Abstract
One hundred thirty-four blaCMY-2 plasmids from Salmonella and Escherichia coli strains from animals and food in Canada were characterized. Five plasmid groups were identified based on replicon type and restriction profiles. Three groups contained E. coli plasmids only. IncA/C plasmids included most multiresistant plasmids and all those of bovine origin.
TEXT
In North America, Salmonella enterica serovar Newport (8) and Salmonella enterica serovar Heidelberg (1, 7) carrying the plasmid-borne blaCMY-2 gene have been sources of concern in animal production and public health. However, these plasmids have also been found in isolates of other Salmonella enterica serovars (8) and Escherichia coli (2, 3). Previous studies in Canada have characterized blaCMY-2 plasmids in bacteria from human, cattle, and environmental sources (12–14, 16). The objective of this study was to characterize blaCMY-2 plasmids in S. enterica and E. coli isolates from a broader spectrum of food and food-producing animals in Canada.
Bacterial isolates were obtained by passive and active surveillance from poultry, cattle, swine, and related meat products. S. enterica isolates, collected between 1999 and 2007 by the World Organization for Animal Health (OIE) Reference Laboratory for Salmonellosis at the Laboratory for Food-borne Zoonoses and by the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) (10), served as a basis for this study. E. coli isolates obtained by CIPARS between 2003 and 2007 completed this collection (10).
Antimicrobial susceptibility testing was performed on OIE Reference Laboratory isolates (1999 to 2004) by agar dilution (18, 19) and on CIPARS isolates (2003 to 2007) by broth microdilution (6, 10). Isolates with a cefoxitin MIC of ≥16 mg/liter were eligible for inclusion. Sixty-nine cefoxitin-resistant Salmonella isolates were selected to include the broadest possible diversity of serovars and of sources. Forty-nine cefoxitin-resistant E. coli isolates were randomly selected. Eight pairs of cefoxitin-resistant S. enterica and E. coli isolates from the same chicken ceca were included.
blaCMY was detected by PCR (17, 21), and plasmid DNA (Qiagen plasmid kit; Qiagen, Hilden, Germany) was electroporated into E. coli ElectroMAX DH10B (Invitrogen, Carlsbad, CA). Transformants were selected on Mueller-Hinton agar (BD, Franklin Lakes, NJ) containing 8 mg/liter ceftiofur (Sigma-Aldrich, St. Louis, MO).
Transformants underwent PCR and antimicrobial susceptibility testing (described above), as well as replicon typing (5) and plasmid restriction analysis using BglII (New England BioLabs, Ipswich, MA). Restriction fragments of ≥3 kb were identified, and profiles were clustered using Dice similarity coefficients and the unweighted pair group method using arithmetic means (BioNumerics version 5.1; Applied Maths, Belgium).
Sixty-four restriction profiles were identified and grouped into five clusters based on replicon type (clusters A to E in Fig. 1). Sequencing of the blaCMY PCR product of one transformant of each profile confirmed that all were blaCMY-2.
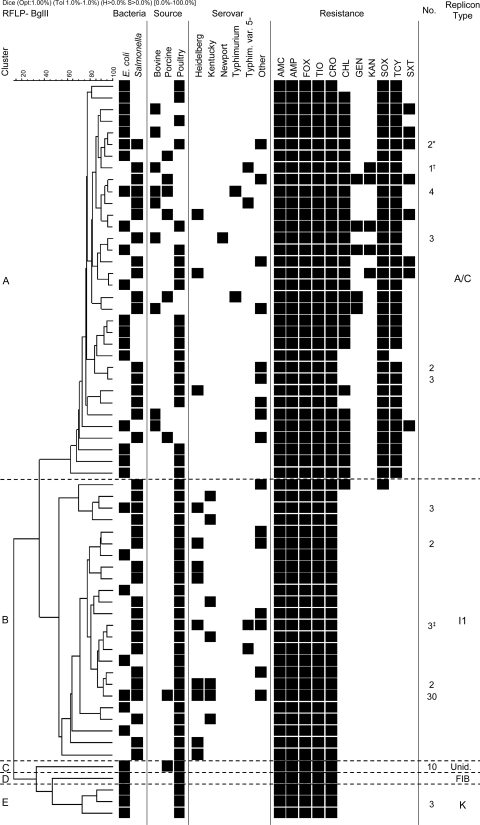
Fig 1.
Restriction fragment length polymorphism (RFLP) analysis of blaCMY-2 plasmids, associated characteristics, and source data. Salmonella enterica serovars other than those represented included Agona, Bredeney, Derby, Enteritidis, Infantis, Mbandaka, Reading, and Thompson and monophasic variants I:4,(5),12:r:−, I:4,12:i:−, I:6,8:eh:−, I:RoughO:r:1,2, and I:RoughO:fgs:−. Resistance to streptomycin was not assessed, as the recipient strain was resistant to streptomycin. AMC, amoxicillin-clavulanic acid; AMP, ampicillin; FOX, cefoxitin; TIO, ceftiofur; CRO, ceftriaxone; CHL, chloramphenicol; GEN, gentamicin; KAN, kanamycin; SOX, sulfisoxazole; TCY, tetracycline; SXT, trimethoprim-sulfamethoxazole. Numbers of isolates are indicated where more than one plasmid was identified with the same restriction profile. *, one of these plasmids conferred resistance to SOX but not to TCY and SXT; †, the replicon types of this plasmid were A/C and FIIs; ‡, the replicon type of one of three plasmids in this cluster was unidentified (Unid.).
Plasmids from clusters A and B (repA/C and repI1, respectively) were the most frequent and diverse. These clusters correspond to common groups of blaCMY-2 plasmids previously described in North America (4, 12, 13, 16, 20) and contain plasmids from S. enterica and E. coli, with four restriction profiles found in both species (Fig. 1). Plasmids from the three remaining clusters (repK, repFIB, and unidentified replicon type) were found in E. coli only. Other Canadian studies have identified E. coli isolates from water that contain plasmids from all five clusters (12, 13) and isolates from hospitalized patients that contain plasmids from clusters A, B, C, and E (2). Thus, blaCMY-2 plasmids from these five clusters have disseminated in E. coli from a variety of sources. However, those from clusters C, D, and E may not have spread extensively in S. enterica. No clear association between plasmid types or clusters and time of isolation was visible.
blaCMY-2 plasmids from seven pairs of S. enterica and E. coli isolates originating from the same samples belonged to different replicon types. One pair contained identical repI1 plasmids, and the restriction profiles of this pair of plasmids were indistinguishable and identical to those of 28 other S. enterica and E. coli plasmids (Fig. 1), suggesting the coincident occurrence of two isolates independently acquiring the same plasmid rather than plasmid transfer in vivo.
Plasmids belonging to all five clusters were identified in bacteria from poultry (primarily chickens) and poultry meat. Plasmids encoding resistance to β-lactams only were overwhelmingly found in poultry and poultry products, thus suggesting a possible association with the use of this class of antimicrobial agents (including ceftiofur) in poultry (7). This contrasts with repA/C plasmids, which were found in multiple isolates from all sources investigated. This probably reflects the potential of these multiresistance plasmids to be coselected by the use of antimicrobial agents other than β-lactams. Plasmids in bacteria from cattle and beef were all repA/C (Fig. 1). Together, these data suggest different dynamics and perhaps different selective and coselective pressures for blaCMY-2 in the major animal commodities. Resistance to extended-spectrum cephalosporins (ESCs) is more frequent in poultry than in cattle and swine in Canada (11), and it appears that blaCMY-2 plasmids are also more diverse in poultry than in other animals.
Plasmids from repA/C and repI1 were found in 13 and 10 Salmonella serovars, respectively, and both were found in S. Heidelberg, S. enterica serovar Infantis, and S. enterica serovar Typhimurium variant 5− (Fig. 1). S. enterica serovar Newport is less frequent in Canadian farm animals than in the United States (11, 20), but as expected, blaCMY-2 plasmids from this serovar were all repA/C multiresistance plasmids. The majority of blaCMY-2 plasmids from S. Heidelberg were repI1 plasmids, as was found previously (1). However, repA/C plasmids were also found in this serovar, confirming the repeated acquisition of blaCMY-2 plasmids by S. Heidelberg from poultry (7). In contrast, chicken-associated S. enterica serovar Kentucky contained only repI1 plasmids, possibly reflecting the more recent appearance of ESC resistance in this serovar (9, 11).
As observed by others (4, 13, 14), resistance to agents other than β-lactams was observed in repA/C plasmids only, with one exception (Fig. 1). The most frequent additional resistances were to tetracycline, sulfonamides, and chloramphenicol. One plasmid from a porcine Salmonella strain encoded additional resistance to trimethoprim, kanamycin, and gentamicin. As in other studies (4, 15), chloramphenicol resistance was encoded by the floR gene (data not shown), thus confirming the potential for selection of blaCMY-2 multiresistance plasmids by the use of florfenicol in the animal industry (15).
In conclusion, a large diversity of blaCMY-2 plasmids of five replicon types were identified in S. enterica and E. coli isolates from food animals and derived food products in Canada. These plasmids are similar to those found in bacteria from the environment and from human hospitals. This confirms that, despite a few exceptions, blaCMY-2 plasmids are promiscuous and have diffused across bacterial populations from a wide spectrum of ecological and epidemiological compartments. This should be a source of concern, since a broad and complex reservoir of ESC resistance determinants associated with mobile genetic elements has arisen and may be difficult to control.
ACKNOWLEDGMENTS
We thank the staff of the Salmonella Typing Laboratory and Antimicrobial Resistance Laboratories, the Laboratory for Food-borne Zoonoses, the Canadian Integrated Program for Antimicrobial Resistance Surveillance, and Elizabeth Hillyer for technical assistance.
Footnotes
Published ahead of print 9 December 2011
REFERENCES
- 1. Andrysiak AK, et al. 2008. Genetic characterization of clinical and agri-food isolates of multi drug resistant Salmonella enterica serovar Heidelberg from Canada. BMC Microbiol. 8:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baudry PJ, Mataseje L, Zhanel GG, Hoban DJ, Mulvey MR. 2009. Characterization of plasmids encoding CMY-2 AmpC beta-lactamases from Escherichia coli in Canadian intensive care units. Diagn. Microbiol. Infect. Dis. 65:379–383 [DOI] [PubMed] [Google Scholar]
- 3. Baudry PJ, et al. 2008. Comparison of antimicrobial resistance profiles among extended-spectrum-beta-lactamase-producing and acquired AmpC beta-lactamase-producing Escherichia coli isolates from Canadian intensive care units. Antimicrob. Agents Chemother. 52:1846–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Call DR, et al. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54:590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 6. Centers for Disease Control and Prevention 2009. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2007. US Department of Health and Human Services, CDC, Atlanta, GA [Google Scholar]
- 7. Dutil L, et al. 2010. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 16:48–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frye JG, Fedorka-Cray PJ. 2007. Prevalence, distribution and characterisation of ceftiofur resistance in Salmonella enterica isolated from animals in the U. S. A. from 1999 to 2003. Int. J. Antimicrob. Agents 30:134–142 [DOI] [PubMed] [Google Scholar]
- 9. Government of Canada 2006. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2004. Public Health Agency of Canada, Guelph, ON, Canada [Google Scholar]
- 10. Government of Canada 2009. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2006. Public Health Agency of Canada, Guelph, ON, Canada [Google Scholar]
- 11. Government of Canada 2010. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2007. Public Health Agency of Canada, Guelph, ON, Canada [Google Scholar]
- 12. Mataseje LF, et al. 2010. Comparison of CMY-2 plasmids isolated from human, animal, and environmental Escherichia coli and Salmonella spp. from Canada. Diagn. Microbiol. Infect. Dis. 67:387–391 [DOI] [PubMed] [Google Scholar]
- 13. Mataseje LF, et al. 2009. Characterization of cefoxitin-resistant Escherichia coli isolates from recreational beaches and private drinking water in Canada between 2004 and 2006. Antimicrob. Agents Chemother. 53:3126–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mataseje LF, et al. 2009. Characterization of Canadian cefoxitin-resistant non-typhoidal Salmonella isolates, 2005-06. J. Antimicrob. Chemother. 64:723–730 [DOI] [PubMed] [Google Scholar]
- 15. Meunier D, et al. 2010. Plasmid-borne florfenicol and ceftiofur resistance encoded by the floR and bla CMY-2 genes in Escherichia coli isolates from diseased cattle in France. J. Med. Microbiol. 59:467–471 [DOI] [PubMed] [Google Scholar]
- 16. Mulvey MR, Susky E, McCracken M, Morck DW, Read RR. 2009. Similar cefoxitin-resistance plasmids circulating in Escherichia coli from human and animal sources. Vet. Microbiol. 134:279–287 [DOI] [PubMed] [Google Scholar]
- 17. M'Zali FH, et al. 1997. Transcontinental importation into the UK of Escherichia coli expressing a plasmid-mediated AmpC-type beta-lactamase exposed during an outbreak of SHV-5 extended-spectrum beta-lactamase in a Leeds hospital. J. Antimicrob. Chemother. 40:823–831 [DOI] [PubMed] [Google Scholar]
- 18. National Committee for Clinical Laboratory Standards 1999. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A National Committee for Clinical Laboratory Standards, Villanova, PA [Google Scholar]
- 19. National Committee for Clinical Laboratory Standards 2002. Performance standards for antimicrobial susceptibility testing; eighth informational supplement. NCCLS document M100-S12 National Committee for Clinical Laboratory Standards, Villanova, PA [Google Scholar]
- 20. Poole TL, et al. 2009. Conjugative transferability of the A/C plasmids from Salmonella enterica isolates that possess or lack blaCMY in the A/C plasmid backbone. Foodborne Pathog. Dis. 6:1185–1194 [DOI] [PubMed] [Google Scholar]
- 21. Winokur PL, Vonstein DL, Hoffman LJ, Uhlenhopp EK, Doern GV. 2001. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]