Abstract
The Gram-positive, anaerobic, endospore-forming bacterium Clostridium acetobutylicum has considerable biotechnological potential due to its ability to produce solvents as fermentation products, in particular the biofuel butanol. Its genome contains a putative agr locus, agrBDCA, known in staphylococci to constitute a cyclic peptide-based quorum sensing system. In staphylococci, agrBD is required for the generation of a peptide signal that, upon extracellular accumulation, is sensed by an agrCA-encoded two-component system. Using ClosTron technology, agrB, agrC, and agrA mutants of C. acetobutylicum ATCC 824 were generated and phenotypically characterized. Mutants and wild type displayed similar growth kinetics and no apparent differences in solvent formation under the conditions tested. However, the number of heat-resistant endospores formed by the mutants in liquid culture was reduced by about one order of magnitude. On agar-solidified medium, spore formation was more strongly affected, particularly in agrA and agrC mutants. Similarly, accumulation of the starch-like storage compound granulose was almost undetectable in colonies of agrB, agrA, and agrC mutants. Importantly, these defects could be genetically complemented, demonstrating that they were directly linked to agr inactivation. A diffusible factor produced by agrBD-expressing strains was found to restore granulose and spore formation in the agrB mutant. Furthermore, a synthetic cyclic peptide, designed on the basis of the C. acetobutylicum AgrD sequence, was also capable of complementing the defects of the agrB mutant when added exogenously to the culture. Together, these findings support the hypothesis that agr-dependent quorum sensing is involved in the regulation of sporulation and granulose formation in C. acetobutylicum.
INTRODUCTION
The Gram-positive, endospore-forming bacterium Clostridium acetobutylicum is strictly fermentative and gains energy by converting sugars and starch to organic acids and solvents (5, 17). In the past, it has been exploited for the large-scale production of acetone and butanol, but the classical industrial fermentation process is currently considered uneconomical in most countries, despite increasing oil prices (16). Essential for the engineering of more efficient strains and processes is a more thorough understanding of the organism's physiology and metabolism (22).
The fermentation metabolism of C. acetobutylicum is complex. During exponential growth in batch culture, acetic and butyric acid are produced, which allows the cells to gain more than 3 ATPs per molecule of glucose. However, continued acid production poses a problem as the pH of the medium decreases and undissociated acids diffuse back into the cells, thereby affecting intracellular pH and leading to proton gradient dissipation. To avoid acid death, cells shift their metabolism to solvent formation: some of the produced acids (mainly butyric acid) are taken up again and, together with remaining sugars, converted to butanol, acetone, and also some ethanol (5, 17). This switch to solvent production coincides with the initiation of the complex developmental program of sporulation. As part of these processes, a starch-like storage compound termed granulose is transitorily formed and accumulates in the cytoplasm (24, 35). Both solventogenesis and sporulation are linked via the transcription factor Spo0A, the master regulator of sporulation: Spo0A is essential for the initiation of sporulation in solventogenic clostridia (as in Bacillus subtilis) but also required for high solvent production (8, 34). In contrast to B. subtilis, Spo0A in C. acetobutylicum is not activated via a complex phosphorelay system. Instead, its phosphorylation state is controlled by multiple orphan histidine kinases through direct interaction (37). The whole cascade of sigma factors that regulates the sporulation process downstream of phosphorylated Spo0A in B. subtilis is present in C. acetobutylicum (2, 18) although some differences seem to exist (for instance in the activation of σF by SpoIIE). However, while general conditions for solventogenesis and sporulation are known, and progress is being made in unraveling the underlying regulatory networks, the chemical or physical nature of the initiating signals still remains to be elucidated.
There is, however, some evidence to suggest that cell-cell communication (quorum sensing) systems are widely distributed in Clostridium spp. (43) and that they may play a role in regulating both solventogenesis and sporulation (3, 20, 21). Quorum sensing is a communication mechanism that relies on small, diffusible signal molecules and allows bacteria to coordinate changes in gene expression with cell population density (42). Most Gram-positive quorum sensing systems are based on secreted peptides which can be linear or cyclic and sometimes contain extensive posttranslational modifications (23, 39). Relatively little is known about the operation of such systems in clostridial species, but for the solventogenic Clostridium sacharoperbutylacetonicum, an as-yet-unidentified, self-generated signal in the culture supernatant has been reported to induce solvent formation in a “low-solvent” mutant, suggesting that the metabolic shift from acid to solvent production might be subject to quorum sensing control (20). Furthermore, in silico analysis suggests that agr-type quorum sensing systems may exist in many of the sequenced members of the class Clostridia (reference 43 and unpublished data from this laboratory) although thus far, experimental evidence has been provided for only three species. In Clostridium sporogenes, group I Clostridium botulinum, and Clostridium perfringens, agr-homologous systems are required for efficient sporulation. In group I C. botulinum and C. perfringens, the system is also involved in the control of toxin production (3, 21, 30, 41).
The agr system was first discovered in Staphylococcus aureus, where it comprises five genes: agrB, agrD, agrC and agrA (which make up the actual cell-cell signaling system) and RNAIII (the main effector of target gene regulation) (see reference 28 for an excellent review). A cyclic, so-called autoinducing peptide (AIP) is derived from an internal AgrD fragment, through the action of the membrane-associated protein AgrB, and acts as a signal molecule. Upon extracellular accumulation, the AIP is sensed by a two-component system consisting of the histidine kinase AgrC and the response regulator AgrA. Phosphorylated AgrA then further activates the agrBDCA operon and induces RNAIII expression but also controls a subset of target genes independently of RNAIII (33).
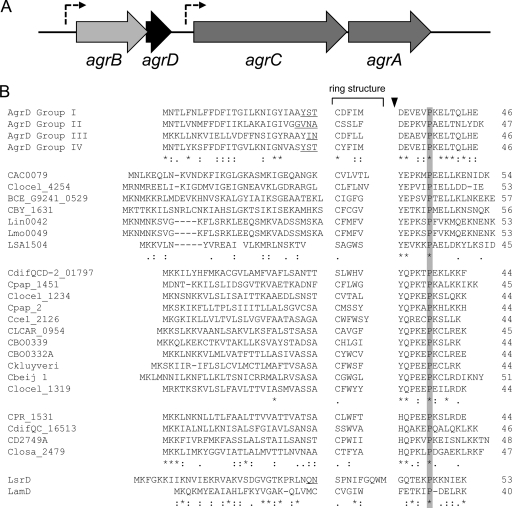
Interestingly, C. acetobutylicum is one of the members of the genus that appears to possess a complete agrBDCA cluster (Fig. 1A), although a regulatory RNA similar to RNAIII has not yet been identified. In contrast to the staphylococcal system, agrBD is predicted to be transcribed independently of agrCA (31), and this is experimentally supported by a gene array time course series that revealed strong increases in agrB and agrD transcript levels when the cells entered stationary phase, i.e., just before the initiation of solvent formation and sporulation, whereas agrCA expression remained relatively constant (analysis of microarray data provided in the supplemental material of reference 1).
Fig 1.
Schematic representation of the C. acetobutylicum agrBDCA cluster (A) and alignment of confirmed and putative AgrD sequences (B). (A) The C. acetobutylicum agrBDCA cluster is predicted to comprise two independent transcriptional units (indicated by the dashed arrows), agrBD and agrCA. (B) The alignment shows the experimentally confirmed AgrD proteins from S. aureus (AgrD group I to IV), E. faecalis (LsrD), and L. plantarum (LamD), as well as selected putative AgrD sequences from other species. The latter included the following (locus tags are given in parentheses, if available, and identify the respective sequence in the alignment): C. acetobutylicum (CAC0079), Clostridium cellulovorans 743B (Clocel_1234, Clocel_1319, and Clocel_4254), Bacillus cereus G9241 (BCE_G9241_0529), Listeria innocua Clip11262 (Lin0042), Listeria monocytogenes EGD-e (Lmo0049), Clostridium butyricum 5521 (CBY_1631), Lactobacillus sakei subsp. sakei 23K (LSA1504), Clostridium difficile QCD-23m63 (CdifQCD-2_01797), Clostridium papyrosolvens DSM 2782 (Cpap_1451 and Cpap 2, the latter not annotated), Clostridium cellulolyticum H10 (Ccel_2126), Clostridium carboxidivorans P7 (CLCAR_0954), Clostridium botulinum ATCC 3502 (CBO0332A and CBO0339), Clostridium kluyveri (Ckluyveri; not annotated), C. beijerninckii NCIMB 8052 (Cbeij 1; not annotated), Clostridium perfringens SM101 (CPR_1531), C. difficile QCD-66c26 (CdifQC_16513), C. difficile 630 (CD2749A), Clostridium saccharolyticum WM1 (Closa_2479). The black triangle indicates confirmed and proposed processing sites downstream of the ring-encoding part of AgrD. A highly conserved proline, present in all sequences, is shaded gray. Note the sequence conservation immediately downstream of the proposed cleavage site and upstream of the conserved proline. For each AIP group, identities (*), conserved (:), and semiconserved substitutions (.) are displayed. Amino acids constituting the N-terminal tail of confirmed AIPs are underlined. Numbers indicate the total length of the AgrD sequence.
Here we report the mutational analysis of the agr locus in C. acetobutylicum ATCC 824.
MATERIALS AND METHODS
Bacterial strains and media.
C. acetobutylicum ATCC 824 was grown at 37°C in an anaerobic cabinet (MG1000 Anaerobic Work Station, Don Whitley Scientific) containing an atmosphere of 80% nitrogen, 10% hydrogen, and 10% carbon dioxide. Clostridial strains were cultured either in clostridial basal medium (CBM) (29) or 2× YTG (tryptone, 16 g/liter; yeast extract, 10 g/liter; NaCl, 5g/liter; glucose, 10g/liter; adjusted to pH 5.8 for agar-solidified medium and pH 5.2 for broth). Escherichia coli TOP10 (used as a host for ClosTron and complementation plasmids) was grown in Luria-Bertani medium (37) at 37°C. Antibiotics were used at the following concentrations: chloramphenicol, 25 μg/ml; erythromycin, 10 μg/ml (CBM) or 40 μg/ml (2× YTG); thiamphenicol, 15 μg/ml.
Plasmids, primers, DNA techniques.
Oligonucleotides were synthesized by Eurofins MWG Operon, Germany (see Table S1 in the supplemental material). PCR amplifications were carried out using the FailSafe PCR system (Epicentre) or Taq DNA polymerase (New England BioLabs). Electroporation of C. acetobutylicum was performed as described previously (38). Plasmid isolation and genomic DNA preparations were carried out using the QIAprep miniprep kit (Qiagen, United Kingdom) and the DNeasy blood and tissue kit (Qiagen), respectively. Restriction enzymes were supplied by New England BioLabs and were used according to the manufacturer's instructions. Standard methods were used for agarose gel electrophoresis and transformation of E. coli (37).
Construction of mutants using ClosTron technology.
Construction of mutants was based on a modified group II intron (the ClosTron) which carries an erythromycin resistance marker and has been retargeted so that it integrates into specific sites of the host's chromosome. Mutants were constructed in C. acetobutylicum ATCC 824 targeting genes agrB (cac0078), agrC (cac0080), and agrA (cac0081), using the standard methodology described by Heap et al. (10, 12). IBS, EBS1d, and EBS2 primers used for retargeting the intron are listed in Table S1 (in the supplemental material). Numbers in the primer names indicate the retargeting site used. PCR-amplified retargeted HindIII/BsrGI fragments were cloned into the pMTL007C-E2 vector. Genomic DNA from putative agr mutants (agrB::CTermB, agrC::CTermB, and agrA::CTermB) was subjected to two PCR screens. First, primers flanking the insertion site were employed to confirm that the intron was present in the desired target gene (primer pairs, CAC0078-F/CAC0078-R, CAC0080-F/CAC0080-R, and CAC0081-F/CAC0081-R for agrB, agrC, and agrA, respectively). To confirm the orientation in which the intron was present, primers amplifying across the intron-exon junction were used (primer pairs, CAC0078-R/EBS universal, CAC0080-F/EBS universal, and CAC0081-R/EBS universal for agrB, agrC, and agrA, respectively).
Generation of complementation vectors.
Complementation vectors for the mutant strains encompassed the complete coding sequences of agrA, agrBD, agrC, and agrCA together with their respective putative promoter regions. For the agrBD cluster, the cognate intergenic upstream region was assumed to harbor the promoter. For the agrCA cluster, the 224-bp region upstream of the start codon of agrC was assumed to contain the necessary regulatory elements. Coding sequences, including their putative promoter regions, were PCR amplified from genomic DNA. The agrBD and agrC regions were amplified using primer pairs agrBD-F/agrBD-R and agrCA-F/agrC-R, respectively. The resulting PCR fragments were digested with XhoI and BamHI and subcloned into the pMTL85141 vector (11) linearized with the same enzymes. The agrCA cluster, including the 224-bp upstream region, was amplified as two fragments, using primer pairs agrCA-F/agrC-R and agrCA2-F/agrA-R. Taking advantage of a SacI recognition site in the agrC gene, the two fragments were digested with XhoI/SacI and SacI/BamHI, respectively, and ligated into the XhoI/BamHI-linearized pMTL85141 vector. For expression of agrA, the putative promoter region of the agrCA cluster was amplified using primer pair agrCA-F/agrCProm-R (introducing an NdeI site at the start codon of agrC) and the agrA gene was amplified using primer pair agrA-F/agrA-R (introducing an NdeI site at the start codon of agrA). The XhoI/NdeI-digested promoter region and the NdeI/BamHI-digested agrA fragment were introduced into XhoI/BamHI-linearized pMTL85141 in a three-fragment ligation step. All complementation plasmids were verified by sequencing.
Spore assays.
C. acetobutylicum strains were grown in 5 ml CBM broth containing 0.5% CaCO3 and 5% glucose (CBM-S) to enable sporulation. After 5 days, a 200-μl sample of culture was heated to 80°C for 10 min. Serial dilutions were carried out and 10-μl aliquots of the heat-treated cell suspension plated onto CBM agar. Colonies were enumerated after 24 h of incubation.
Detection of granulose.
To assess the accumulation of granulose, C. acetobutylicum strains were grown on agar-solidified CBM containing 5% glucose (CBM-G) to promote granulose accumulation. After incubation for 24 h to 48 h, plates were exposed to iodine vapor, staining granulose-containing colonies dark brown (36).
Detection of diffusible signals by cross-streaking.
Wild-type C. actetobuytlicum and agr mutants were precultured in CBM broth until visible growth was detectable. An inoculation loop was used to streak these cell suspensions on CBM-G plates, generating streaks approximately 7 mm wide and taking care that streaks were physically separated. After incubation for 48 h, plates were subjected to iodine staining as described above.
Chemical complementation of the agrB mutant and detection of AIPs.
Cyclic AIPs were synthesized and high-performance liquid chromatography (HPLC) purified by Peptide Protein Research Ltd. (Fareham, United Kingdom); the purity was estimated to range from 78 to 90%, depending on the AIP. Lyophilized peptides were dissolved in dimethyl sulfoxide (DMSO) to obtain 1 mM stock solutions.
For complementation studies in broth, the agrB::CTermB strain was grown in CBM-S to an optical density at 600 nm (OD600) of 0.4 to 0.5. The culture was then divided into 3-ml aliquots and supplemented with an appropriate volume of AIP stock solutions to obtain final AIP concentrations of 2 and 20 μM. DMSO controls were included in all experiments. After 5 days of incubation, spore assays were performed as described above. For plate-based complementation assays, filter discs (6-mm diameter; Whatman) were placed either on the surface of CBM-G plates whose surface had been spread with an appropriate dilution of an early-exponential-phase culture of agrB::CTermB or next to the streaks of mutant strains. Samples (5 μl) of the appropriate AIP stock solutions were placed on the filter discs. Plates were incubated for 48 h and subjected to iodine staining to visualize granulose formation as described above.
Statistical analysis.
The data were analyzed in GraphPad Prism using the unpaired t test. Significance levels are indicated by asterisks in graphs.
RESULTS
Insertional inactivation of agr genes using ClosTron technology.
A putative agr quorum sensing system is present in the genome of C. acetobutylicum ATCC 824 and also in that of the recently sequenced EA 2018 strain (13, 27). It consists of a gene cluster comprising the genes agrB, agrD, agrC, and agrA (Fig. 1A). Using ClosTron technology, all agr genes apart from agrD were insertionally inactivated in the ATCC 824 strain, resulting in the mutant strains agrB::CTermB, agrC::CTermB, and agrA::CTermB. A defect in the agrB gene is expected to abolish production of the postulated AIP signal, whereas agrA and agrC mutants should be unable to respond to AIP accumulation. For each gene, four independent mutant clones were isolated and further characterized. This was done to avoid accidental isolation of strains carrying undesired second site mutations: the isolation of ClosTron mutants involves multiple restreaking of candidate colonies and this may increase the risk of obtaining spontaneously “degenerated” strains, typically characterized by a reduced or abolished capacity to form solvents and to sporulate (9, 19).
Correct insertion of ermB-carrying introns into the target genes was confirmed by several PCR screens using primers flanking the insertion site and primers amplifying across the intron-exon junction (data not shown).
Phenotypic characterization of agr mutants.
A preliminary analysis revealed that one of the obtained agrA and one of the obtained agrC mutant clones differed phenotypically from the other three and showed signs of degeneration (data not shown). These clones were therefore excluded from further, more detailed analyses.
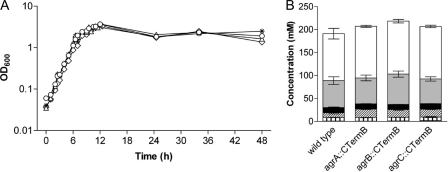
When cultured in CBM-S broth, agrA::CTermB, agrB::CTermB, and agrC::CTermB strains showed no marked differences in their growth kinetics (Fig. 2A) or their ability to form solvents (Fig. 2B) compared with the wild type. The ability to degrade starch and swimming motility were also not affected (data not shown). However, microscopic examination revealed a noticeable reduction in the number of endospores formed by all three mutant strains. This decrease in spore numbers appeared to be more pronounced in samples taken from colonies than in those from liquid cultures.
Fig 2.
Growth and solvent formation of C. acetobutylicum agr mutants. Wild-type C. acetobutylicum and agr mutants were grown in CBM-S broth. (A) Growth was monitored by following the optical density at 600 nm: wild type (asterisks), agrA mutant (triangles), agrB mutant (circles), agrC mutant (diamonds). (B) After 72 h, culture supernatant samples were taken and analyzed for the produced acids (acetate, checks; butyrate, lines) and solvents (butanol, white; acetone, gray; ethanol, black). The values shown are the means ± standard errors of the means of results from six independent experiments. Product concentrations are not significantly different between the wild-type and the mutant strains, except for the difference between butyrate produced by the agrA mutant and the wild type (P = 0.02).
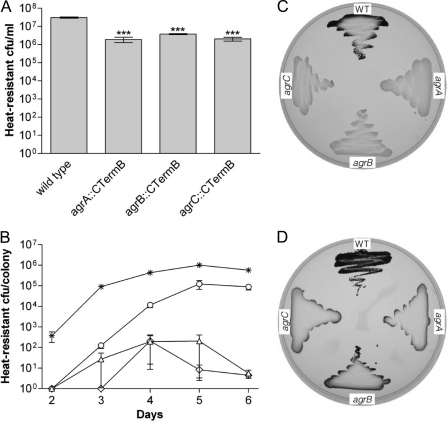
To obtain a more quantitative measure for the differences observed, the number of spores present in a given culture volume (liquid cultures) or in similar-sized colonies was determined (more precisely, this procedure quantifies the number of heat-resistant CFU as a measure for spores that can germinate and grow after a heat treatment for 10 min at 80°C). In liquid cultures, an about 10-fold reduction in the number of spores was observed for all agr mutants compared to the wild-type strain (Fig. 3A). In contrast, colonies of the mutant strains grown on CBM plates contained between 1 and 5 orders of magnitude fewer spores than the wild type. Interestingly, the severity of the observed sporulation defect under these conditions was dependent on the sampling time point and the inactivated agr gene. After 6 days, spore numbers for the agrA and agrC mutant were reduced by about 5 orders of magnitude, whereas for the agrB mutant, sporulation efficiency was reduced by only about 1 order of magnitude compared to that of the wild type (Fig. 3B).
Fig 3.
Sporulation and granulose accumulation in C. acetobutylicum agr mutants. (A) Sporulation assays of wild-type C. acetobutylicum and agr mutants cultured in CBM-S broth for 5 days. The values shown are the means ± standard errors of the means (SEM) of results from ≥3 clones of each strain (each clone representing an independently derived mutant). All values are significantly different from the wild-type strain (***, P < 0.001). (B) The number of heat-resistant endospores formed per colony was determined for C. acetobutylicum (asterisks) and the mutant strains agrA::CTermB (triangles), agrB::CTermB (circles), and agrC::CTermB (diamonds) over a time course of 6 days. The values shown are the means ± SEM of results from 10 colonies of each strain and time point. Mutant strains and wild type are significantly different in all given time points (P < 0.001). (C and D) Granulose assays. Wild-type C. acetobutylicum (WT), agrA::CTermB (agrA), agrB::CTermB (agrB), and agrC::CTermB (agrC) were grown on CBM-G plates and assayed for granulose formation after 24 h (C) and 48 h (D).
Granulose accumulation is known to coincide with the initiation of sporulation, and the simple method of staining granulose with iodine vapor—which stains granulose-containing cells dark brown—appears to be a reliable indicator for sporulation on plates. As shown in Fig. 3C and D, all mutants tested negative for granulose after 24 h (Fig. 3C) and produced only very little granulose along the edges of the streak after 48 h (Fig. 3D). This is in stark contrast to results with the wild-type strain, which started granulose accumulation at around 24 h and, at 48 h, stained intensely brown, suggesting that cells had produced considerable amounts of this glycogen-like storage product.
Genetic complementation of agr mutants.
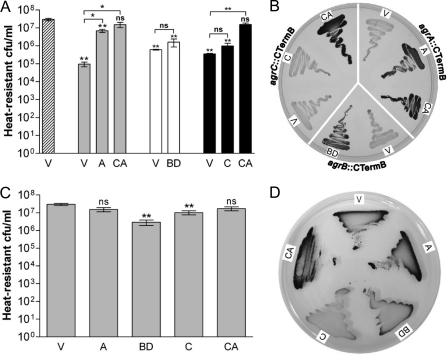
The obtained agr mutants were genetically complemented to confirm that the ClosTron insertions rather than second site mutations were responsible for the observed phenotypes. For these complementation studies, the agr genes under the control of their native promoters were introduced into the modular vector pMTL85141 (11) and transformed into the respective mutant strains by electroporation. Introduction of the agrA complementation vector (pMTL85141-agrA) successfully restored spore formation and granulose production in the agrA::CTermB strain, whereas the agrC complementation vector (pMTL85141-agrC) failed to complement the agrC::CTermB strain (Fig. 4A and B). Since agrC and agrA appear to form an operon, it is possible that the ClosTron insertion in agrC had also negatively affected expression of the downstream located agrA. Therefore, the complete agrCA cluster (pMTL85141-agrCA) was introduced into the agrC mutant and found to restore sporulation and granulose formation to wild-type levels. In view of this result, the complete agrBD cluster was cloned (pMTL85141-agrBD) and transformed into the agrB::CTermB strain. Introduction of pMTL85141-agrBD partially restored spore formation of the agrB mutant in liquid culture (Fig. 4A) and reestablished granulose formation on CBM-G plates (Fig. 4B). Taken together, these results confirmed that the observed phenotypical changes in the agr mutants were directly linked to the inactivation of the respective genes.
Fig 4.
Complementation of C. acetobutylicum agr mutants and effect of overexpression of agr genes. (A and B) For complementation studies, strains agrA::CTermB (gray bars), agrB::CTermB (white bars), and agrC::CTermB (black bars) were transformed with the vector control (V) or the indicated complementation vectors carrying the agrA (A), agrBD (BD), agrC (C), and agrCA (CA) genes. As a control, the wild-type strain carrying the vector control (striped bar) was also included. Sporulation assays were performed after 5 days in CBM-S broth (A) and granulose accumulation was assayed on CBM-G plates after 24 h (B). The data obtained for mutant strains carrying either a complementation vector or the empty plasmid were compared to those for the wild-type strain harboring the empty vector; significance levels are indicated immediately above the bars. The significance levels of the differences between complemented mutants and the corresponding mutant strains transformed with empty vector are indicated above the brackets: **, P < 0.01; *, P < 0.05; ns, not significant. (C and D) Wild-type agr genes were introduced into the parent C. acetobutylicum strain to investigate the effect of overexpression of these genes on sporulation after 5 days (C) and granulose formation after 24 h (D). The values shown are the means ± standard errors of the means of results from ≥3 independent experiments. The data shown in panel C were compared to those for the wild-type strain carrying the empty plasmid, and the significance levels are indicated: **, P < 0.01; ns, not significant.
The constructed complementation plasmids were also introduced into the ATCC 824 wild-type strain to assess the phenotypic consequences of agr gene overexpression. This was motivated by the finding that sporulation was only partially restored in the complemented agrB mutant. As can be seen from Fig. 4C, expression of agrA and agrCA in this background had no marked effect on sporulation in liquid culture compared to the vector control. In contrast, overexpression of agrBD reduced the number of spores about 10-fold and a slight, but significant, reduction was also observed for agrC. A possible explanation for this could be that expression of the agrBD cluster from a multicopy plasmid resulted in too much signal or the wrong timing and thus disturbed the regulatory network in some way. This could also explain why the agrB mutant appeared to be only partially complemented in liquid broth. For granulose formation on plates, a different picture emerged. Again, overexpression of agrA had no obvious effect, but in comparison to the vector control granulose formation was clearly increased in the agrCA overexpressing strain. Furthermore, granulose formation was reduced in wild-type strains overexpressing agrBD or agrC (Fig. 4D and data not shown).
A diffusible factor produced by wild-type C. acetobutylicum restores sporulation and granulose formation in the agrB mutant.
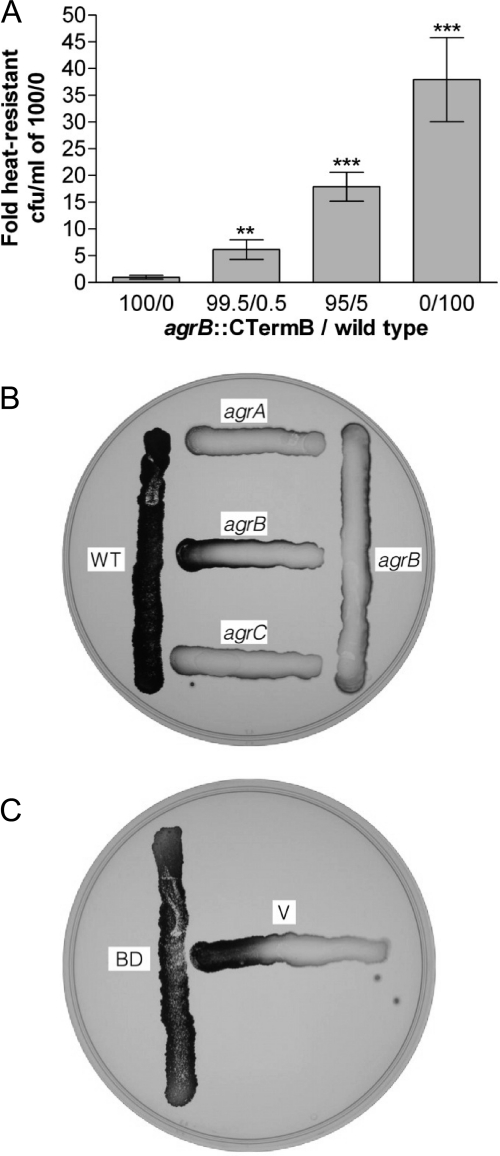
If the C. acetobutylicum agrBDCA cluster represents a true quorum sensing system working in the same way as its staphylococcal counterparts, the agrB mutant's phenotype should be caused by a lack of extracellular AIP. Therefore, several experiments were performed to test whether the defects in sporulation and granulose production in this mutant could be restored by a quorum sensing signal secreted by the wild type. The agrB::CTermB strain was grown in coculture with wild-type C. acetobutylicum in a number of different ratios ranging from 0.5 to 5% wild type. After 5 days, these cultures were heat treated to kill off all vegetative cells and to induce germination of the spores that had been formed. Spore counts were performed on CBM plates containing erythromycin to ensure that only germinating spores derived from the agrB mutant could grow and form colonies (the ClosTron insertion confers erythromycin resistance). Interestingly, the presence of wild-type cells increased spore formation in the agrB::CTermB strain by up to one order of magnitude, even when only present at low ratios (Fig. 5A and data not shown), thus supporting the hypothesis that agrBD is required for the production of a diffusible quorum sensing signal.
Fig 5.
A diffusible factor produced by wild-type C. acetobutylicum restores sporulation and granulose formation in the agrB mutant. (A) Early-exponential-phase cultures of agrB::CTermB were mixed in the indicated ratios with the wild-type strain. Spore assays were performed after 5 days, and heat-treated spores were plated on erythromycin-supplemented CBM plates, which allowed only germintated agrB mutant spores to grow. Wild-type controls (0/100) were counted on nonselective medium. Shown is the fold increase in spore numbers compared to the agrB::CTermB single culture control (100/0). The values given are the means ± standard errors of the means of results from three independent experiments. All values are significantly different from the control 100/0 (***, P < 0.001; **, P < 0.01). (B) agrA::CTermB (agrA), agrB::CTermB (agrB), and agrC::CTermB (agrC) were cross-streaked with wild-type C. acetobutylicum (WT) on CBM-G plates and assayed for granulose accumulation after 48 h. (C) Shown is the granulose stain of a cross-streak of the complemented agrB::CTermB strain (BD) and agrB::CTermB carrying the vector control (V) after 48 h.
To exclude the possibility that the wild type exerted this effect via direct cell-to-cell contact, T-streak experiments on agar plates were also performed. The agrB::CTermB strain was streaked on CBM-G plates next to wild-type C. acetobutylicum, agrA::CTermB, and agrC::CTermB, incubated for 48 h and then assessed for granulose formation. Close vicinity of the wild type restored granulose formation in the agrB mutant but failed to do so in the agrA and agrC mutants (Fig. 5B). Interestingly, T-streaks of the agrA and agrC mutant also failed to restore granulose formation in the agrB mutant, suggesting that they did not produce the required amounts of diffusible signal. As expected, the complemented agrB mutant was capable of restoring granulose formation in the agrB::CTermB strain when streaked next to it (Fig. 5C).
Chemical complementation of the agrB mutant with synthetic peptides.
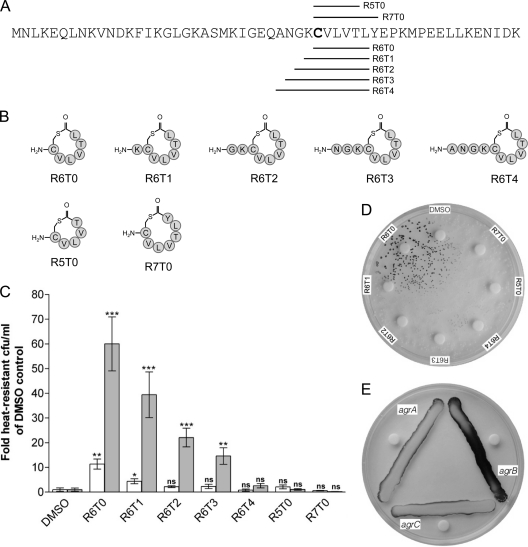
A number of synthetic, cyclic AIPs (Fig. 6A and B) were assessed for their ability to enhance sporulation of the agrB mutant. The sequences for these peptides were derived from the C. acetobutylicum AgrD sequence (Fig. 6A), assuming the formation of five-, six-, or seven-amino-acid ring structures, respectively. The length of the N-terminal tail sequence (0 to 4 amino acids) was based on those found in other known AIPs (25, 28, 40). Cultures of agrB::CTermB were supplemented with these synthetic AIPs (final concentrations, 2 and 20 μM) and assayed for spore formation after 5 days (Fig. 6C). Interestingly, highest activities in our test system were observed for an AIP consisting of a six-amino-acid ring without an N-terminal tail moiety (termed R6T0). Inclusion of such a tail moiety reduced the activity of the resulting AIP with increasing tail length (R6T1 to R6T4 [Fig. 6C]). As a control, cyclic AIPs consisting of five- and seven-membered rings, but no tail moiety, were used (R5T0 and R7T0, respectively) and showed no significant effect on sporulation.
Fig 6.
Effect of synthetic cyclic peptides on sporulation and granulose formation. (A) C. acetobutylicum AgrD sequence displaying the position of peptide stretches (horizontal lines) that were used to design the synthetic, cyclic AIPs shown schematically in panel B. The bold C indicates the conserved cysteine required for thiolactone bond formation. (C) The number of heat-resistant endospores formed by the C. acetobutylicum agrB::CTermB strain was analyzed after 5 days of growth in CBM-S broth in the presence of 2 μM (white bars) and 20 μM (gray bars) concentrations of the indicated peptides and compared to a DMSO control. The values shown are the means ± standard errors of the means of results from three independent experiments. Significance levels of differences between AIP-supplemented cultures and corresponding DMSO controls are indicated above the bars: ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant. (D) Granulose stain of agrB::CTermB grown on CBM-G plates supplemented with the indicated AIPs on filter discs and assayed after 48 h. (E) Granulose stain of agrA::CTermB (agrA), agrB::CTermB (agrB), and agrC::CTermB (agrC) grown next to filter discs impregnated with R6T0 AIP stock solution.
The synthetic AIPs were also tested for their ability to induce granulose formation in the agrB mutant. Filter discs were placed on agar plates freshly inoculated with the agrB mutant and impregnated with 5 μl AIP stock solution (1 mM). Granulose formation was assessed after 48 h. Under these conditions, the R6T0 AIP restored granulose formation, whereas the other six-membered AIPs containing an N-terminal tail (R6T1-R6T4) had no visible effect. Again, the five- and seven-membered control AIPs without N-terminal tail (R5T0 and R7T0, respectively) showed no activity (Fig. 6D). None of the synthetic AIPs was able to restore granulose formation in the agrA and agrC mutant (Fig. 6E and data not shown).
DISCUSSION
In this study, the function of the C. acetobutylicum agrBCDA gene cluster was investigated. The results suggest that it is a true quorum sensing system involved in the cell density-dependent regulation of granulose and endospore formation. All genes of the cluster could be inactivated, apart from agrD, for which, due to its small size, no suitable ClosTron insertion site could be identified. However, agrB and agrD inactivation are predicted to have the same consequence: a lack of AIP production. Thus, mutants were constructed that failed to either produce (agrB::CTermB) or respond to (agrC::CTermB and agrA::CTermB) the presumed AIP quorum sensing signal.
One of the problems encountered during agr mutant construction was that the organism is inherently prone to degeneration, characterized by a loss of several key features, in particular the ability to sporulate and to produce acetone and butanol (9, 19). The ClosTron mutagenesis procedure employed (10) involved frequent restreaking of colonies and thus carried a risk of enriching and isolating degenerate mutant strains. It was therefore absolutely essential to ensure that the phenotypes of isolated mutants were caused by the ClosTron insertion rather than second site mutations. This was achieved by (i) screening several independent mutant clones for each gene and eliminating potential outliers and (ii) subsequent genetic complementation.
So what is the evidence that the C. acetobutylicum agr cluster acts as a functional quorum sensing unit responsible for production and detection of an extracellular AIP? First, inactivation of the putative AIP signal-generating (AgrB) and -receiving (AgrCA) parts of the cluster resulted in almost identical phenotypes, suggesting that they are part of the same regulatory circuit. Second, wild-type characteristics could be restored when growing the signal-deficient agrB mutant in close vicinity to the wild type, which suggests AgrB-dependent production of a diffusible signaling compound by the latter. Third, this wild-type-produced diffusible signal could not complement agrC and agrA mutants, suggesting that the agrCA-encoded two-component system is required for signal reception and transmission. Fourth, restoration of both granulose formation and sporulation by the agrB mutant was also achieved with a synthetic AIP, whose sequence was based on a relevant section of the putative signal precursor polypeptide, AgrD. Again, the AgrCA system was required for the perception of this signal. Taken together, the results provide strong support for the hypothesis that the C. acetobutylicum agrBD cluster is required for the production of a cyclic AIP, which accumulates extracellularly and is sensed by the AgrCA two-component system. Although agrCA are predicted to form a separate transcriptional unit (31) and, in contrast to agrBD, are not drastically upregulated during entry into stationary phase (analysis of microarray data provided in the supplemental material of reference 1), they are required for upregulation of AIP production, as agrA and agrC mutants both failed to chemically complement the agrB mutant in the cross-streaking experiments.
No AIP structures have been published for any of the clostridial Agr systems investigated so far. However, the results obtained here for a set of synthetic AIPs suggest that the native C. acetobutylicum AIP has a thiolactone ring structure that, unlike its staphylococcal counterparts, consists of six instead of five amino acids (CVLVTL; cyclized by thiolactone bond formation between the SH group of cysteine and the carboxy group of the N-terminal leucine) (Fig. 6B). This finding was not entirely unexpected, given the sequence conservation at the C. acetobutylicum AgrD C-terminal end (Fig. 1B). It was reasonable to assume that the N-terminal end of the AIP ring structure is represented by the conserved cysteine. The C-terminal end was predicted to be a leucine, located five amino acids downstream of the conserved cysteine, for several reasons. First, this leucine is located immediately adjacent to a tyrosine which is conserved in a large subgroup of AgrD proteins (Fig. 1B) and therefore likely to belong to the C-terminal part of AgrD which is not involved in ring formation. Second, the proposed cleavage site between this leucine and the adjacent tyrosine is in the same position, relative to a highly conserved proline, as the respective cleavage sites in all other AgrD homologues for which corresponding AIPs have been identified, i.e., staphylococcal AgrD, Lactobacillus plantarum LamD, and Enterococcus faecalis FsrD (14, 15, 25, 26, 40). Whether the R6T0 peptide, which showed the strongest effect in the sporulation assays and was the only one to restore granulose formation on plates, represents the true structure of the native signaling peptide remains to be seen. However, given the lack of activity for the related five- and seven-amino acid peptide rings (R5T0 and R7T0), it seems likely that the native AIP will contain a six-membered amino acid ring structure, identical or very similar to that of R6T0. Although the AgrD-derived AIPs produced by staphylococci and Lactobacillus plantarum all contain pentapeptide ring structures (14, 15, 40), larger rings are not without precedent: the E. faecalis FsrD-derived AIP contains a lacton ring structure consisting of nine amino acids (25). In this context it is also of interest that the LamD-derived AIP from L. plantarum consists only of a cyclic thiolactone pentapetide without N-terminal tail moiety (40).
For many years, the factors that trigger sporulation in clostridial species have remained elusive. There is now an increasing body of evidence to suggest that agr-dependent quorum sensing, and thus presumably cell density, plays an important role in this process. Earlier work from this laboratory has shown that agrBD systems are required for efficient sporulation in Clostridium sporogenes and group I Clostridium botulinum (3), and a recent study on Clostridium perfringens also established drastically reduced endospore numbers in agrB mutant cultures (21). Furthermore, almost all currently sequenced members of the genus Clostridium possess at least one set of agrBD homologous genes (reference 43 and unpublished data from this laboratory). An interesting question is, therefore, what advantage Clostridium spp. might gain by linking cell-cell communication (and thus information on cell density) with endospore formation. At least for C. acetobutylicum it seems that activation of agr quorum sensing at high cell densities is not an absolute prerequisite for sporulation to take place, as spores are still formed when the system is rendered nonfunctional, particularly under conditions of liquid culture. This is similar to the situation in B. subtilis, where the role of quorum sensing in the regulation of sporulation is very well established (32). Nutrient limitation is an essential trigger for sporulation in this organism, and cell density is thought to play a modulatory role because B. subtilis quorum sensing mutants still sporulate, albeit with much reduced efficiency. Information on high cell density or crowding could be valuable in this context, as it might indicate the degree of competition for scarce nutrients (7) and allow the population to adjust sporulation levels accordingly. For C. acetobutylicum, however, this explanation is unlikely to apply, as nutrient limitation does not appear to be a major factor in clostridial sporulation (4). It is also not clear whether granulose formation is directly regulated by quorum sensing or indirectly as part of the ongoing sporulation process. Direct regulation in response to cell density might allow cells to increase sugar uptake and initiate granulose formation before external resources run out. This would not only take the sugar away from potential competitors but also ensure that C. acetobutylicum cells can complete the sporulation process with only limited dependence on external carbon and energy sources. The last point could be important given that high butanol levels are known to affect membrane-associated functions, including transport processes.
How the C. acetobutylicum Agr system integrates with the regulatory cascade(s) that controls endospore formation will be an interesting aspect of future studies. An attractive hypothesis could be that AgrA influences the phosphorylation status of Spo0A, either directly or indirectly, by controlling the expression of the recently described Spo0A-phosphorylating/dephosphorylating orphan histidine kinases (38). Alternatively, the system could control spo0A expression levels, as has been suggested for C. perfringens (21). However, given that Spo0A is important for both spore formation and solvent production (spo0A inactivation drastically reduces butanol and acetone production [8]), it is difficult to explain with these hypotheses why the latter remained unaffected, at least in liquid culture, for all C. acetobutylicum agr mutants. It is possible that differing levels of phosphorylated Spo0A are needed for the initiation of solventogenesis and sporulation, respectively (similar to the situation in B. subtilis, where increased levels appear to be required for the initiation of sporulation [6]). Another explanation could be that the C. acetobutylicum Agr system acts by modulating the expression of key sporulation factors downstream of phosphorylated Spo0A. For instance, Jones et al. (18) recently found that inactivation of σF in C. acetobutylicum abolishes sporulation before cells form granulose and undergo asymmetric cell division but does not block solvent formation. Thus, Agr-dependent regulation of an early key sporulation factor such as σF may allow the organism to produce solvents without committing to the sporulation pathway. Clearly, Agr-dependent quorum sensing in C. acetobutylicum is part of a complex regulatory network that, depending on other environmental and internal stimuli, will place more or less emphasis on cell density-dependent inputs, as illustrated by the differences observed for agr-independent sporulation on solid and in liquid medium.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Biotechnology and Biological Sciences Research Council UK through SysMO and project grants BB/F003390/1, BB/I004475M, and BB/G016224/1.
We thank John Heap for helpful discussions and Ann-Kathrin Kotte for establishing the best conditions for granulose assays.
Footnotes
Published ahead of print 16 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alsaker KV, Papoutsakis ET. 2005. Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum. J. Bacteriol. 187:7103–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bi C, Jones SW, Hess DR, Tracy BP, Papoutsakis ET. 2011. SpoIIE is necessary for asymmetric division, sporulation, and expression of σF, σE, and σG but does not control solvent production in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 193:5130–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cooksley C, et al. 2010. Regulation of neurotoxin production and sporulation by a putative agrBD signaling system in proteolytic Clostridium botulinum. Appl. Environ. Microbiol. 76:4448–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dürre P, Hollergschwandner C. 2004. Initiation of endospore formation in Clostridium acetobutylicum. Anaerobe 10:69–74 [DOI] [PubMed] [Google Scholar]
- 5. Dürre P. 2005. Formation of solvents in clostridia, p 671–693 In Dürre P. (ed), Handbook on clostridia. CRC Press, Boca Raton, FL [Google Scholar]
- 6. Fujita M, Losick R. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19:2236–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grossman AD. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477–508 [DOI] [PubMed] [Google Scholar]
- 8. Harris LM, Welker NE, Papoutsakis ET. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:3586–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hartmanis MGN, Ahlman H, Gatenbeck S. 1986. Stability of solvent formation in Clostridium acetobutylicum during repeated subculturing. Appl. Microbiol. Biotechnol. 23:369–371 [Google Scholar]
- 10. Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452–464 [DOI] [PubMed] [Google Scholar]
- 11. Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78:79–85 [DOI] [PubMed] [Google Scholar]
- 12. Heap JT, Cartman ST, Kuehne SA, Cooksley C, Minton NP. 2010. ClosTron-targeted mutagenesis. Methods Mol. Biol. 646:165–182 [DOI] [PubMed] [Google Scholar]
- 13. Hu S, et al. 2011. Comparative genomic and transcriptomic analysis revealed genetic characteristics related to solvent formation and xylose utilization in Clostridium acetobutylicum EA 2018. BMC Genomics 12:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ji G, et al. 2005. Staphylococcus intermedius produces a functional agr autoinducing peptide containing a cyclic lactone. J. Bacteriol. 187:3139–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ji G, Beavis RC, Novick RP. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. U. S. A. 92:12055–12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones DT. 2001. Applied acetone-butanol fermentation, p 125–168 In Bahl H., Dürre P. (ed), Clostridia. Biotechnology and medical applications. Wiley-VCH Verlag GmbH, Weinheim, Germany [Google Scholar]
- 17. Jones DT, Woods DR. 1986. Acetone-butanol fermentation revisited. Microbiol. Rev. 50:484–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones SW, Tracy BP, Gaida SM, Papoutsakis ET. 2011. Inactivation of σF in Clostridium acetobutylicum ATCC 824 blocks sporulation prior to asymmetric division and abolishes σE and σG protein expression but does not block solvent formation. J. Bacteriol. 193:2429–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kashket ER, Cao Z-Y. 1995. Clostridial strain degeneration. FEMS Microbiol. Rev. 17:307–316 [Google Scholar]
- 20. Kosaka T, Nakayama S, Nakaya K, Yoshino S, Furukawa K. 2007. Characterization of the sol operon in butanol-hyperproducing Clostridium saccharoperbutylacetonicum strain N1-4 and its degeneration mechanism. Biosci. Biotechnol. Biochem. 71:58–68 [DOI] [PubMed] [Google Scholar]
- 21. Li J, Chen J, Vidal JE, McClane BA. 2011. The Agr-like quorum-sensing system regulates sporulation and production of enterotoxin and beta2 toxin by Clostridium perfringens type A non-food-borne human gastrointestinal disease strain F5603. Infect. Immun. 79:2451–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lütke-Eversloh T, Bahl H. 2011. Metabolic engineering of Clostridium acetobutylicum: recent advances to improve butanol production. Curr. Opin. Biotechnol. 22:634–647 [DOI] [PubMed] [Google Scholar]
- 23. Lyon GJ, Novick RP. 2004. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25:1389–1403 [DOI] [PubMed] [Google Scholar]
- 24. Meinecke B, Bahl H, Gottschalk G. 1984. Selection of an asporogenous strain of Clostridium acetobutylicum in continuous culture under phosphate limitation. Appl. Environ. Microbiol. 48:1064–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakayama J, et al. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145–154 [DOI] [PubMed] [Google Scholar]
- 26. Nakayama J, et al. 2006. Revised model for the Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal AgrD. J. Bacteriol. 188:8321–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nölling J, et al. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Novick RP, Geisinger E. 2008. Quorum-sensing in staphylococci. Annu. Rev. Genet. 42:541–564 [DOI] [PubMed] [Google Scholar]
- 29. O'Brien RW, Morris JG. 1971. Oxygen and the growth and metabolism of Clostridium acetobutylicum. J. Gen. Microbiol. 68:307–318 [DOI] [PubMed] [Google Scholar]
- 30. Ohtani K, et al. 2009. Virulence gene regulation by the agr system in Clostridium perfringens. J. Bacteriol. 191:3919–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paredes CJ, Rigoutsos I, Papoutsakis ET. 2004. Transcriptional organization of the Clostridium acetobutylicum genome. Nucleic Acids Res. 32:1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pottathil M, Lazazzera BA. 2003. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front. Biosci. 8:32–45 [DOI] [PubMed] [Google Scholar]
- 33. Queck SY, et al. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32:150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ravagnani A, et al. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol. Microbiol. 37:1172–1185 [DOI] [PubMed] [Google Scholar]
- 35. Reysenbach AL, Ravenscroft N, Long S, Jones DT, Woods DR. 1986. Characterization, biosynthesis, and regulation of granulose in Clostridium acetobutylicum. Appl. Environ. Microbiol. 52:185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robson RL, Robson RM, Morris JG. 1974. The biosynthesis of granulose by Clostridium pasteurianum. Biochem. J. 144:503–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 38. Steiner E, et al. 2011. Multiple orphan histidine kinases interact directly with Spo0A to control the initiation of endospore formation in Clostridium acetobutylicum. Mol. Microbiol. 80:641–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sturme MHJ, et al. 2002. Cell to cell communication by autoinducing peptides in Gram-positive bacteria. Antonie Van Leeuwenhoek 81:233–243 [DOI] [PubMed] [Google Scholar]
- 40. Sturme MHJ, et al. 2005. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 187:5224–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vidal JE, Chen J, Li J, McClane BA. 2009. Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13. PLoS One 4:e6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Williams P, Winzer K, Chan WC, Camara M. 2007. Look who's talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. B 362:1119–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wuster A, Babu MM. 2008. Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J. Bacteriol. 190:743–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.