Abstract
Karstic caves represent one of the most important subterranean carbon storages on Earth and provide windows into the subsurface. The recent discovery of the Herrenberg Cave, Germany, gave us the opportunity to investigate the diversity and potential role of bacteria in carbonate mineral formation. Calcite was the only mineral observed by Raman spectroscopy to precipitate as stalactites from seepage water. Bacterial cells were found on the surface and interior of stalactites by confocal laser scanning microscopy. Proteobacteria dominated the microbial communities inhabiting stalactites, representing more than 70% of total 16S rRNA gene clones. Proteobacteria formed 22 to 34% of the detected communities in fluvial sediments, and a large fraction of these bacteria were also metabolically active. A total of 9 isolates, belonging to the genera Arthrobacter, Flavobacterium, Pseudomonas, Rhodococcus, Serratia, and Stenotrophomonas, grew on alkaline carbonate-precipitating medium. Two cultures with the most intense precipitate formation, Arthrobacter sulfonivorans and Rhodococcus globerulus, grew as aggregates, produced extracellular polymeric substances (EPS), and formed mixtures of calcite, vaterite, and monohydrocalcite. R. globerulus formed idiomorphous crystals with rhombohedral morphology, whereas A. sulfonivorans formed xenomorphous globular crystals, evidence for taxon-specific crystal morphologies. The results of this study highlighted the importance of combining various techniques in order to understand the geomicrobiology of karstic caves, but further studies are needed to determine whether the mineralogical biosignatures found in nutrient-rich media can also be found in oligotrophic caves.
INTRODUCTION
Recent interest in the role of microbial processes in biogeochemical cycles and the largely unexplored subsurface diversity has spurred research on the geomicrobiology of deep marine and terrestrial environments. Calculations indicate that the total amount of carbon in intraterrestrial organisms may equal that of all terrestrial and marine plants (55). An important part of this biomass is within subsurface microbial ecosystems deep inside the earth (6). Although, in the last 20 years, numerous studies have started to investigate microbial biodiversity in a wide range of habitats, including pristine and contaminated groundwater, marine subsurface habitats, sedimentary and magmatic terrestrial environments, and various caves (31, 37, 52, 70, 72), the physiological and biochemical features of these communities are still awaiting exploration (55).
Caves provide a window into the subsurface and are a prime habitat for investigating subsurface microbial life (1). The majority of previous cave research focused on cave systems where chemical energy fuels microbial communities, such as the ferromanganese deposits of the Lechuguilla Cave in New Mexico (21, 53), the sulfidic Frasassi cave system in Italy or the Movile Cave in Romania (18, 47), the nitrate/nitrite-dominated Nullarbor Cave in Australia (40), or a number of caves receiving allochthonous organic matter input (34). Karstic areas are of specific interest because these carbonate-dominated habitats represent one of the most important natural subterranean carbon reservoirs on Earth (22). Caves provide easy access to karstic environments. However, if caves are open to the public, it can be difficult to differentiate between the pristine indigenous microbial communities and those introduced into the cave by visiting humans or animals. To date, only a limited number of studies have examined the diversity and activity of microorganisms colonizing karstic habitats (56, 60, 69). Other studies focused on microbial isolates obtained from caves, which are capable of carbonate mineral formation (for example, see references 13, 15, 24, 35, and 63).
Our multidisciplinary study took advantage of the recently discovered Herrenberg Cave, which was discovered in the Thuringian forest in Germany during the excavation of a new rail tunnel. This karstic cave was not affected by human activity before 2008 and appeared to have no contact with migrating higher animals, e.g., bats. Thus, it provided a unique pristine habitat for elucidating the active microbial community in a karstic system. Our goal was to achieve a first glimpse into the geomicrobiology of this pristine cave by combining a broad variety of techniques, including phylogenetic analyses, microscopic techniques, and cultivation-based methods. We had the opportunity to sample stalactite material and fluvial sediments some hours before the cave was permanently closed, and construction work proceeded. With this pristine material, we aimed to (i) elucidate the hidden bacterial diversity and activity, (ii) evaluate the potential of bacterial isolates for carbonate mineralization, and (iii) to study in detail different phases of mineral formation with scanning electron microscopy and energy-dispersive X-ray spectroscopy (SEM-EDX) and confocal laser scanning microscopy (CLSM).
MATERIALS AND METHODS
Study site and sampling.
Herrenberg Cave (situated between 40 and 90 m below the ground) is located in Thuringia, Germany (R-value 4430286, H-value 5588500, DHDN/Gauss-Kruger zone 4, 11°01′007,5′′E, 50°25′41,6′′N) at the southern foreland of the Thuringian Forest. It was formed during Tertiary and Quarternary times in Middle Triassic limestones and marls of the Lower Muschelkalk formation (mu), i.e., in the main aquifer (Fig. 1). The cave is between 8 to 0.5 m wide and up to 20 m high. Parts of the cave lie under the regional groundwater level characterized by 1.2 to 2.7 mg liter−1 dissolved organic carbon (DOC), which flows mainly north to south. Weakly mineralized water (30 to 100 mg liter−1 total dissolved salts [TDS] of the Ca-Mg-SO4 type) from quartzite-rich shales enters the limestones of the main aquifer, resulting in intensive solution of calcite and strong mineralization of groundwater [up to 600 mg liter−1 TDS of the Ca-(Mg)-HCO3 type]. A small creek flows from north to southwest into a cave lake near the human-made entrance. The average nitrate concentrations were 19.5 mg liter−1 in groundwater and 13.7 mg liter−1 in seepage water collected from stalactites. The ammonium concentration was 0.01 to 0.02 mg liter−1 or below the detection limit. Water and air temperatures in the cave varied between 7 and 9°C.
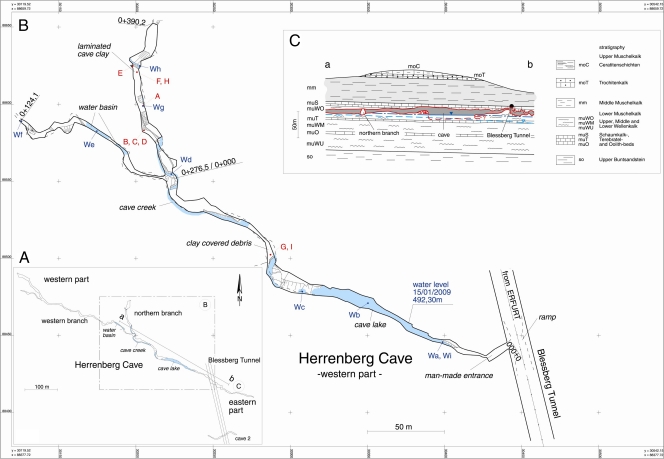
Fig 1.
Herrenberg Cave. (A) Map of the Herrenberg Cave located in southwest Thuringia, Germany, with profile line a-b. (B) Sample points in the western part of the cave (samples from sites A to I are solid samples, while Wa to Wi are water samples). A, stalactite; B, brown straw stalactite; C, white straw stalactite; D, eccentric straw stalactite; E, cave wall material; F, entrance sediment material; G, H, and I, fluvial sediments. (C) Geological profile of the area. (Courtesy of the Thuringian State Institute of Environment and Geology, Thuringia, Germany.)
Samples from different sites of the cave were obtained from the cave in January 2009 using aseptic techniques (Fig. 1) stalactite (site A samples) and straw stalactites (site B samples, brown straw stalactite; site C samples, white straw stalactite; site D samples, excentric straw stalactite), black-colored clay wall material (site E samples), mud at the human-made entrance of the cave (site G samples), and fluvial sediments (site F, H, and I samples). All samples were stored at 4°C until return to the laboratory.
Determination of nitrate, ammonium, total metal concentrations, and C/N analysis.
Water samples were filtered through glass microfiber filters (Whatman) and were analyzed for NO3− (12) and NH4− (10) concentrations. Sediment samples for total Al, Mg, Ca, Fe, Mn, and Na analyses were dried at 100°C, powdered, completely extracted with aqua regia (38), and then analyzed with an inductively coupled plasma mass spectrometer (ICP-MS) (PQ3S; Thermo Electron, United Kingdom). Sediment samples were ground to <100 μm with a ball mill and analyzed for total carbon and nitrogen by dry combustion with a CN analyzer (Vario Max; Elementar Analysensysteme GmbH, Germany). Inorganic carbon was determined by measuring the total amount of carbon after removal of organic carbon (OC) by ignition of samples for 4 h at 550°C (36). OC concentrations were then calculated from the difference between total and inorganic carbon concentrations.
XRD.
Powder X-ray diffraction (XRD) patterns were obtained with a Seifert-FPM XRD 7, equipped with a graphite monochromator, using Cu Kα radiation at 40 kV and 30 mA. Step scanning was done from 10° to 52° 2ϴ with increments of 0.02° 2ϴ and a counting time of 10 s per step. Sediment samples 6 and 9 were mounted on glass slides, while precipitates from culture experiments were prepared on Si slides. For clay mineral analysis, the clay fractions of samples 6 and 9 were separated by sedimentation and analyzed as (i) oriented specimens on ceramic tiles, (ii) saturated with glycol, and (iii) after heating to 550°C.
Infrared and Raman spectroscopy.
Raman spectra were recorded with a Fourier transform (FT)-Raman spectrometer (Bruker MultiRAM) in microscopic and macroscopic mode with a spectral resolution of 2 cm−1. The instrument was equipped with a Nd:YAG laser (excitation wavelength [λexc] of 1,064 nm) as the excitation source and a liquid nitrogen-cooled germanium detector. FT-infrared (IR) spectra were measured using a Bruker IFS66 spectrometer equipped with a doped triglycerin sulfate (DTGS) detector and 4-cm−1 spectral resolution.
Bacterial community analysis. (i) DNA and RNA extraction, cDNA synthesis, and PCR amplification.
Genomic DNA and RNA were isolated and purified from environmental samples using the RNA PowerSoil kit combined with the RNA PowerSoil DNA elution accessory kit (Mo Bio Laboratories, Carlsbad, CA) according to the manufacturer's instructions except for the stalactite sample. In this case, a piece of stalactite (∼0.5-cm2 surface area) was mixed with the bead solution from the kit's bead tube and vortexed to remove surface cells. The liquid was then returned to the kit's bead tube to proceed with the manufacturer's protocol. RNase-free DNase I (Fermentas) was used for DNase treatment of isolated RNA. cDNA was synthesized using the RevertAid first-strand cDNA synthesis kit (Fermentas) according to the manufacturer's instructions. PCR amplification of 16S rRNA gene fragments from isolated genomic DNA or cDNA was performed using the universal eubacterial forward fd1 and reverse rp2 primers (51) in a Peqlab advanced Primus 96 thermocycler. The PCR mixtures contained 1 U Taq DNA polymerase in the manufacturer's buffer (Jena Bioscience, Jena, Germany), 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates (dNTPs), and 0.3 μM (each) primer. Samples were first denatured at 95°C for 5 min, followed by 30 cycles, with 1 cycle consisting of denaturation at 95°C for 30 s, annealing at 55°C for 45 s, and extension at 72°C for 1.5 min, followed by a final extension step at 72°C for 10 min. PCR products were purified using the NucleoSpin extract II kit (Macherey-Nagel, Germany) according to the manufacturer's instructions.
(ii) Construction and analysis of clone libraries.
Purified 16S rRNA amplicons were cloned into the pCR4-TOPO vector using the TOPO TA cloning kit for Sequencing with One Shot TOP10 chemically competent Escherichia coli cells according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Ligation reactions originating from the stalactite DNA (site A samples) and sediment RNA (site F samples) were shipped to the Genome Center at Washington University (St. Louis, MO) for transformation and bidirectional sequencing with vector-specific primers (T3/T7). The rest of the clone libraries were constructed and screened in the laboratory. Plasmids were extracted by boiling a loopful of bacterial cells in 50 μl water for 10 min and pelleting the debris by centrifugation (2 min at 15,000 × g). The supernatant was transferred into fresh tubes. The inserts were amplified with vector-specific M13f/M13r primers (73). 16S rRNA gene fragments were sequenced by Macrogen (Seoul, South Korea) and AGOWA (Berlin, Germany). Clones were grouped with amplified ribosomal DNA restriction analysis (ARDRA) using the enzymes BsuRI and MspI (Fermentas) as described previously (50).
(iii) Phylogenetic analysis.
Sequences were assembled, and vector sequences flanking the 16S rRNA gene inserts were removed using Geneious Pro version 4.6.4 (Biomatters, Auckland, New Zealand). Operational taxonomic units (OTUs) were determined with 0.15 distance cutoff using the Aligner and Complete Linkage Clustering Tool of the Ribosomal Database Project (RDP) (http://pyro.cme.msu.edu). All sequences were checked for possible chimeric artifacts with the Chimera Check Tool from RDP (19). Percent coverage was calculated according to standard equations (5), and rarefaction analysis was performed using Analytic Rarefaction 1.3 (39). The Shannon diversity index was calculated using the EstimateS software program (http://viceroy.eeb.uconn.edu). Sequence similarity was determined using the Basic Local Alignment Search Tool (BLAST) algorithm against the GenBank database available from the National Center for Biotechnology Information (2).
(iv) Quantitative PCR for quantification of bacterial 16S rRNA genes.
The copy numbers of bacterial 16S rRNA genes were determined in triplicate for each sediment sample (site F to I samples) and stalactite sample (site A samples) by quantitative PCR (qPCR) using the 331F-797R primers/bactprobe combination (46) and Maxima Probe qPCR MasterMix (Fermentas) on an Mx3000P instrument (Stratagene). Cycling conditions were 95°C for 10 min and 40 cycles, with 1 cycle consisting of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s (46). Standard curves were prepared using serial dilutions of mixtures of plasmids containing 16S rRNA genes originating from clones FC36, FC63, FC13, FC29, OMA08, FC75, FC6, FC83, OME09, and FC11, representing the most frequently observed phyla. Standard curves were linear from 4.7 × 108 to 4.7 ×103 16S rRNA gene copies with an efficiency of 104.7%.
Cultivation and identification of isolates.
Serial dilutions from sediments were spread on B4 medium containing 2.5 g Ca acetate, 10 g glucose, 4 g yeast extract, 15 g agar liter−1 (pH 8.0) (9) and incubated at room temperature or 30°C for 4 weeks in the dark to allow for growth of heterotrophic bacteria. Bacteria were isolated on B4 medium, and isolates were periodically examined by light microscopy to determine the presence of crystals. For microscopic analyses, isolates were transferred to liquid B4 medium and modified B4 (B4MLY) (2.5 g Ca acetate, 0.5 g glucose, and 0.5 g yeast extract liter−1 [pH 8.0]) medium and incubated at room temperature and at 30°C for 4 weeks. Genomic DNA from pure cultures was isolated by boiling a loopful of bacterial biomass for 10 min in 100 μl of 5% CHELEX 100 solution (Bio-Rad). After 2 min of centrifugation at 15,000 × g (5415C table centrifuge; Eppendorf, Germany), the supernatant was used as a template for PCR amplification. 16S rRNA genes were amplified, sequenced, and analyzed as described above.
CLSM.
Upon return to the laboratory, stalactite samples were fixed in 4% formaldehyde in 1× phosphate-buffered saline (PBS) prior to confocal laser scanning microscopy (CLSM). Liquid cultures in B4 medium of the CaCO3-precipitating bacterial isolates, Arthrobacter sulfonivorans (SCM3) and Rhodococcus globerulus (SCM4), were examined directly without fixation. Samples were stained directly with SYTOX Green (nucleic acid-specific dye), SYPRO Orange (protein staining dye), and Ca-green (free Ca2+-ion-specific dye) (Molecular Probes Inc., Eugene, OR). Staining was carried out on fully hydrated aggregates and directly on the surface of the stalactite sample according to the manufacturer's instructions.
Cell aggregates and the stalactite sample were analyzed by CLSM using a TCS SP5X, controlled by the LAS 2.1.0 software program (Leica, Heidelberg, Germany), equipped with an upright microscope and a super continuum light source. Images were collected with a 63× water immersion lens with a numerical aperture (NA) of 1.2 and a 63× water immersible lens with an NA of 0.9. Scanning was carried out by simple viewing from the top. The samples were examined selecting the excitation lines at 485 nm (reflection and SYPRO), 504 nm (reflection and SYTOX), and 506 nm (reflection and Ca-green). Fluorescence emission signals were recorded from 500 to 650 nm (proteins) for cell aggregates, 516 to 574 nm (nucleic acids), and 516 to 560 nm (free Ca2+ ions) for the stalactite sample. In addition, the reflection signals were recorded at the point of excitation. Optical sections of aggregates were recorded in a single scan, and stalactites were recorded at 0.5-μm step size. Images were visualized by using the microscope software (Leica) for maximum-intensity projections and Imaris, version 7.1.0 (Bitplane, Zürich, Switzerland), for XYZ projections. Image data of microbial isolates were subjected to deconvolution using Huygens version 3.6.0 (SVI, The Netherlands) calculated with the CMLE algorithm.
Scanning electron microscopy (SEM) and energy-dispersive X-ray spectroscopy (EDX).
A droplet of bacterial suspension from liquid cultures was prepared for SEM by air drying on an electrically conductive, adhesive tag (Leit-Tab; Plano GmbH, Wetzlar, Germany). In order to prevent loss of inorganic material, critical point drying was not performed. The samples were then sputter coated with platinum (thickness of approximately 8 nm) using a SCD005 sputter coater (BAL-TEC, Liechtenstein) to avoid surface charging. Finally, the specimens were investigated with a field emission (FE) SEM LEO-1530 Gemini (Carl Zeiss NTS GmbH, Oberkochen, Germany) at an electron energy of 8 keV using the secondary electron detector. For material contrast, the built-in semiconductor-based back-scattered electron detector was used. For correlation with topography, a secondary electron image was measured simultaneously. The samples used for material contrast SEM were used for EDX, as well. Measurements were carried out using a LEO-1450 instrument (Carl Zeiss NTS GmbH, Oberkocheņ Germany) equipped with an EDX system Quantax 200 with an XFlash 5030 detector (Bruker AXS, Berlin, Germany).
Nucleotide sequence accession numbers.
The sequences of representative clones were deposited in the GenBank database under accession numbers FR734304 to FR734402. The GenBank accession numbers for strains SCM4 (Rhodococcus globerulus) and SCM3 (Arthrobacter sulfonivorans) are FR669673 and FR669674, respectively.
RESULTS
Geochemical characteristics and mineralogy of the Herrenberg Cave.
In January 2009, we investigated the 800-m-long western branch of the Herrenberg Cave (Fig. 1). Some parts of the walls, roof, and cave floor were covered with magnificent speleothems, e.g., stalagmites and stalactites (site A to D samples), including rare 2-m-long soda straw stalactites. Cave wall material (site E samples) and fluvial sedimentary deposits consisted of mostly mud, clay, and gravel (site F to I samples) and had pH values of 7.2 to 8.0. They contained small amounts of organic carbon (0.5%), nitrogen (0.06%), various amounts of total Al (63.6 to 114.9 mg g−1), Ca (10.8 to 40.5 mg g−1), Mg (15.8 to 28.6 mg g−1), and Fe (31 to 62 mg g−1). Raman microspectroscopic analyses showed a homogeneous distribution of calcite across the surface of the stalactite and straw stalactite samples. All sediment samples contained muscovite, while the content of quartz and carbonates (e.g., dolomite, calcite, and/or aragonite) was considerably higher in site E, G, and I samples, as shown by IR spectroscopy. Two sediments (site F and I samples) were further investigated using XRD, which showed that they contained quartz, dolomite, micaceous minerals, and clays. Clay minerals included illite, kaolinite, and small amounts of vermiculite or chlorite (diffractograms not shown). Additional calcite was found in site F samples.
Distribution, diversity, and activity of cave bacteria.
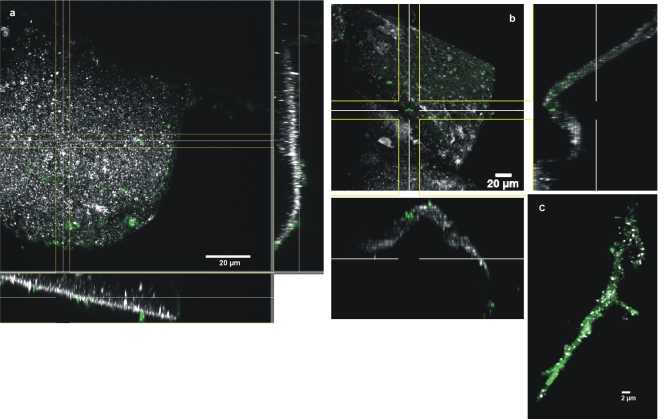
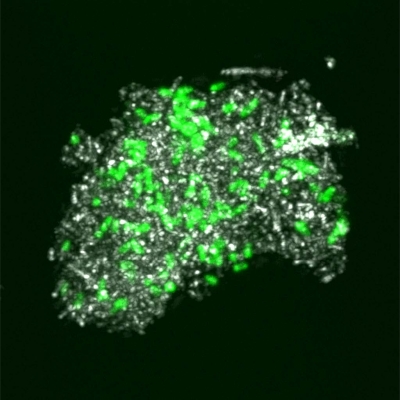
The copy numbers of bacterial 16S rRNA genes determined for fluvial sediments (site F to I samples) and one stalactite (site A samples) yielded bacterial cell counts of approximately 1.5 × 107, 2.5 × 104, 4.6 × 107, and 2.6 × 104 g−1 (wet weight) sediment for samples 6, 7, 8, and 9, respectively, and 8.5 × 102 cm−2 stalactite surface (site A samples). CLSM of a nucleic acid-stained stalactite sample provided information about the distribution and density of bacterial cells on its surface. On the outer surface, no biofilm formation was observed, with low cell densities observed on the stalactite surface (Fig. 2a). Although a delocalization of cells from the outer surface to the interior surface during sample transport prior to microscopic analysis is unlikely but cannot be completely ruled out, it is interesting to note that bacteria were also detected on the outer surface as well as on the interior surface of the stalactite (Fig. 2b). In addition, numerous bacteria were observed on the surface of a hypha-like structure originating from the stalactite sample (Fig. 2c). Hypha-like structures were also observed in other stalactite samples, confirming the presence of fungal members of the microbial community inhabiting this cave.
Fig 2.
Confocal laser scanning microscopy (CLSM) of a stalactite sample taken in the Herrenberg Cave. (a and b) XYZ projection showing the distribution of bacteria on the outer surface (a) and inner surface (b) (cut surface by sampling) of a stalactite surface after staining with SYTOX Green for nucleic acids. (c) CLSM image of a hypha-like structure originating from the Herrenberg Cave, colonized by bacterial cells after staining with SYTOX Green for nucleic acids. Nucleic acids are shown in green, while the reflection signal is shown in white.
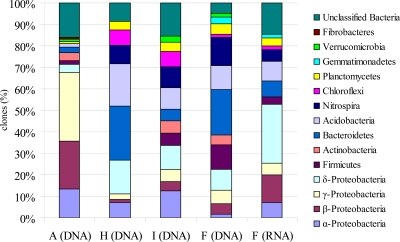
Microbial community characterization was performed on only the stalactite samples (site A samples) and the three sediment samples (site F, H, and I samples), as a limited amount of DNA was obtained from the straw stalactites. A total of 371 16S rRNA gene sequences were analyzed for the 4 samples. The 112 stalactite-derived clones grouped into 37 OTUs, and the library had 37.8% coverage. OTUs related to unclassified bacteria with unclarified phylogenetic affiliations comprised 16% of total clone sequences. The majority of clones (71%) were identified as members of the Proteobacteria (Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria subclasses) with a dominance of the Gammaproteobacteria subclass (including the genera Pseudomonas and Acinetobacter). A minor number of clone sequences were related to Gram-positive bacteria (Firmicutes and Actinobacteria), such as the Actinobacteria genus Arthrobacter. In addition, members of the phyla Bacteroidetes, Acidobacteria, Planctomycetes, Verrucomicrobia, and Fibrobacteres were shown to be present in the bacterial communities (Fig. 3; see Table S1 in the supplemental material).
Fig 3.
Frequencies of bacterial phylogenetic lineages detected in 16S rRNA gene and 16S rRNA clone libraries from stalactite and sediment samples taken in the Herrenberg Cave. Calculations were made based on the total number of clones associated with phylotypes of sequenced representatives. Sample A, stalactite; samples F, H, and I, fluvial sediments.
From the three fluvial sediments (site F, H, and I samples), 204 clones were analyzed, and the clones grouped into 35 (site F samples), 22 (site H samples), and 27 (site I samples) OTUs. Only 18 OTUs were detected in more than one of the three sediment samples. Coverage approximated 20.0, 63.6, and 77.8% for site F, H, and I samples, respectively. Bacterial community composition with respect to the majority of taxonomic groups represented in the sediment libraries overlapped at the phylum level (Fig. 3); however, the relative proportions of the different phyla in the three sediment libraries differed. A number of clone sequences (5 to 15%) were related to yet unclassified bacteria with unclarified phylogenetic affiliation. Other sequences grouped within the bacterial phyla Proteobacteria (Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria subclasses), Firmicutes and Actinobacteria, as well as Bacteroidetes, Acidobacteria, Nitrospira, Chloroflexi, Planctomycetes, Gemmatimonadetes, and Verrucomicrobia (Fig. 3). The majority of clones (>90%) shared high sequence similarity (95 to 99%) with as yet uncultured organisms, detected in subsurface environments (deep-sea sediments, caves, groundwater), soils, aquatic habitats (fresh- and seawater), as well as plant-associated environments or cold-adapted environments (arctic and tundra soil) (8, 17, 64, 66, 67, 68, 74) (see Table S1 in the supplemental material).
To identify the active fraction of cave bacterial communities, RNA was extracted and used for clone library construction. RNA extracts from stalactites were below the concentrations needed for cDNA amplification. A total of 55 sequences were analyzed for the RNA-derived clone library of sediment sample F (Fig. 3), and these sequences grouped into 30 OTUs with a sampling coverage of 40%. The Shannon index was 3.56. The 16S rRNA gene- and 16S rRNA sequence-based results showed great overlap on the phylum level (Fig. 3), pointing to the activity of most bacterial groups detected within the samples. DNA-based cloning showed that the Gammaproteobacteria and Deltaproteobacteria were the dominant proteobacterial groups, whereas RNA-based cloning showed that the Deltaproteobacteria subgroup was the most abundant within the active bacterial community. Some taxa, e.g., the phyla Acidobacteria and Nitrospira, exhibited lower abundance in the active community than in the DNA-based clone libraries. More than half of the metabolically active bacteria (53% of total RNA-derived clones) were related to members of the Alpha-, Beta-, Gamma-, and Deltaproteobacteria, which was similar to the observed predominance of these groups in the stalactite DNA-based library but contrasted with the proportions of these groups in the sediment DNA-based libraries (Fig. 3). Representatives of some phyla (e.g., Verrucomicrobia) were not detected as active members of the fluvial sediments (Fig. 3).
Isolation of carbonate-precipitating bacteria.
Plating from serial dilutions of sediment samples was followed by random isolation. A total of 32 bacterial strains were obtained on alkaline B4 carbonate-precipitating medium containing Ca acetate, glucose, and yeast extract. Because liquid cultures are more suitable for microscopic analyses, colonies were transferred to liquid B4 medium and a modified B4 medium (B4MLY), which contained reduced amounts of glucose and yeast extract. Nine isolates were able to grow in liquid medium and could be maintained under laboratory conditions. Isolates were identified as Pseudomonas putida (100% sequence similarity), Serratia plymuthica (99%), Flavobacterium hercynium (98%), Flavobacterium johnsoniae (2 isolates) (99%), Stenotrophomonas rhizophila (99%), Rhodococcus fascians (99%), Rhodococcus globerulus (100%), and Arthrobacter sulfonivorans (100%). The latter two isolates showed the most intense formation of precipitates in liquid media within 4 weeks of incubation at room temperature. Therefore, these two isolates were chosen for further studies and were cultivated in B4 and B4MLY media at 15°C, at room temperature, and at 30°C. B4MLY medium was used to investigate biomineralization under less-carbon-rich conditions. Biomineralization was also studied in the nutrient-rich B4 medium, because this was used in numerous previous studies investigating carbonate-precipitating cave isolates (13, 35, 63).
Phases of biomineral formation by Arthrobacter sulfonivorans strain SCM3 and Rhodococcus globerulus strain SCM4.
Crystalline material was detected by light microscopy in media inoculated with either A. sulfonivorans or R. globerulus after 20 days of incubation at room temperature or 30°C. Since only slow growth occurred at 15°C within 4 weeks of incubation, we focused on higher temperatures. No mineral formation was observed in any of the uninoculated and autoclaved controls. The pH decreased from pH 8.0 to a pH of 7.29 to 7.84 in the first 24 to 72 h of growth. At the end of the incubation, the pH increased up to 8.21.
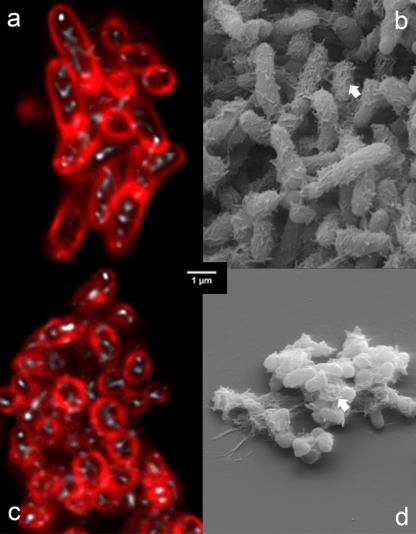
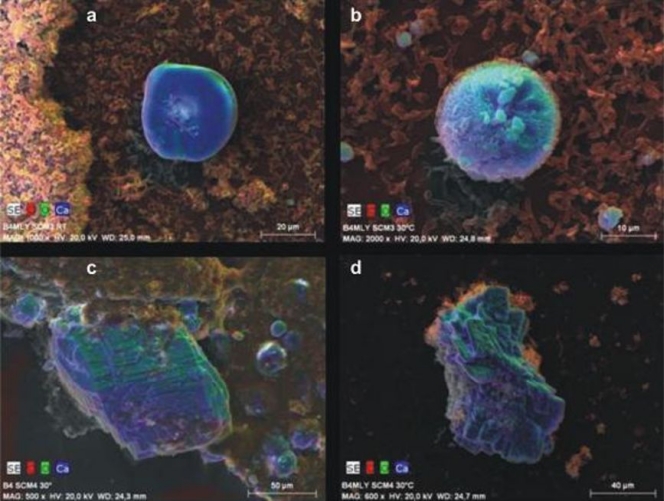
Bacteria grew predominantly as aggregates, as seen by SEM of precipitated material (Fig. 4b and d). This observation was confirmed for both isolates using the noninvasive CLSM technique (Fig. 4a and c), which permits analysis of fully hydrated aggregates without fixation and dehydration. Aggregates observed by CLSM showed cell surface protein staining and a cell internal reflection signal (Fig. 4a and c). Extracellular polymeric substances (EPS) were observed on SEM images of cell aggregates from both isolates (Fig. 4b to d). Ca-green staining showed the accumulation of free Ca2+ ions around/within the aggregates (Fig. 5). As the precipitated CaCO3 could not be visualized and investigated by laser scanning microscopy, SEM-EDX, XRD, and Raman spectroscopy were used to identify the precipitated materials. EDX verified that precipitates produced by both isolates were composed of Ca, C, and O, and the atomic percent ratios indicated the presence of CaCO3 (Fig. 6a to d; EDX spectra not shown). XRD and Raman analyses showed that both isolates produced calcite or a mixture of calcite and vaterite. In some samples, a single broad peak (corresponding to a d-spacing of 0.435 to 0.439 nm) pointed to the additional formation of monohydrocalcite. Differences were shown in two cases, both at 30°C. In B4MLY medium, the Arthrobacter isolate produced not only calcite, as seen for the Rhodococcus strain, but also vaterite. In addition, in B4 medium, the Rhodococcus strain produced not only calcite and vaterite but monohydrocalcite as well. Interestingly, the morphology of the crystals differed depending on the bacterial species. The crystalline material produced by A. sulfonivorans showed xenomorphous, spherical particles, whereas the precipitates of R. globerulus contained idiomorphous, often rhombohedral crystals. Spherical crystals were not detected in cultures of R. globerulus (Fig. 7a to d).
Fig 4.
(a and c) Confocal laser scanning microscopy of Arthrobacter sulfonivorans (a) and Rhodococcus globerulus (c) cell aggregates after deconvolution. The cells scanned once after they were stained with SYPRO Orange for proteins. Protein is shown in red, while the reflection signal is shown in white. Note the intracellular reflection signals. (b and d) Scanning electron microscopy of A. sulfonivorans (b) and R. globerulus (d) cell aggregates and EPS (extracellular polymeric substance) material (white arrows). Due to fixation and dehydration, bacterial cells appear smaller than in laser microscopy images. The scale bar shown in the center of the figure applies to all four panels.
Fig 5.
Confocal laser scanning microscopy (CLSM) of Arthrobacter sulfonivorans cell aggregates after staining with Ca-green for free Ca2+ ions. Free calcium ions are shown in green, while the reflection signal is shown in white.
Fig 6.
EDX mapping of bacterial calcite minerals of Arthrobacter sulfonivorans (SCM3) (a and b) and Rhodococcus globerulus (SCM4) (c and d). RT, room temperature; B4, B4 liquid medium (28); B4MLY, modified B4 medium. The SEM images show false colors of red, green, and blue for the local occurrence of the elements carbon, oxygen, and calcium, respectively. A gray level SEM image is underlying each. The shape of the blue-green carbonate minerals depend on the strain; it is round if precipitated in the presence of Arthrobacter sulfonivorans but idiomorphous when formed by Rhodococcus globerulus. Note the differences in magnification and scale bars.
Fig 7.
Comparison of topography-dependent secondary electron (SE) images to material-dependent back-scattered (BS) electron images of agglomerates produced by Arthrobacter sulfonivorans (SCM3) (a and b) and Rhodococcus globerulus (SCM4) (c and d). The darker gray color of the BS signal indicates lower atomic numbers, i.e., organic material. The lighter gray color indicates higher atomic numbers, i.e., inorganic material. Note the differences in magnification and scale bars. RT, room temperature; B4, B4 liquid medium; B4MLY, modified B4 medium.
DISCUSSION
Stalactite and sediment bacterial assemblages of the Herrenberg Cave.
Our geological and geochemical data showed that the Herrenberg Cave is a typical karstic habitat (29) including light-limited conditions for microbial life. Mineralogical investigations indicated that calcite precipitates from seepage water as stalactites and accumulates along with insoluble silicates in sediment deposits at the bottom of the cave. In contrast to other diversity studies on karstic caves or karst pools (27, 41, 69), this cave was not accessible to migrating animals (higher animals) or humans in the past. However, a glimpse into this pristine, karstic cave showed that the microbial communities present were not strikingly unique compared to other cave systems, suggesting that the microbial assemblages in karstic environments may be stable and resistant to minor or occasional human activities.
Within the bacterial communities, a high percentage of clone sequences detected were related to unclassified bacteria with no clear phylogenetic affiliation. This is similar to previous studies in karstic aquifers which could identify only one third of the 16S rRNA gene sequences lower than the domain Bacteria, indicating autochthonous bacterial communities (27). However, the detection of sequences in our clone libraries that were related to Bacteria derived from plant-associated environments, soils, and freshwater/seawater habitats suggested that aboveground and subsurface communities were related, likely due to connections between the two habitats via karstic water. This was seen in another study investigating shallow pools of a different karstic cave system, which found that microbial communities were related to organisms from groundwater or karst water habitats, freshwater, marine, or soil environments (69). The hypha-like structures observed in our samples indicated that fungi are members of the Herrenberg Cave microbial community, similar to what was found in the moonmilk deposits of Ballynamintra Cave in Ireland (62). These organisms can be involved in the transformation of various substrates and in biomineral formation (32, 33).
The stalactite bacterial community was dominated by the physiologically diverse Proteobacteria (44). Such dominance was also observed in other caves, e.g., the Lascaux Cave (4), the Tito Bustillo Cave (64), or the Altamira Cave (57, 59). Sequences related to those of Acidobacteria were detected in all clone libraries of the Herrenberg Cave, similar to the presence of Acidobacteria in the other caves, such as the Altamira Cave (Santillana del Mar, Spain) (65) and Wind Cave, North Dakota (17). The fact that there are only four cultivated species of this phylum limits our understanding of their potential metabolic function in caves. Some evidence exists that Acidobacteria are capable of methylotrophic growth (42), which may be an ecological advantage in an oligotrophic habitat with low inputs of organic matter. A number of cultivation-based investigations reported that diverse groups of Gram-positive bacteria inhabit cave environments (13, 26, 35, 45). However, the two Gram-positive phyla (Firmicutes and Actinobacteria) in our libraries were detected at a relatively low abundance. These contrasting results may arise from the differences in selectivity of the cultivation-based and nucleic acid-based approaches. It is known that a number of biases affect nucleic acid template amplification during PCR (43). PCR-based methods also rely on DNA extractions which can be biased due to differential lysis of cell wall structures that leads to distortions in detecting different bacterial groups in a given environmental sample (30, 49).
The pure cultures obtained from sediments were affiliated with representatives of various genera of the Gammaproteobacteria group, as well as the phyla Bacteroidetes and Actinobacteria. Bacteria from the genera Pseudomonas, Stenotrophomonas, Serratia, Flavobacterium, Arthrobacter, and Rhodococcus have also been isolated from other caves or karst environments, such as the Kartchner Caverns in Arizona in the United States or from karstic water samples (20, 41). Some of these genera were also detected in the stalactite clone library (see Table S1 in the supplemental material). The presence of Arthrobacter-related sequences in the stalactite clone library emphasizes the potential role of these bacteria in calcite mineralization, since members of the genus Arthrobacter are known to be capable of carbonate formation (14, 35).
Biomineral formation by A. sulfonivorans and R. globerulus.
The activity of our bacterial strains resulted in carbonate crystal formation. We observed only slow bacterial growth over 4 weeks of incubation at 15°C, suggesting that the rate of metabolic activity was affected. Nevertheless, long-term metabolic transformations may carry significant potential for carbonate precipitation processes and reflect slow transformations that occur in situ. The changes in pH observed during incubation are likely due to the consumption of carbon sources: Arthrobacter and Rhodococcus species are known to utilize acetate, which would result in an increase in the pH of the medium. The latter is an important factor supporting carbonate precipitation (16). Although members of the Arthrobacter and Rhodococcus genera are known to produce calcite and vaterite (14, 35), the A. sulfonivorans and R. globerulus species have not been reported previously to precipitate calcite. Furthermore, these bacteria might also be able to form other carbonates, e.g., dolomite.
The formation of vaterite seemed to be favored at higher temperatures, while monohydrocalcite formation was restricted to cultivation in B4 medium. These minerals were not detected in samples by XRD, suggesting that the low in situ temperature and small amount of available nutrients in the cave did not favor formation of these minerals. Although the different crystal structures of the newly formed minerals could not be explained completely by the experimental conditions, the cultivation conditions (61), e.g., available nutrients (58), may have influenced which type of CaCO3 mineral was formed. The stable forms of CaCO3, calcite, and aragonite are known to be the most common microbial carbonates (23). In vitro bacterial precipitation of the metastable vaterite form has also been reported (14, 63), suggesting that this biogenic mineral structure is more common than previously proposed. Bacteria have been implicated in creating secondary mineral formations, such as speleothems, in studies of caves in which a combination of cultivation and electron microscopic analyses using B4 medium were used (3, 13). Future analyses using portable Raman spectroscopy equipment should allow in situ mineralogical and microbiological investigations of caves similar to analyses in studies of prehistoric cave paintings (54). In comparison with other analytical techniques, Raman spectroscopy is a nondestructive method and requires extremely small amounts of sample material. The latter was an important aspect of our study due to the limited amount of samples available. Raman spectroscopy has been shown to be a powerful tool for mineralogical investigations of moonmilk deposits in karstic habitats (48).
Imaging of A. sulfonivorans and R. globerulus cultures using strategic microscopic techniques allowed us to visualize the early phases of carbonate mineralization. CLSM in particular showed us that after aggregation, free Ca2+ ions were entrapped and provided a surface for nucleation sites for crystal formation. In the later phases of mineral formation, species-dependent mineral morphologies were visualized with SEM-EDX. The production of EPS, cell aggregation, and Ca2+ aggregation are known to be important factors in the precipitation process (59, 71). The production of specific bacterial outer structures and their chemical nature might also be crucial for the bacterial crystallization process, influencing the mineralogy and morphology of calcium carbonate crystals (11). Calcite formation by two different Bacillus species yielded different mineral morphologies independent of the chemical nature of capsular polysaccharides and EPS produced (25). Another work (7) provided evidence of differences in the cell wall properties of Arthrobacter and Rhodococcus species. The cell walls of Arthrobacter spp. lacked mycolic acids in its type B peptidoglycan, whereas Rhodococcus globerulus and R. erythropolis strains produced mycolic acids (with 34 to 56 C atoms). The presence or absence of mycolic acids influences the cell wall surface properties resulting in a more hydrophilic (Arthrobacter) and relatively hydrophobic (Rhodococcus) cell surface. In addition, the amount of peptidoglycan and the degree of amidation of free carboxyl groups or other compounds (e.g., phosphate-containing teichoic acids as cell wall accessory polymers of Arthrobacter species) may also influence the cell surface charge (7, 28). Such differences may have caused the observed differences in calcium carbonate mineral morphology of the two isolates (Fig. 7).
The recent discovery of the Herrenberg Cave, with its extraordinary long straw stalactites, and its lack of human disturbance offered an opportunity to reveal the hidden microbial diversity of a karstic environment. The detection of soil-associated microorganisms in the Herrenberg Cave ecosystem provides evidence for a connection to surface environments. This suggests that we have to be cautious about what is considered a microbial contaminant when characterizing the microbiology of highly accessed cave systems. This detected link between surface and subsurface microbial communities has important implications to critical zone research which aims to understand how such linked communities impact terrestrial biogeochemical cycling (1). Further work should attempt to detect carbonate biosignatures in stalactites by means of noninvasive high spatial mineralogical investigations. Combined with the results of pure culture studies and microscopic analyses, the results will allow a better understanding of the geomicrobiology of the Earth's hidden belowground carbon storage.
Supplementary Material
ACKNOWLEDGMENTS
We thank Martina Herrmann and Susanne Grube for their technical support with qPCR analyses. We also thank Martina Hermann and Ulrich Bläß for helpful discussions.
This study is part of the research project AquaDiv@Jena funded by the ProExcellence Initiative of the federal state of Thuringia, Germany.
Footnotes
Published ahead of print 16 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Akob DM, Küsel K. 2011. Where microorganisms meet rocks in the Earth's Critical Zone. Biogeosciences 8:2523–2562 [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Banks ED, et al. 2010. Bacterial calcium carbonate precipitation in cave environments: a function of calcium homeostasis. Geomicrobiol. J. 27:444–454 [Google Scholar]
- 4. Bastian F, Alabouvette C, Saiz-Jimenez C. 2009. Bacteria and free-living amoeba in the Lascaux Cave. Res. Microbiol. 160:38–40 [DOI] [PubMed] [Google Scholar]
- 5. Begon M, Harper JL, Townsend CR. 1990. Ecology: individuals, populations and communities. Blackwell Scientific Publications, Oxford, England [Google Scholar]
- 6. Ben-Ari ET. 2002. Microbiology and geology: solid marriage made on Earth. ASM News 68:13–18 [Google Scholar]
- 7. Bendinger B, Rijnaarts HHMK, Altendorf Zehnder AJB. 1993. Physicochemical cell surface and adhesive properties of coryneform bacteria related to the presence and chain length of mycolic acids. Appl. Environ. Microbiol. 59:3973–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bottos EM, Vincent WFCW, Greer Whyte LG. 2008. Prokaryotic diversity of arctic ice shelf microbial mats. Environ. Microbiol. 10:950–966 [DOI] [PubMed] [Google Scholar]
- 9. Bouquet E, Boronat A, Ramos-Cormenzana A. 1973. Production of calcite (calcium carbonate) crystals by soil bacteria is a general phenomenon. Nature 246:527–529 [Google Scholar]
- 10. Bower CE, Holm-Hansen T. 1980. A salicylate-hypochlorite method for determining ammonia in seawater. Can. J. Fish. Aquat. Sci. 37:794–798 [Google Scholar]
- 11. Braissant O, Cailleau GC, Dupraz Verrecchia AP. 2003. Bacterially induced mineralization of calcium carbonate in terrestrial environments: the role of exopolysaccharides and amino acids. J. Sediment. Res. 73:485–490 [Google Scholar]
- 12. Braman RS, Hendrix SA. 1989. Nanogram nitrite and nitrate determination in environmental and biological materials by vananadium(III) reduction with chemiluminescence detection. Anal. Chem. 61:2715–2718 [DOI] [PubMed] [Google Scholar]
- 13. Cacchio P, Contento RC, Ercole G, Cappucio MP, Martinez Lepidi A. 2004. Involvement of microorganisms in the formation of carbonate speleothems in the Cervo Cave (Làquila-Italy). Geomicrobiol. J. 21:497–509 [Google Scholar]
- 14. Cacchio P, Ercole CG, Cappuccio Lepidi A. 2003. Calcium carbonate precipitation by bacterial strains isolated from limestone cave and from a loamy soil. Geomicrobiol. J. 20:85–98 [Google Scholar]
- 15. Canaveras JC, Sanchez-Moral SV, Soler Saiz-Jimenez C. 2001. Microorganisms and microbially induced fabrics in cave walls. Geomicrobiol. J. 18:223–240 [Google Scholar]
- 16. Castanier S, Le Metayer-Levrel G, Perthuisot JP. 1999. Ca-carbonates precipitation and limestone genesis — the microbiogeologist point of view. Sediment. Geol. 126:9–23 [Google Scholar]
- 17. Chelius MK, Moore JC. 2004. Molecular phylogenetic analysis of Archaea and Bacteria in Wind Cave, South Dakota. Geomicrobiol. J. 21:123–134 [Google Scholar]
- 18. Chen Y, et al. 2009. Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J. 3:1093–1104 [DOI] [PubMed] [Google Scholar]
- 19. Cole JR, et al. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cousin S, Brambilla E, Yang J, Stackebrandt E. 2008. Culturable aerobic bacteria from the upstream region of a karst water rivulet. Int. Microbiol. 11:91–100 [PubMed] [Google Scholar]
- 21. Cunningham KI, Northup DE, Pollastro RM, Wright WG, LaRock EJ. 1995. Bacteria, fungi and biokarst in Lechuguilla Cave, Carlsbad Caverns National Park, New Mexico. Environ. Geol. 25:2–8 [Google Scholar]
- 22. Ehrlich HL. 1998. Geomicrobiology: its significance for geology. Earth-Sci. Rev. 45:45–60 [Google Scholar]
- 23. Ehrlich HL. 2002. Geomicrobiology, 4th ed. Marcel Dekker, New York, NY [Google Scholar]
- 24. Engel AS, Stern LA, Bennett PC. 2004. Microbial contributions to cave formation: new insights into sulfuric acid speleogenesis. Geology 32:369–372 [Google Scholar]
- 25. Ercole C, Cacchio P, Botta AL, Centi V, Lepidi A. 2007. Bacterially induced mineralization of calcium carbonate: the role of exopolysaccharides and capsular polysaccharides. Microsc. Microanal. 13:42–50 [DOI] [PubMed] [Google Scholar]
- 26. Ercole C, Cacchio PG, Cappucio Lepidi A. 2001. Deposition of calcium carbonate in karst caves: role of bacteria in Stiffe's Cave. Int. J. Speleol. 30:69–79 [Google Scholar]
- 27. Farnleitner AH, et al. 2005. Bacterial dynamics in spring water of alpine karst aquifers indicates the presence of stable autochthonous microbial endokarst communities. Environ. Microbiol. 7:1248–1259 [DOI] [PubMed] [Google Scholar]
- 28. Fiedler F, Schäffer MJ. 1987. Teichoic acids in cell walls of strains of the “nicotianae” group of Arthrobacter: a chemotaxonomic marker. Syst. Appl. Microbiol. 9:16–21 [Google Scholar]
- 29. Ford DC, Williams PW. 2007. Karst hydrogeology and geomorphology. John Wiley and Sons, Chichester, England [Google Scholar]
- 30. Frostegård Å, et al. 1999. Quantification of bias related to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 65:5409–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fry JC, et al. 2007. Prokaryotic populations and activities in an interbedded coal deposit, including a previously deeply buried section (1.6–2.3 km) above ∼150 Ma basement rock. Geomicrobiol. J. 26:163–178 [Google Scholar]
- 32. Gadd GM. 2004. Mycotransformation of organic and inorganic substrates. Mycologist 18:60–70 [Google Scholar]
- 33. Gadd GM. 2007. Geomycology: biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation. Mycol. Res. 111:3–49 [DOI] [PubMed] [Google Scholar]
- 34. Groth I, Saiz-Jimenez C. 1999. Actinomycetes in hypogean environments. Geomicrobiol. J. 16:1–8 [Google Scholar]
- 35. Groth I, Schumann P, Laiz L, Sanchez-Moral S, Canaveras JC, Saiz-Jimenez C. 2001. Geomicrobiological study of the Grotta di Cervi, Porto Badisco, Italy. Geomicrobiol. J. 18:241–258 [Google Scholar]
- 36. Grüneberg E, Schöning I, Kalkoa EKV, Weisser WW. 2010. Regional organic carbon stock variability: a comparison between depth increments and soil horizons. Geoderma 155:426–433 [Google Scholar]
- 37. Hazen TC, Jimenez L, Devictoria GL. 1991. Comparison of bacteria from deep subsurface sediment and adjacent groundwater. Microbiol. Ecol. 22:293–304 [DOI] [PubMed] [Google Scholar]
- 38. Hoffman G. 1991. Methodenbuch. Die Untersuchung von Böden, vol. 1. VDLUFA-Verlag, Darmstadt, Germany [Google Scholar]
- 39. Holland SM. 2003. Analytical rarefaction 1.3. User's guide and application. UGA Stratigraphy Lab, University of Georgia, Athens, GA: http://www.uga.edu/strata/software/anRareReadme.html [Google Scholar]
- 40. Holmes AJ, et al. 2001. Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ. Microbiol. 3:256–264 [DOI] [PubMed] [Google Scholar]
- 41. Ikner LA, Toomey RSG, Nolan JW, Neilson BM, Pryor Maier RM. 2007. Culturable microbial diversity and the impact of tourism in Kartcher Caverns, Arizona. Microbiol. Ecol. 53:30–42 [DOI] [PubMed] [Google Scholar]
- 42. Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. 2008. Real-time detection of actively metabolizing microbes by redox sensing as applied to methylotroph populations in Lake Washington. ISME J. 2:696–706 [DOI] [PubMed] [Google Scholar]
- 43. Kanagawa T. 2003. Bias and artifacts in multitemplate Polymerase Chain Reactions (PCR). J. Biosci. Bioeng. 96:317–323 [DOI] [PubMed] [Google Scholar]
- 44. Kersters K, De Vos P, Gillis M, Swings J, Vandamme P, Stackebrandt E. 2006. Introduction to the Proteobacteria, p 3–37 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes. Springer Verlag, New York, NY [Google Scholar]
- 45. Laiz L, Groth I, Gonzalez I, Saiz-Jimenez C. 1999. Microbiological study of the dripping waters in Altamira Cave (Santillana del Mar, Spain). J. Microbiol. Methods 36:129–138 [DOI] [PubMed] [Google Scholar]
- 46. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics, John Wiley and Sons Ltd, London, England [Google Scholar]
- 47. Macalady JL, Jones DS, Lyon EH. 2007. Extremely acidic, pendulous cave wall biofilms from the Frasassi cave system, Italy. Environ. Microbiol. 9:1402–1414 [DOI] [PubMed] [Google Scholar]
- 48. Martínez-Arkarazo I, Angulo M, Zuloaga O, Usobiaga A, Madariaga JM. 2007. Spectroscopic characterisation of moonmilk deposits in Pozalagua tourist cave (Karrantza, Basque Country, North of Spain). Spectrochim. Acta A 68:1058–1064 [DOI] [PubMed] [Google Scholar]
- 49. Martin-Laurent F, et al. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Massol-Deya AA, Odelson DA, Hickey RF, Tiedje JM. 1995. Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal DNA sequences and restriction endonuclease analysis (ARDRA), p 3.3.2:1–3.3.2:8 In Akkermans ADL, van Elsas JD, deBruijn FJ. (ed), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 51. Nadkarni MA, Martin FE, Jacques NA, Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad range (universal) probe and primer set. Microbiology 148:257–266 [DOI] [PubMed] [Google Scholar]
- 52. Nealson KH, Inagaki F, Takai K. 2005. Hydrogen-driven subsurface lithoautotrophic microbial ecosystems (SLiMEs): do they exist and why should we care? Trends Microbiol. 13:405–410 [DOI] [PubMed] [Google Scholar]
- 53. Northup DE, et al. 2003. Diverse microbial communities inhabiting ferromanganese deposits in Lechuguilla and Spider Caves. Environ. Microbiol. 5:1071–1086 [DOI] [PubMed] [Google Scholar]
- 54. Ospitali F, Smith DC, Lorblanchet M. 2006. Preliminary investigations by Raman microscopy of prehistoric pigments in the wall-painted cave at Roucadour, Quercy, France. J. Raman Spectrosc. 37:1063–1071 [Google Scholar]
- 55. Pedersen K. 2000. Exploration of deep intraterrestrial microbial life: current perspectives. FEMS Microbiol. Lett. 185:9–16 [DOI] [PubMed] [Google Scholar]
- 56. Personné JC, Poty F, Mahler BJ, Drogue C. 2004. Colonization by aerobic bacteria in karst: laboratory and in situ experiments. Ground Water 63:3367–3373 [DOI] [PubMed] [Google Scholar]
- 57. Portillo MC, Gonzalez JM, Saiz-Jimenez C. 2008. Metabolically active microbial communities of yellow and gray colonizations on the walls of Altamira Cave, Spain. J. Appl. Microbiol. 104:681–691 [DOI] [PubMed] [Google Scholar]
- 58. Portillo MC, Porca E, Cuezva S, Canaveras JC, Sanchez-Moral S, Gonzalez JM. 2009. Is the availability of different nutrients a critical factor for the impact of bacteria on subterraneous carbon budgets? Naturwissenschaften 96:1035–1042 [DOI] [PubMed] [Google Scholar]
- 59. Portillo MC, Saiz-Jimenez C, Gonzalez JM. 2009. Molecular characterization of total and metabolically active bacterial communities of “white colonizations” in the Altamira Cave, Spain. Res. Microbiol. 160:41–47 [DOI] [PubMed] [Google Scholar]
- 60. Pronk M, Goldscheider N, Zopfi J. 2009. Microbial communities in karst groundwater and their potential use for biomonitoring. Hydrogeol. J. 17:37–48 [Google Scholar]
- 61. Rivadeneyra MA, Delgado G, Ramos-Cormenzana A, Delgado R. 1998. Biomineralization of carbonates by Halomonas eurihalina in solid and liquid media with different salinities: crystal formation sequence. Res. Microbiol. 149:277–287 [DOI] [PubMed] [Google Scholar]
- 62. Rooney DC, Hutchens E, Clipson N, Baldini J, McDermott F. 2010. Microbial community diversity of moonmilk deposits at Ballynamintra Cave, Co. Waterford, Ireland. Microb. Ecol. 60:753–761 [DOI] [PubMed] [Google Scholar]
- 63. Sanchez-Moral S, Canaveras JC, Laiz L, Saiz-Jimenez C, Bedoya J, Luque L. 2003. Biomediated precipitation of calcium carbonate metastable phases in hypogean environments: a short review. Geomicrobiol. J. 20:491–500 [Google Scholar]
- 64. Schabereiter-Gurtner C, Saiz-Jimenez C, Pinar G, Lubitz W, Rölleke S. 2002. Phylogenetic 16S rRNA analysis reveals the presence of complex and partly unknown bacterial communities in Tito Bustillo cave, Spain, and on its palaeolithic paintings. Environ. Microbiol. 4:392–400 [DOI] [PubMed] [Google Scholar]
- 65. Schabereiter-Gurtner C, Saiz-Jimenez C, Pinar G, Lubitz W, Rölleke S. 2002. Altamira cave paleolithic paintings harbour partly unknown bacterial communities. FEMS Microbiol. Lett. 211:7–11 [DOI] [PubMed] [Google Scholar]
- 66. Schauer R, Bienhold C, Ramette A, Harder J. 2010. Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J. 4:159–170 [DOI] [PubMed] [Google Scholar]
- 67. Schloss PD, Handelsman J. 2006. Toward a census of bacteria in soil. PLoS Comput. Biol. 2:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schmalenberger A, Tebbe CC. 2002. Bacterial community composition in the rhizosphere of a transgenic, herbicide-resistant maize (Zea mays) and comparison to its non-transgenic cultivar Bosphore. FEMS Microbiol. Ecol. 40:29–37 [DOI] [PubMed] [Google Scholar]
- 69. Shabarova T, Pernthaler J. 2010. Karst pools in subsurface environments: collectors of microbial diversity or temporary residence between habitat types. Environ. Microbiol. 12:1061–1074 [DOI] [PubMed] [Google Scholar]
- 70. Stevens TO, McKinley JP. 1995. Lithoautotrophic microbial ecosystems in deep basalt aquifers. Science 270:450–454 [Google Scholar]
- 71. Van Lith Y, Warthmann R, Vasconcelos C, Mckenzie JA. 2003. Sulphate-reducing bacteria induce low-temperature Ca-dolomite and high Mg-calcite formation. Geobiology 1:71–79 [Google Scholar]
- 72. van Waasbergen LG, Balkwill DL, Crocker FH, Bjornstad BN, Miller RV. 2000. Genetic diversity among Arthrobacter species collected across a heterogeneous series of terrestrial deep-subsurface sediments as determined on the basis of 16S rRNA and recA gene sequences. Appl. Environ. Microbiol. 66:3454–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhou J, Davey ME, Figueras JB, Rivkina E, Gilichinsky D, Tiedje JM. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil. Microbiology 143:3913–3919 [DOI] [PubMed] [Google Scholar]
- 74. Zinger L, Shahnavaz B, Baptist F, Geremia RA, Choler P. 2009. Microbial diversity in alpine tundra soils correlates with snow cover dynamics. ISME J. 3:850–859 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.