage of 100%, three clusters were found. Overall, T. durum sourdoughs (clusters I and II) were correlated with (i) a high percentage of obligately heterofermentative LAB species, (ii) a low percentage of facultatively heterofermentative and obligately homofermentative LAB species, and (iii) a low cell density of yeasts. However, within T. durum sourdoughs, Modizzosu (Modizzosu sourdough batches are designated with an alphanumeric code beginning with Q), Pane Carasau (R), Pane di Lentini (T), and Pagnotta del Dittaino PDO (U), grouped in cluster II, can be distinguished from the others for the lower concentration in serine and total FAA, especially for T and U sourdoughs. Modizzosu (Q) and Pane Carasau (R) were also characterized by the highest numbers of yeast strains and LAB species.
Fig 3.
Score and loading plots of first and second principal components and clusters after principal component analysis and cluster analysis based on composition of flour (maltose, glucose, and fructose and total and individual free amino acids) and microbial community (cell densities of lactic acid bacteria and yeasts, number of species and strains isolated from each sourdough, and percentage of obligately homofermentative and obligately and facultatively heterofermentative lactic acid bacteria) data from sourdoughs used for the manufacture of traditional/typical Italian breads. Clusters I, II, and III are indicated by dotted ellipses and correspond to those shown in Fig. 4. Sourdoughs are indicated by the following alphanumeric codes: Pane di Altamura PDO, A_1, A_2, A_3; Pane di Laterza, B_1, B_2, B_3; Pane di Matera PGI, C_1, C_2, C_3; Pane di Montecalvo Irpino, D_1, D_2, D_3; Pane Casereccio di Reggio Calabria, E_1, E_2, E_3; Pane Casereccio del Molise, F_1, F_2, F_3; Pane Casareccio di Genzano PGI, G_1, G_2, G_3; Bozza Pratese, H_1, H_2, H_3; Pane di Altopascio Tradizionale, I_1, I_2, I_3; Pane di Terni, L_1, L_2, L_3; Bastone di Padova, M_1, M_2, M_3; Coppia Ferrarese PGI, N_1, N_2, N_3; Pane Casereccio Marchigiano, O_1, O_2, O_3; Pane di Cappelli, P_1, P_2, P_3; Moddizzosu, Q_1, Q_2, Q_3; Pane Carasau, R_1, R_2, R_3; Pane nero di Castelvetrano, S_1, S_2, S_3; Pane di Lentini, T_1, T_2, T_3; Pagnotta del Dittaino PDO, U_1, U_2, U_3. Sourdoughs based on Triticum durum flour are indicated by a black circle, whereas sourdoughs based on Triticum aestivum flour are indicated by a white circle. Total FAA, total free amino acids; Lactic acid bacteria, cell density of lactic acid bacteria; Yeasts, cell density of yeasts; LabSpp, number of lactic acid bacterial species; LabStr, number of lactic acid bacterial strains; YSpp, number of yeast species; YStr, number of yeast strains; HOF, obligately homofermentative lactic acid bacteria; HEFfa, facultatively heterofermentative lactic acid bacteria; HEFob, obligately heterofermentative lactic acid bacteria.
Fig 4.
Dendrogram obtained from cluster analysis based on composition of flour (maltose, glucose, and fructose and total and individual free amino acids) and microbial community (cell densities of lactic acid bacteria and yeasts, number of species and strains isolated from each sourdough, and percentage of obligately homofermentative and obligately and facultatively heterofermentative lactic acid bacteria) data from sourdoughs used for the manufacture of traditional/typical Italian breads. Sourdoughs are indicated by the following alphanumeric codes: Pane di Altamura PDO, A_1, A_2, A_3; Pane di Laterza, B_1, B_2, B_3; Pane di Matera PGI, C_1, C_2, C_3; Pane di Montecalvo Irpino, D_1, D_2, D_3; Pane Casereccio di Reggio Calabria, E_1, E_2, E_3; Pane Casereccio del Molise, F_1, F_2, F_3; Pane Casareccio di Genzano PGI, G_1, G_2, G_3; Bozza Pratese, H_1, H_2, H_3; Pane di Altopascio Tradizionale, I_1, I_2, I_3; Pane di Terni, L_1, L_2, L_3; Bastone di Padova, M_1, M_2, M_3; Coppia Ferrarese PGI, N_1, N_2, N_3; Pane Casereccio Marchigiano, O_1, O_2, O_3; Pane di Cappelli, P_1, P_2, P_3; Moddizzosu, Q_1, Q_2, Q_3; Pane Carasau, R_1, R_2, R_3; Pane nero di Castelvetrano, S_1, S_2, S_3; Pane di Lentini, T_1, T_2, T_3; Pagnotta del Dittaino PDO, U_1, U_2, U_3.
Within T. aestivum sourdoughs, some differences were found. Pane Casereccio di Reggio Calabria (E), Pane Casereccio del Molise (F), Bozza Pratese (H), and Pane Casareccio di Genzano PGI (G) were characterized by the highest concentrations of total FAA. Furthermore, Pane Casereccio di Reggio Calabria (E), Pane Casereccio del Molise (F), and Bozza Pratese (H) sourdoughs were characterized by the highest percentages of obligately heterofermentative LAB among T. aestivum sourdoughs. In contrast, Pane di Altopascio Tradizionale (I) and Pane Casereccio Marchigiano (O) were characterized by the lowest concentrations of total FAA.
PCA analysis on ingredients other than the composition of the flour (percentage of NaCl, percentage of baker's yeast, and percentage of sourdough used during back slopping), technology parameters (time and temperature of back slopping), and the sourdough microbiotas showed that the yeast cell density and the percentage of baker's yeast were correlated, whereas both were inversely correlated with the LAB cell density. No other correlations were found (data not shown).
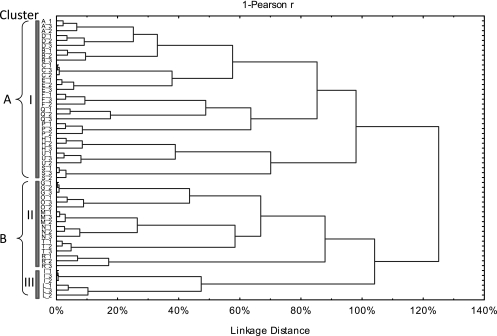
Regarding the correlation between microbial communities and biochemical characteristics of sourdoughs, the two PCs explained ca. 55.25% of the total variance of the data (Fig. 5). Cluster analysis grouped the sourdoughs in two main clusters (A and B) linked together at 125% (Fig. 6). At a linkage distance of 100%, the sourdoughs were grouped in three clusters. Cluster I grouped together most sourdoughs, showing a high percentage of obligately heterofermentative LAB and correlated with a high concentration of acetic acid (Fig. 5 and 6). Cluster I included all T. durum sourdoughs, with the only two exceptions being Pane di Lentini (T) and Pane Carasau (R), as well as three T. aestivum sourdoughs (Pane Casereccio di Reggio Calabria, E, Pane Casereccio del Molise, F, and Bozza Pratese, H).
Fig 5.
Score and loading plots of first and second principal components and clusters after principal component analysis and cluster analysis based on the microbial community (cell densities of lactic acid bacteria and yeasts, percentage of obligately homofermentative and obligately and facultatively heterofermentative lactic acid bacteria) and biochemical (pH, organic acids, quotient of fermentation, and ethanol) data from sourdoughs used for the manufacture of traditional/typical Italian breads. Clusters I, II, and III are indicated by dotted ellipses and correspond to those shown in Fig. 6. Sourdoughs are indicated by the following alphanumeric codes: Pane di Altamura PDO, A_1, A_2, A_3; Pane di Laterza, B_1, B_2, B_3; Pane di Matera PGI, C_1, C_2, C_3; Pane di Montecalvo Irpino, D_1, D_2, D_3; Pane Casereccio di Reggio Calabria, E_1, E_2, E_3; Pane Casereccio del Molise, F_1, F_2, F_3; Pane Casareccio di Genzano PGI, G_1, G_2, G_3; Bozza Pratese, H_1, H_2, H_3; Pane di Altopascio Tradizionale, I_1, I_2, I_3; Pane di Terni, L_1, L_2, L_3; Bastone di Padova, M_1, M_2, M_3; Coppia Ferrarese PGI, N_1, N_2, N_3; Pane Casereccio Marchigiano, O_1, O_2, O_3; Pane di Cappelli, P_1, P_2, P_3; Moddizzosu, Q_1, Q_2, Q_3; Pane Carasau, R_1, R_2, R_3; Pane nero di Castelvetrano, S_1, S_2, S_3; Pane di Lentini, T_1, T_2, T_3; Pagnotta del Dittaino PDO, U_1, U_2, U_3. Sourdoughs based on Triticum durum flour are indicated by a black circle, whereas sourdoughs based on Triticum aestivum flour are indicated by a white circle. QF, quotient of fermentation; HOF, obligately homofermentative lactic acid bacteria; HEFfa, facultatively heterofermentative lactic acid bacteria; HEFob, obligately heterofermentative lactic acid bacteria; FAA, total free amino acids.
Fig 6.
Dendrogram obtained from cluster analysis based on the microbial community (cell densities of lactic acid bacteria and yeasts, percentage of obligately homofermentative and obligately and facultatively heterofermentative lactic acid bacteria) and biochemical (pH, organic acids, quotient of fermentation, and ethanol) data from sourdoughs used for the manufacture of traditional/typical Italian breads. Sourdoughs are indicated by the following alphanumeric codes: Pane di Altamura PDO, A_1, A_2, A_3; Pane di Laterza, B_1, B_2, B_3; Pane di Matera PGI, C_1, C_2, C_3; Pane di Montecalvo Irpino, D_1, D_2, D_3; Pane Casereccio di Reggio Calabria, E_1, E_2, E_3; Pane Casereccio del Molise, F_1, F_2, F_3; Pane Casareccio di Genzano PGI, G_1, G_2, G_3; Bozza Pratese, H_1, H_2, H_3; Pane di Altopascio Tradizionale, I_1, I_2, I_3; Pane di Terni, L_1, L_2, L_3; Bastone di Padova, M_1, M_2, M_3; Coppia Ferrarese PGI, N_1, N_2, N_3; Pane Casereccio Marchigiano, O_1, O_2, O_3; Pane di Cappelli, P_1, P_2, P_3; Moddizzosu, Q_1, Q_2, Q_3; Pane Carasau, R_1, R_2, R_3; Pane nero di Castelvetrano, S_1, S_2, S_3; Pane di Lentini, T_1, T_2, T_3; Pagnotta del Dittaino PDO, U_1, U_2, U_3.
Overall, Coppia Ferrarese PGI (N), Bastone di Padova (M), Pane Casereccio Marchigiano (O), Pane Casareccio di Genzano PGI (G), Pane di Lentini (T), and Pane Carasau (R) sourdoughs were clustered together (cluster II) because they shared (i) a high percentage of facultatively heterofermentative LAB strains, (ii) a low percentage of obligately heterofermentative LAB, and (iii) a high yeast cell density. Pane di Altopascio Tradizionale (I) and Pane di Terni (L) sourdoughs (cluster III) were characterized by the highest percentage of obligately homofermentative LAB strains. Pane di Altopascio Tradizionale also showed a high concentration of lactic acid and the highest QF value (Fig. 5 and 6).
DISCUSSION
This study describes the LAB and yeast microbiotas of 19 sourdoughs used for the manufacture of traditional/typical Italian breads. Viable cell counts differed depending on the medium, especially for LAB (32, 42). The ratio between LAB and yeasts of the 19 Italian sourdoughs ranged from 100:1 to 10:1, which is common for sourdoughs (for reviews, see references 19 and 22). The only two exceptions were Pane di Matera PGI (ca. 1,000:1) and Pane Casareccio di Genzano PGI (ca. 1:1) sourdoughs. The number of strains varied from 5 to 17. Although the number of isolates was probably not sufficient to describe all the species diversity of the sourdoughs analyzed, the number of LAB species also varied markedly among the Italian sourdoughs. There was no LAB species common to all sourdoughs. Except for Pane di Montecalvo Irpino and Pane Casereccio del Molise, all sourdoughs had a peculiar composition of the LAB species. Numerically, the most frequent isolates belonged to the species L. sanfranciscensis (ca. 28% of the total LAB isolates), L. plantarum (ca. 16%), and L. paralimentarius (ca. 14%), which were identified in 12, 10, and 5 Italian sourdoughs, respectively. L. sanfranciscensis is considered one of the most important species of type I sourdoughs (16). L. plantarum is a ubiquitous species, found in several food ecosystems (7), including sourdoughs (19). Overall, the stable persistence of L. sanfranciscensis and L. plantarum in sourdoughs is explained through (i) the unique central metabolism and/or transport of sourdough-related carbohydrates (15, 20), (ii) activated proteolytic enzymes (20), (iii) particular stress adaptation responses (12), and (iv) synthesis of antimicrobial compounds (18). Previous characterization of the Pane di Matera PGI sourdough shows the predominance of L. plantarum and L. mesenteroides (50). L. plantarum and L. brevis are the species most frequently isolated from sourdoughs of the Molise region (33). Several years ago, the Pane di Altamura PDO sourdough was characterized, with L. plantarum found to be the dominant species (33a). The results of this study were in part different from some of the previous findings. In particular, W. cibaria and W. confusa were the dominant species found in Pane di Altamura PDO sourdough. These differences may be related to the following factors: (i) bakeries and methodologies of analysis (e.g., numbers of samples and isolates, temperature, pH, and culture media) (35, 36), (ii) contamination by bakery environment (36), and especially, (iii) instability of the sourdough microbiotas during daily back slopping as affected by the type of flour and related autochthonous LAB (29, 38). Indeed, a stable microbiota over a long period of time has been described only in a few sourdoughs that are used as the sole leavening agent (4, 18, 37).
S. cerevisiae was the species of yeast most frequently isolated. These results were in agreement with previous studies on sourdoughs from the center and south of Italy (31, 39). The dominance of a few strains of S. cerevisiae can be related to contamination by baker's yeast (45). The dominance of C. humilis has been shown in Pagnotta del Dittaino PDO sourdough (21). K. barnettii and K. exigua were isolated only from Pane nero di Castelvetrano sourdough. K. exigua has previously been isolated from a number of sourdoughs (16), whereas the presence of K. barnettii in wheat sourdoughs is reported only in one previous paper (45).
This study showed that the type of flour (T. durum or T. aestivum), as well as the concentration of some flour nutrients required by the microorganisms dominating the sourdough ecosystem, may have a key role for selecting the population of LAB. The main distinguishing features of sourdoughs made with T. durum flour were the highest concentrations of maltose, glucose, fructose, and FAA correlated with the sole or main presence of obligately heterofermentative LAB, the lowest number of facultatively heterofermentative LAB, and the low cell density of yeasts. As previously reported by other authors (34, 36), the microbiotas of six out of nine T. aestivum sourdoughs was dominated by facultatively heterofermentative LAB. Only in three T. aestivum sourdoughs (Pane Casereccio di Reggio Calabria, Pane Casereccio del Molise, and Bozza Pratese) were obligately heterofermentative LAB found as the dominating LAB species, in agreement with previous papers (16). Because these sourdoughs (along with that used for Pane Casareccio di Genzano PGI) were based on flours characterized by the highest concentrations of fermentable carbohydrates and total FAA among T. aestivum flours, it may be hypothesized that in flours characterized by lower concentrations of such nutrients (e.g., Bastone di Padova, Coppia Ferrarese PGI, and Pane Casereccio Marchigiano), obligately heterofermentative LAB are less competitive than facultatively heterofermentative and/or obligately homofermentative LAB. Amino acid transamination reactions and/or the arginine deiminase pathway are metabolic activities that favor energy production, cofactor (re)cycling, and/or tolerance toward acid stress and, hence, contribute to the competitiveness and dominance of certain species of LAB found in sourdoughs (16). In addition, FAA and GABA produced by LAB may increase the nutritional value of the breads. For instance, the amount of GABA in 150 g of Pane di Matera PGI represents the minimum effective daily dose to get positive effects in humans (25). Overall, the addition of baker's yeast negatively affected the final cell density of LAB. However, further studies are necessary to highlight the specific effect of each technological parameter (percentage of NaCl, percentage of baker's yeast, percentage of sourdough used during back slopping, and time and temperature of back slopping) on the composition of the related microbiotas (46).
This study gave a comprehensive and comparative view of the LAB and yeast microbiotas of numerous sourdoughs used for the manufacture of traditional/typical Italian breads. The microbiota structure distinguished almost all sourdoughs studied. This in turn affected the biochemical features (concentration of lactic, acetic acids, ethanol, and FAA) of sourdoughs. The results of this study could be helpful to choose specific starter cultures. This should guarantee a high reproducibility of the quality characterizing the traditional and/or typical breads.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Beldem S.A. (Andenne, Belgium).
We thank Francesco Mazzacane (Dipartimento di Biologia e Chimica Agro-Forestale ed Ambientale, Università degli Studi di Bari Aldo Moro, Italy) for technical assistance. We also thank all of the bakeries for kind collaboration.
Footnotes
Published ahead of print 9 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Andrade MJ, Rodrìguez M, Sànchez BE, Aranda Cordoba JJ. 2006. DNA typing methods for differentiation of yeasts related to dry-cured meat products. Int. J. Food Microbiol. 107:48–58 [DOI] [PubMed] [Google Scholar]
- 3. Barnett JA, Payne RW, Yarrow D. 2000. Yeast: characteristics and identification, 3rd ed Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 4. Böcker G, Vogel RF, Hammes WP. 1990. Lactobacillus sanfrancisco als stabiles Element in einem Reinzucht-Sauerteig-Präparat. Getreide Mehl Brot 44:269–274 [Google Scholar]
- 5. Cardinali G, Bolano A, Martini A. 2001. A DNA extraction and purification method for several yeast genera. Ann. Microbiol. 51:121–130 [Google Scholar]
- 6. Reference deleted.
- 7. Corsetti A, Gobbetti M. 2002. Lactobacillus plantarum, p 1501–1507 In Proginsli H, Fuquay JW, Fox PF. (ed), Encyclopedia of dairy science. Academic Press Ltd., New York, NY [Google Scholar]
- 8. Corsetti A, Settanni L, Valmorri S, Mastrangelo M, Suzzi G. 2007. Identification of subdominant sourdough lactic acid bacteria and their evolution during laboratory-scale fermentations. Food Microbiol. 24:592–600 [DOI] [PubMed] [Google Scholar]
- 9. D'Amico A. 2004. The enhancement of the typical products value: from commodity to experience. The case of Esperya.com. Br. Food J. 106:793–805 [Google Scholar]
- 10. De Angelis M, et al. 2001. Characterization of nonstarter lactic acid bacteria from Italian ewe cheeses based on phenotypic, genotypic, and cell wall protein analyses. Appl. Environ. Microbiol. 67:2011–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Angelis M, et al. 2007. Molecular and functional characterization of Lactobacillus sanfranciscensis strains isolated from sourdoughs. Int. J. Food Microbiol. 114:69–82 [DOI] [PubMed] [Google Scholar]
- 12. De Angelis M, Gobbetti M. 2004. Environmental stress responses in Lactobacillus: a review. Proteomics 4:106–122 [DOI] [PubMed] [Google Scholar]
- 13. De Angelis M, et al. 2006. Selection of potential probiotic lactobacilli from pig feces to be used as additives in pelleted feeding. Res. Microbiol. 157:792–801 [DOI] [PubMed] [Google Scholar]
- 14. de Los Reyes-Gavilán CG, Limsowtin GKY, Tailliez P, Séchaud L, Accolas JP. 1992. A Lactobacillus helveticus-specific DNA probe detects restriction fragment length polymorphisms in this species. Appl. Environ. Microbiol. 58:3429–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Vuyst L, Vancanneyt M. 2007. Biodiversity and identification of sourdough lactic acid bacteria. Food Microbiol. 24:120–127 [DOI] [PubMed] [Google Scholar]
- 16. De Vuyst L, Vrancken G, Ravyts F, Rimaux T, Weckx S. 2009. Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol. 26:666–675 [DOI] [PubMed] [Google Scholar]
- 17. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gänzle MG, Vogel RF. 2003. Contribution of reutericyclin production to the stable persistence of Lactobacillus reuteri in an industrial sourdough fermentation. Int. J. Food Microbiol. 80:31–45 [DOI] [PubMed] [Google Scholar]
- 19. Gobbetti M. 1998. Interactions between lactic acid bacteria and yeasts in sourdoughs. Trends Food Sci. Technol. 9:267–274 [Google Scholar]
- 20. Gobbetti M, De Angelis M, Corsetti A, Di Cagno R. 2005. Biochemistry and physiology of sourdough lactic acid bacteria. Trends Food Sci. Technol. 16:57–69 [Google Scholar]
- 21. Gullo M, Romano AD, Pulvirenti A, Giudici P. 2003. Candida humilis dominant species in sourdoughs for the production of durum wheat bran flour bread. Int. J. Food Microbiol. 80:55–59 [DOI] [PubMed] [Google Scholar]
- 22. Hammes WP, et al. 2005. Microbial ecology of cereal fermentations. Trends Food Sci. Technol. 16:4–11 [Google Scholar]
- 23. Iacumin L, et al. 2009. Description of the microflora of sourdoughs by culture-dependent and culture-independent methods. Food Microbiol. 26:128–135 [DOI] [PubMed] [Google Scholar]
- 24. Istituto Nazionale di Sociologia Rurale 2000. Atlante dei prodotti tipici: il pane, p 13 Agra-Rai Eri, Rome, Italy [Google Scholar]
- 25. Kajimoto O, et al. 2004. Hypotensive effect of fermented milk containing γ-aminobutyric acid (GABA) in subjects with high normal blood pressure. Nippon Shokuhin Kagaku Kogaku Kaishi 51:79–86 [Google Scholar]
- 26. Kurtzman CP, Robnett CJ. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371 [DOI] [PubMed] [Google Scholar]
- 27. Meroth CB, Hammes WP, Hertel C. 2003. Identification and population dynamics of yeasts in sourdough fermentation processes by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:7453–7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meroth CB, Walter J, Hertel C, Brandt MJ, Hammes WP. 2003. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Minervini F, et al. 2010. Robustness of Lactobacillus plantarum starters during daily propagation of wheat flour sourdough type I. Food Microbiol. 27:897–908 [DOI] [PubMed] [Google Scholar]
- 29a.Ministero per le Politiche Agricole. 1999. Decreto ministeriale 8 Settembre 1999, no. 350. Gazzetta Officiale della Repubblica Italiana 12 Ottobre 1999, no. 240.
- 30. Naser SM, et al. 2005. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 151:2141–2150 [DOI] [PubMed] [Google Scholar]
- 31. Pulvirenti A, Solieri L, Gullo M, De Vero L, Giudici P. 2004. Occurrence and dominance of yeast species in sourdough. Lett. Appl. Microbiol. 38:113–117 [DOI] [PubMed] [Google Scholar]
- 32. Randazzo CL, Heilig H, Restuccia C, Giudici P, Caggia C. 2005. Bacterial population in traditional sourdough evaluated by molecular methods. J. Appl. Microbiol. 99:251–258 [DOI] [PubMed] [Google Scholar]
- 33. Reale A, et al. 2011. Identification of lactobacilli isolated in traditional ripe wheat sourdoughs by using molecular methods. World J. Microbiol. Biotechnol. 27:237–244 [Google Scholar]
- 33a. Ricciardi A, Parente E, Piraino P, Paraggio M, Romano P. 2005. Phenotypic characterization of lactic acid bacteria from sourdoughs for altamura bread produced in Apulia (southern Italy). Int. J. Food Microbiol. 98:63–72 [DOI] [PubMed] [Google Scholar]
- 34. Robert H, Gabriel V, Fontagné-Faucher C. 2009. Biodiversity of lactic acid bacteria in French wheat sourdough as determined by molecular characterization using species-specific PCR. Int. J. Food Microbiol. 135:53–59 [DOI] [PubMed] [Google Scholar]
- 35. Scheirlinck I, et al. 2007. Influence of geographical origin and flour type on diversity of lactic acid bacteria in traditional Belgian sourdoughs. Appl. Environ. Microbiol. 73:6262–6269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scheirlinck I, et al. 2008. Taxonomic structure and stability of the bacterial community in Belgian sourdough ecosystems assessed by culturing and population fingerprinting. Appl. Environ. Microbiol. 74:2414–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scheirlinck I, Van der Meulen R, De Vuyst L, Vandamme P, Huys G. 2009. Molecular source tracking of predominant lactic acid bacteria in traditional Belgian sourdoughs and their production environments. J. Appl. Microbiol. 106:1081–1092 [DOI] [PubMed] [Google Scholar]
- 38. Siragusa S, et al. 2009. Taxonomic structure and monitoring of the dominant population of lactic acid bacteria during wheat flour sourdough type I propagation using Lactobacillus sanfranciscensis starters. Appl. Environ. Microbiol. 75:1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Succi M, et al. 2003. Presence of yeasts in southern Italian sourdoughs from Triticum aestivum flour. FEMS Microbiol. Lett. 225:143–148 [DOI] [PubMed] [Google Scholar]
- 40. Torriani S, Felis GE, Dellaglio F. 2001. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 67:3450–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Van der Meulen R, et al. 2007. Population dynamics and metabolite target analysis during laboratory fermentations of wheat and spelt sourdoughs. Appl. Environ. Microbiol. 73:4741–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vera A, Rigobello V, Demarigny Y. 2009. Comparative study of culture media used for sourdough lactobacilli. Food Microbiol. 26:728–733 [DOI] [PubMed] [Google Scholar]
- 43. Vogel RF, Ehrmann MA. 2008. Sourdough fermentations, p. 119–144 In Cocolin L, Ercolini D. (ed), Molecular techniques in the microbial ecology of fermented foods. Springer, Heidelberg, Germany [Google Scholar]
- 44. Vogelmann SA, Hertel C. 2011. Impact of ecological factors on the stability of microbial associations in sourdough fermentation. Food Microbiol. 28:583–589 [DOI] [PubMed] [Google Scholar]
- 45. Vrancken G, et al. 2010. Yeast species composition differs between artisan bakery and spontaneous laboratory sourdoughs. FEMS Yeast Res. 10:471–481 [DOI] [PubMed] [Google Scholar]
- 46. Vrancken G, Rimaux T, Weckx S, Leroy F, De Vuyst L. 2011. Influence of temperature and backslopping time on the microbiota of a type I propagated laboratory wheat sourdough fermentation. Appl. Environ. Microbiol. 77:2716–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reference deleted.
- 48. Ward LJH, Timmins MJ. 1999. Differentiation of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus by polymerase chain reaction. Lett. Appl. Microbiol. 29:90–92 [DOI] [PubMed] [Google Scholar]
- 49. Weckx S, et al. 2010. Lactic acid bacteria community dynamics and metabolite production of rye sourdough fermentations share characteristics of wheat and spelt sourdough fermentations. Food Microbiol. 27:1000–1008 [DOI] [PubMed] [Google Scholar]
- 50. Zotta T, Piraino P, Parente E, Salzano G, Ricciardi A. 2008. Characterization of lactic acid bacteria isolated from sourdoughs for cornetto, a traditional bread produced in Basilicata (southern Italy). World J. Microbiol. Biotechnol. 24:1785–1795 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.