Abstract
Chlorinated solvents are among the most prevalent groundwater contaminants in the industrialized world. Biodegradation with Dehalococcoides-containing mixed cultures is an effective remediation technology. To elucidate transcribed genes in a Dehalococcoides-containing mixed culture, a shotgun metagenome microarray was created and used to investigate gene transcription during vinyl chloride (VC) dechlorination and during starvation (no chlorinated compounds) by a microbial enrichment culture called KB-1. In both treatment conditions, methanol was amended as an electron donor. Subsequently, spots were sequenced that contained the genes most differentially transcribed between the VC-degrading and methanol-only conditions, as well as spots with the highest intensities. Sequencing revealed that during VC degradation Dehalococcoides genes involved in transcription, translation, metabolic energy generation, and amino acid and lipid metabolism and transport were overrepresented in the transcripts compared to the average Dehalococcoides genome. KB-1 rdhA14 (vcrA) was the only reductive dehalogenase homologous (RDH) gene with higher transcript levels during VC degradation, while multiple RDH genes had higher transcript levels in the absence of VC. Numerous hypothetical genes from Dehalococcoides also had higher transcript levels in methanol-only treatments, indicating that many uncharacterized proteins are involved in cell maintenance in the absence of chlorinated substrates. In addition, microarray results prompted biological experiments confirming that electron acceptor limiting conditions activated a Dehalococcoides prophage. Transcripts from Spirochaetes, Chloroflexi, Geobacter, and methanogens demonstrate the importance of non-Dehalococcoides organisms to the culture, and sequencing of identified shotgun clones of interest provided information for follow-on targeted studies.
INTRODUCTION
Chlorinated solvents are among the most prevalent groundwater contaminants in industrialized countries, and their presence poses risks to human and environmental health. KB-1 is an anaerobic microbial community dominated by Dehalococcoides organisms that efficiently dechlorinates tetrachloroethene (PCE) and trichloroethene (TCE) to ethene (7, 8, 38). Dehalococcoides grows slowly in isolation and often has complex nutritional requirements, whereas Dehalococcoides growing in mixed communities grows and dechlorinates more rapidly, producing very robust cultures. This suggests beneficial relationships between nondechlorinating organisms and Dehalococcoides. It is therefore important to identify Dehalococcoides and non-Dehalococcoides genes within these communities that are expressed during dechlorination.
There have been a few published microarray studies involving Dehalococcoides. In the first, Johnson et al. (14) examined transcription in Dehalococcoides strain 195 along a timeline from exponential to stationary phase in an effort to understand factors that limit the growth of this slow-growing isolate. They also used the arrays to investigate transcriptional response to different concentrations and forms of corrinoid cofactors (14). Another study used comparative genomics to compare DNA from a Dehalococcoides-containing enrichment culture (ANAS) to arrayed oligonucleotides representing the strain 195 genome (39). Genes that were not detected by array in ANAS were predominantly located in insertion elements (IEs), with 88% of the genes located in IEs in strain 195 not detected in the ANAS culture. In addition 13 of the 19 strain 195 reductive dehalogenase homologous (RDH) genes were not detected. These results, in combination with the importance of genomic islands for conferring strain-specific genes (20), highlight the shortcomings of using an array designed for a specific Dehalococcoides strain to analyze DNA from a different Dehalococcoides strain. Recent studies utilizing pangenus arrays demonstrated robust coverage of Dehalococcoides genomes (12, 16) but do not provide information for organisms beyond Dehalococcoides within mixed cultures. Transcriptional analyses for a Dehalococcoides-containing mixed culture have not been reported, and there have been few reports of using microarrays to investigate transcription in mixed microbial communities (11, 23).
In order to identify genes within the KB-1 community that are transcriptionally active during the final critical dechlorination step from vinyl chloride (VC) to ethene, we created and used shotgun metagenome microarrays. We hypothesized that without sequence information we could still detect important genes by identifying transcripts from an arrayed short-insert library (Fig. 1). Analysis of microarray data identified spots containing sequences of functional interest, and subsequent sequencing of the clones identified genes of physiological importance from within the culture's metagenome. Replicate array experiments allowed evaluation of the reproducibility of the shotgun array format. We compared VC-dechlorinating treatments (VC plus methanol [MeOH]) to electron donor-only treatments (MeOH only). Comparison of these two treatments highlighted genes with high transcription levels in Dehalococcoides during VC degradation as well as genes active during electron acceptor deprivation or starvation. In addition, these treatments highlighted genes in non-Dehalococcoides organisms exhibiting high transcript levels during VC dechlorination.
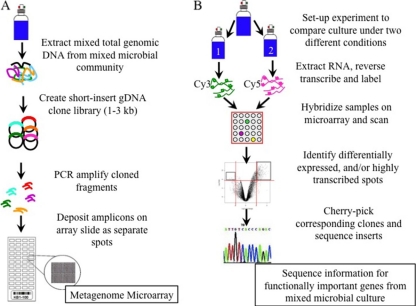
Fig 1.
Experimental work flow. (A) Illustration of the construction of shotgun metagenome microarray from a short-insert gDNA library from a mixed microbial culture. The final array comprised 19,200 spots arrayed in 48 subgrids of 400 spots. (B) Illustration of how these arrays can be interrogated to identify genes within this mixed community that exhibit different transcript levels under certain conditions.
MATERIALS AND METHODS
Shotgun metagenome microarray construction.
Briefly, two shotgun short-insert metagenomic DNA libraries were constructed using genomic DNA (gDNA) from the mixed culture KB-1 and used to generate PCR products that were spotted on the microarray. For each library 1- to 3-kilobase genomic fragments were cloned and transformed into Escherichia coli. Individual bacterial colonies were grown overnight; the inserts were then PCR amplified, purified, and quantified. The PCR products were spotted on GAPS II coated slides (Corning). The final array comprised 19,200 spots arrayed in 48 subgrids of 400 spots. A detailed description of the microarray construction can be found in the supplemental material.
Vinyl chloride degradation experiments.
Culture growth conditions are described in the supplemental material. For each experiment a KB-1 culture was purged with N2-CO2 (80:20 [vol%]) to remove trace chlorinated compounds and left to starve for 4 days to promote degradation of mRNA. Subsequently, 1.2 liters of culture was split into two equal treatments of 600 ml in 1-liter bottles (Pyrex; VWR, Mississauga, ON, Canada) in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI). Into one treatment 5 ml (200 μmol) of vinyl chloride gas was added as the electron acceptor and 15 μl (370 μmol) of methanol as the electron donor (VC plus MeOH treatment). In the second treatment only the electron donor, methanol, was added (MeOH only). Chlorinated ethenes, methane, and ethene were measured by injecting a 300-μl headspace sample onto a Hewlett-Packard 5890 Series II gas chromatograph fitted with a GS-Q column (30-m by 0.53-mm [inner diameter] porous layer open tubular [PLOT] column; J&W Scientific, Folsom, CA) and a flame ionization detector as described by Duhamel et al. (8).
For the first set of arrays, RNA was extracted from both treatments after 3 h when 35 μmol of ethene had been produced with the VC plus MeOH treatment. A second set of experiments was performed a month later with the same set-up. For these, RNA was extracted after 3.5 h when 50 μmol of ethene had been produced with the VC plus MeOH treatment. For each biological replicate, dye-swapped arrays were conducted. Technical replicates (hybridization of the same labeled RNA samples onto two different slides) were conducted for the first biological sample. A total of 12 arrays were utilized: 6 arrays for each condition (VC plus MeOH or MeOH only). The six arrays per condition comprise four from the first RNA sample, with replicate arrays for each dye (Cy3 and Cy5), and two from the later RNA sample, with only one array for each dye conducted.
RNA extraction, reverse transcription, labeling, and hybridization.
For RNA extraction, 50-ml samples were withdrawn from the two culture bottles inside an anaerobic chamber, for a total of four RNA samples. Cells were harvested by centrifugation at 9,900 rpm for 25 min at 4°C under anaerobic conditions. After lysis of the cell pellet with SDS and bead beating, RNA was extracted using an ice-cold acid–phenol-chloroform–isoamyl alcohol method. After purification and DNA removal the RNA was assessed for quantity and quality. The RNA was labeled using an indirect method in which amino allyl-dUTPs were incorporated during reverse transcription (RT) and then monoreactive cyanine dyes were coupled to the aminoallyl cDNA in a separate reaction. First-strand cDNA synthesis was carried out using 10 to 20 μg of total RNA and 10 ng control RNA (Arabidopsis thaliana chlorophyll synthetase gene). The Cy3- or Cy5-labeled cDNA was purified and quantified to ensure that an appropriate amount of dye was incorporated at an appropriate frequency. The labeled cDNA was then hybridized to the slide at 37°C for 16 to 18 h. The slides were then washed, dried, and scanned with an Axon scanner using GenePix Pro software. More details are available in the supplemental material.
Array data analysis.
Data analysis was performed in the R programming language with programs available from Bioconductor such as Linear Analysis of Microarrays (Limma) and gplots (32). The intensities were first corrected for background; then the log ratios (M values) were normalized within and between the arrays (31, 33). A linear model was fit to the data from the six different arrays, and Bayesian analysis was performed to estimate the statistical significance using P values and the B statistic (empirical Bayes log odds of differential expression) of the average M values (log2 [VC + MeOH/MeOH only]) for each spot (31). In addition, spots that had high intensities in each treatment were identified. To do this, the intensities for each channel were corrected for background intensity and then ranked. Subsequently, spots that ranked in the top 5% intensities for the same treatment on all 6 arrays were designated either “top VC plus MeOH” or “top MeOH only.”
Sequencing of selected spots.
Sequencing was performed at The Atlantic Genome Center (TAGC) (Halifax, Canada). Clones of interest were selected from the genomic libraries, and DNA from each clone was amplified using TempliPhi DNA polymerase. DNA sequencing was performed using ET terminator chemistry and capillary DNA sequencers. A series of Basic Local Alignment Search Tool (BLAST) searches (1) were performed to ascribe putative functions and phylogenies to the sequences. Databases that were used include the NCBI nonredundant protein and nucleotide databases, the STRING protein database and the corresponding mappings to clusters of orthologous groups (COGs) (13), and the Greengenes 16S rRNA database (6).
Representation of COGs.
Sequences were ascribed to certain COGs based on a BLASTx search against proteins from the STRING database and mapping to the corresponding COG (13). To determine if the Dehalococcoides genes that were differentially transcribed between the treatments were enriched in sequences belonging to certain COGs, an enrichment ratio was calculated for each COG category in the identified differentially expressed Dehalococcoides genes and compared to the abundance of that COG in an average Dehalococcoides genome. A ratio of 1 indicates that there is the same abundance of that COG in the differentially transcribed genes as in a Dehalococcoides genome. Statistical analysis was performed to evaluate if the overrepresentation was statistically significant based on the sample sizes using a hypergeometric distribution (17, 35). More-detailed methods are available in the supplemental material.
Determination of KB-1 prophage-like region.
After data analysis of the selected shotgun metagenome clones of interest, The Joint Genome Institute (JGI) sequenced, assembled, and annotated a KB-1 community metagenome. The contig sequences and predicted protein sequences are available from the IMG/M website (19) under the microbiome name “aquatic dechlorinating community (KB-1) (sample 10166).” To identify phage-like proteins within the KB-1 metagenome, a custom database was constructed by obtaining all of the annotated bacteriophage proteins from the NCBI database. Then BLASTP searches of the KB-1 metagenome protein sequences were performed against this database. In addition, profile hidden Markov model (HMM) searches were performed using hmmer 2.3.2 (9) on all the KB-1 proteins against a database of Pfam and J. Craig Venter Institute HMMs previously identified as associated with phage proteins. The boundaries of the prophage were defined as the largest region bounded by phage homologous genes, including a core of at least six genes homologous to phage structural genes (including genes encoding at least one phage head protein and one phage tail protein), with no more than 15 open reading frames (ORFs) between coding sequences for proteins that had homologues in the phage database. The results were also manually examined to ensure that the predicted prophage region did not contain known bacterial operons or nonphage housekeeping genes. Functional annotations within the prophage region were assigned based on sequence similarity to known phages, HMM matches, and predicted secondary structural conservation as determined by HHpred (34).
The contigs and predicted proteins from the metagenome of the Dehalococcoides-containing ANAS dechlorinating culture were accessed from the IMG/M website [microbiome name, “ANAS dechlorinating bioreactor (sample 196)”] (28). Prophage-like regions in the ANAS assembly were identified as described above. A previously identified putative prophage in Dehalococcoides strain 195 (DET) was downloaded from NCBI (30). Putative prophage regions in KB-1, ANAS, and DET were compared by performing BLASTP searches of the KB-1 proteins against the ANAS and DET proteins. In addition, the genomic fragments were aligned using Mauve (5).
Phage extraction.
To mimic conditions used for the arrays, the KB-1 culture was starved for 4 to 5 days after completing a dechlorination cycle. Absence of chlorinated compounds was confirmed with gas chromatography as described above. For phage extraction, a 500-ml aliquot of culture was transferred to an anaerobic culture bottle. Following a modified polyethylene glycol (PEG) precipitation protocol, the phage particles were harvested by centrifugation, followed by a phenol-chloroform DNA extraction of this phage fraction. This DNA was tested using PCR for the presence of phage-specific genes (head, tail, and integrase genes), the tceA-homologous gene, hypothetical protein genes near the putative phage genome boundaries, and bacterial 16S rRNA genes. PCR products were visualized on a 1% agarose gel stained postelectrophoresis in an ethidium bromide bath. Complete methods for virus purification, DNA extractions, and PCRs can be found in the supplemental material.
Accession numbers.
The raw sequences from the selected sequenced clones have been deposited in the GSS database of GenBank with accession numbers GS884142 to GS885177 and GS885207 to GS885247. The data from the microarray experiments have been deposited in the GEO database with the accession number GSE20041.
RESULTS AND DISCUSSION
Array composition and results.
We designed a shotgun metagenomic microarray for the KB-1 dechlorinating microbial consortium and examined the microbial community's response under vinyl chloride degradation conditions (VC plus MeOH treatment) compared to electron donor-only conditions (MeOH-only treatments). The array consists of a total of 19,200 spots from short-insert (1- to 3-kb) clone libraries. Given an approximation for the total length of KB-1 community gDNA that was spotted on the array and estimates of the species composition of KB-1 from quantitative PCR (qPCR) surveys, we estimated the fold coverage of the genomes within the KB-1 community (Table 1). From this, it is clear that Dehalococcoides, the dominant organism in the KB-1 culture, is significantly better represented on the array than the other community members.
Table 1.
Population structure of KB-1 and estimated coverage of the different genomes on the shotgun arraya
| KB-1 OTUb | % of population |
Representative genome size (Mb) | Estimated depth of genome coverage on array |
||
|---|---|---|---|---|---|
| Low | High | Low | High | ||
| Dehalococcoides | 48.1 | 70.2 | 1.4 | 12.5 | 18.2 |
| Chlorobi SJA-28 | 6.0 | 14.9 | 2.6 | 0.8 | 2.1 |
| Acetobacterium | 2.7 | 12.0 | 3.3 | 0.3 | 1.3 |
| Spirochaetes SA-8 | 2.8 | 8.3 | 2.8 | 0.4 | 1.1 |
| Sporomusa | 1.2 | 11.5 | 4.4 | 0.1 | 0.9 |
| Methanomicrobiales | 3.5 | 5.8 | 2.7 | 0.5 | 0.8 |
| OP5 | 2.5 | 5.0 | 3.2 | 0.3 | 0.6 |
| Geobacter | 3.0 | 4.4 | 4.4 | 0.2 | 0.4 |
| Methanosarcina | 0.1 | 1.0 | 4.5 | <0.01 | 0.1 |
| Bacteroidales 1 | <0.1 | 0.8 | 5.5 | <0.01 | 0.1 |
| Syntrophobacter | <0.1 | 0.2 | 5.0 | <0.01 | <0.1 |
Microorganisms that were detected in KB-1 by 16S rRNA genes are listed. Low and high estimates of the percentages of the population represent the range of abundance measured by qPCR over the time period that the arrays were constructed and experiments were conducted (see reference 37 for methods). The genome coverage is based on an average amplicon size of 2 kb for 18,042 amplicons. The representative genome size is an average from closely related organisms.
OTU, operational taxonomic unit.
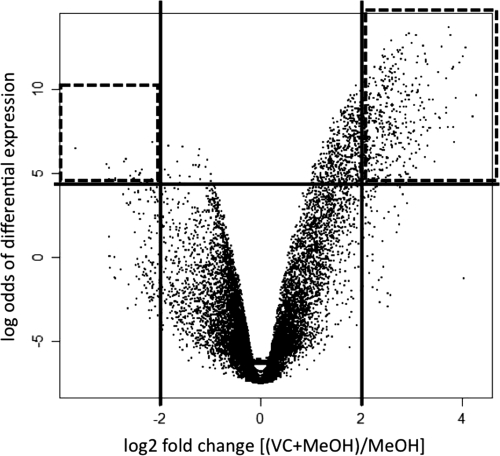
Data visualization suggested that the intensity data were high quality, with even dye distributions within arrays (see Fig. S1 in the supplemental material). Analysis of the data indicated that on average 83% of the spots on the array had a signal-to-noise ratio greater than 2 and that minor trends in background fluorescence were accounted for by normalization (see Fig. S2 in the supplemental material). The Pearson correlation coefficient between M values for the two biological replicates was 0.7059, for the dye-swapped replicates the values were 0.6770 and 0.8983, and for the technical replicates the values were 0.9235 and 0.9150, indicating good agreement between replicate samples. Statistical analysis of the six arrays produced a volcano plot that identified 452 spots with significantly higher intensity levels in the VC plus MeOH treatments than in the MeOH-only treatments (log2 fold change > 2 and B statistic > 4.6) and 95 spots with significantly higher intensity levels in the MeOH-only treatments (log2 fold change < −2, B statistic > 4.6) (Fig. 2). These spots, as well as the 100 highest-intensity spots from each treatment (top VC plus MeOH and top MeOH only), were sequenced, for a total of 747 spots sequenced. Before sequencing all spots of interest, nine spots were selected and sequenced for independent validation of the array hybridization using quantitative PCR. The qPCR data showed good agreement between the array and qPCR-based quantification of specific transcripts within the two treatments' cDNA samples (see Fig. S3 in the supplemental material).
Fig 2.
Volcano plot indicating spots with differential intensity values. The x axis shows values for all spots averaged over the six arrays. A value of 2 indicates that the spot had 4 times the intensity in the VC treatment. The y axis is the B statistic, where 4.6 corresponds to a 99% likelihood that the spot exhibits differential intensity levels. Spots in the upper left and upper right quadrants were chosen as the statistically significant spots to be sequenced.
From a series of BLAST searches, putative functions and taxonomic affiliations were assigned to the spot sequences. For approximately 44% of all sequenced spots, sequence data were gathered from both ends of the clone. As shotgun clones may contain two different genes, these sequences were treated separately in the BLAST analyses. Of the spots sequenced in both directions in the differential analysis, 29.9% had best BLAST matches to different genes at either end of the clone; however, approximately two-thirds of these genes are subunits of the same complex or belong in the same operon. Comparison of multiple shotgun spots containing rdhA5 with cloned control spots containing only the rdhA5 gene revealed that spots exhibiting differential intensity levels typically contained a single gene, while spots containing two different genes did not show differential intensity values (see Fig. S5 in the supplemental material). It is therefore likely that most of the sequences identified here as having different transcript levels between treatments comprise one gene, one transcription unit, or, less likely, separately transcribed genes exhibiting similar responses to the two treatments.
Of the 764 sequences from the 452 spots that had significantly higher intensity levels in the VC plus MeOH treatment, 710 had a significant BLASTx hit to a protein of annotated function, 38 had a hit to a hypothetical protein, and 16 had no hits from the database. In contrast, for the 155 sequences from the 95 spots that had significantly higher intensity levels in MeOH-only treatments, 51 had hits to hypothetical proteins and 20 had no database hits, indicating that, for over 45% of the sequences, no function could be easily assigned. Moreover, hypothetical proteins of Dehalococcoides origin constituted 90% of the hypothetical proteins detected in the MeOH-only sequences, suggesting that a large fraction of the Dehalococcoides genes that are active in the absence of a chlorinated electron acceptor have unknown functions. Detected transcription of genes encoding these hypothetical proteins validates their identification as genes and suggests a role in the starvation response or cell maintenance. We have nominated many of these genes of interest for a heterologous expression, functional screening, and protein crystallization pipeline in order to assess their function. To date, 36 proteins have been expressed from heterologous expression vectors and 16 vectors have produced soluble protein products, 9 of which are hypothetical proteins. These expression vector clones are available on request (for more details see Table S1 in the supplemental material).
In addition to BLASTx hits to proteins, some of the sequences had significant BLASTn hits to rRNA genes (small subunit [SSU] and large subunit [LSU]), including many genes from the sequence sets from spots exhibiting the highest intensity levels (32 and 23% for top MeOH only and top VC plus MeOH, respectively). Taxa that were identified by significant BLASTn matches to rRNA genes include Aminanaerobia, Bacteroidales, Dehalococcoides, Desulfovibrio, Methanoregula, Methanosarcina, Methanomethylovorans, and Treponemataceae SA-8 (see Tables S1 and S3 in the supplemental material).
Functional analysis of Dehalococcoides sequences in selected spots.
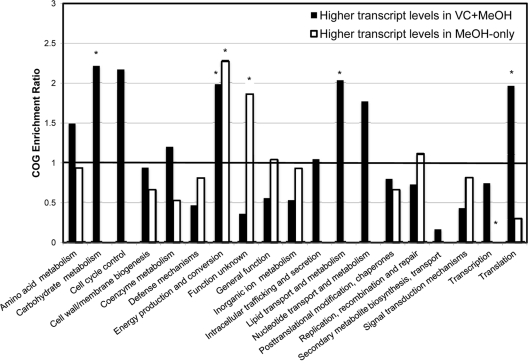
For information on Dehalococcoides genes that showed differential transcript levels between the two treatments, see Tables S4 and S5 in the supplemental material. In order to determine which Dehalococcoides genes were over- or underrepresented in the transcribed gene sets relative to a complete Dehalococcoides genome, we compared the abundances of the COGs in the transcribed gene sets (VC plus MeOH and MeOH only) to the abundance of the COGs in Dehalococcoides genomes. The genes with higher intensity levels in the VC dechlorinating treatments were about 2-fold enriched in COGs involving transcription, translation, and energy production and conversion (Fig. 3). This was expected, as KB-1 Dehalococcoides obtains energy for growth from VC dechlorination (7). The genes that had higher transcript levels in the dechlorinating treatment were also enriched in COGs involving amino acid transport and metabolism, carbohydrate transport and metabolism, and lipid transport and metabolism. Sequences in the lipid metabolism COG include genes involved in terpenoid biosynthesis as well as fatty acid biosynthesis. White et al. (40) suggested that phospholipid furan acid fatty acids that are unique to Dehalococcoides might protect against free radicals that are generated in the process of reductive dechlorination.
Fig 3.
Representation of clusters of orthologous group (COG) categories in Dehalococcoides genes exhibiting different transcript levels between treatments. On the x axis are COGs as defined by the eggNOG database. On the y axis is an enrichment ratio representing the relative proportion of each COG category in the sequenced spots compared to the average COG abundance in Dehalococcoides genomes according to the equation enrichment ratio = proportion of COG X in Dehalococoides sequences with higher transcript levels/proportion of COG X in Dehalococcoides genomes. A ratio of 1 indicates that there is the same proportion of that COG in the sequenced spots as in an average Dehalococcoides genome. COGs which have statistically significant enrichment ratios are marked with an asterisk. P values were calculated using a hypergeometric distribution.
Reductive dechlorination is coupled to energy conservation in Dehalococcoides and is carried out by reductive dehalogenases, which catalyze the removal of chlorine atoms and replacement with hydrogen atoms. Of the 110 energy-related sequences that had higher transcript levels in the VC plus MeOH treatments, only 1 reductive dehalogenase homologous sequence was detected, on four separate spots containing KB-1 rdhA14 or rdhB14, which are homologs of the vinyl chloride reductase gene vcrAB in Dehalococcoides strain VS (22, 38). Spots containing rdhA14 were among the most intense detected on the array. Conversely, the only sequences assigned to the energy COG with higher transcript levels in the MeOH-only treatment all had hits to Dehalococcoides reductive dehalogenase protein homologs or associated transcriptional regulators. Four putative RDH genes were identified: KB-1 rdhA5 and KB-1 rdhA1 on multiple spots and KB-1 rdhA12 and rdhA13 as single sequences. Specifically, the transcription level of KB-1 rdhA5 was among the highest in the MeOH-only treatments. The increased expression of rdhA5 in the absence of VC is consistent with the expression profile of its ortholog in Dehalococcoides strain 195 (DET1545), which is upregulated in stationary phase (14). Identification of four reductive dehalogenases with higher transcript levels in the MeOH-only treatments agrees with reports of upregulation of multiple RDH genes in the absence of chlorinated compounds or in the late stages of dechlorination in other Dehalococcoides cultures (14, 24).
Sequences that had higher transcript levels during dechlorination included those encoding four of the five types of Dehalococcoides hydrogenases (15, 30), only lacking the Hyc hydrogenase (Ni/Fe). In contrast to the reductive dehalogenase genes, there were no hydrogenase genes with higher transcript levels in MeOH-only treatments or in the topmost intensity category for MeOH only (top MeOH only), confirming that hydrogenase genes are transcribed only in response to active respiration (25) and therefore may be better biomarkers for active growth-associated dechlorination than dehalogenase genes.
There were 22 sequences with hits to Dehalococcoides hypothetical proteins whose genes had higher transcript levels during VC degradation. Four sequences had a top BLASTx hit to the hypothetical protein cbdbA1240. The STRING database predicts that this hypothetical protein interacts with aspartyl-tRNA synthetase and cbdbA1241, a fasciclin domain protein. There were two sequences in the top VC plus MeOH and top MeOH-only lists with a hit to the fasciclin domain protein (cbdbA1241). Fasciclin domain proteins contain a cell adhesion domain found in plants, animals, and bacteria (36) and may contribute to the known tendency for Dehalococcoides to associate with other cells in bioflocs (29).
Of the Dehalococcoides sequences that had higher transcript levels in MeOH-only treatments, 48% of the sequences had top BLASTx hits to hypothetical proteins. There were a few hypothetical proteins encoded on multiple spots; for example there were nine separate spots with sequences that had top BLASTx hits to cbdbA1485. Analysis of genome context and gene co-occurence profiles from the STRING database suggests an interaction with the recombinase protein cbdbA77 as well as a CRISPR-associated protein. Other differentially expressed sequences suggest active genetic rearrangement, with hits to Dehalococcoides site-specific recombinases, a Dehalococcoides transposase, and a Dehalococcoides resolvase. There is also evidence for general recombination, as there were six hits to the Holliday junction helicase (RuvB), as well as hits to the recombination proteins RecJ and RecR.
Functional analysis of non-Dehalococcoides sequences from spots of interest.
For a table describing data for all of the spots containing differentially transcribed non-Dehalococcoides genes see Table S6 in the supplemental material. It was difficult to assign many of the sequences from the non-Dehalococcoides spots to a specific taxon or function with confidence. This was due to the fact that many of the members of KB-1 (as identified by 16S rRNA gene analysis) belong to phylogenetic clades with few or no sequenced representatives, and hence the NCBI databases contain no close homologs. Nevertheless, multiple array sequences with hits to Spirochaetes, Chloroflexi, Geobacter, methanogens, and phage indicate the importance of these groups of organisms. In particular, the Geobacter sequences were more robustly assignable given the presence of a closely related genome in the database. Sequences with higher transcript levels in the presence of VC included two sequences with significant hits to metabolic genes from Geobacter lovleyi SZ: beta-ketoacyl synthase and CDP-alcohol phosphatidyltransferase genes.
After sequences from the Dehalococcoides prophage region discussed below were removed, there were only 18 non-Dehalococcoides sequences that had higher transcript levels in the MeOH-only treatment. Of these, five (28%) are most likely of Geobacter origin, one (6%) seems to be related to Methanoregula boonei, and 66% cannot be attributed to a specific taxon. Although a Geobacter is known to dechlorinate PCE and TCE to cis-1,2-dichloroethene (cDCE) in KB-1 (7), it is interesting that Geobacter appeared to be active in the presence of VC as well as in the absence of chlorinated compounds. Despite relatively low coverage of the non-Dehalococcoides genomes on the array, many of the available non-Dehalococcoides genes were differentially transcribed between the two treatments, indicating that their corresponding organisms respond to the presence of VC or its degradation by Dehalococcoides.
Characterization of a Dehalococcoides prophage.
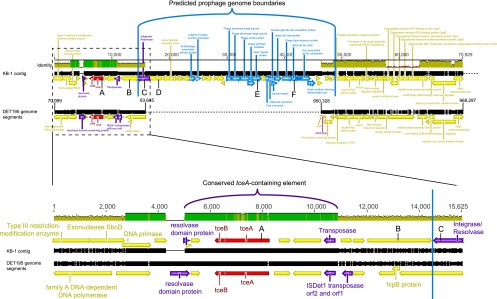
Initially, 50% of the genes that had higher transcript levels in MeOH-only treatments were thought to be non-Dehalococcoides based on the phylogeny of the top BLASTx hit. A BLASTn search of these “non-Dehalococcoides” sequences against the KB-1 metagenome assembly via the IMG/M website (19) (sample 10166) revealed nearly identical hits to a long (140,000-bp) Dehalococcoides contig (DCKB-1_c338). The majority of this contig has high similarity and synteny with other Dehalococcoides genomes; however, the region that had higher transcript levels in the MeOH-only treatment contains a large proportion of hypothetical genes with no significant hits to other Dehalococcoides genomes and contains many annotated phage genes (Fig. 4). Bioinformatic analyses suggested that this genomic region encodes a functional prophage based on its length (∼30 kb), the repertoire and order of phage structural genes, and the lack of intervening bacterial operons or housekeeping genes (see Table S7 in the supplemental material). All but one of the minimum proteins (a receptor binding protein) needed to form a functional phage were definitively annotated, and there are a number of lower-confidence annotations with the right predicted features to fulfill this missing function. The phage lacks a phage-type DNA polymerase gene, but this is not a required gene for phage function; notably, the enterobacterial phage λ also lacks a viral-type DNA polymerase gene (NCBI genome NC_001416). The complete phage is predicted to span the genes from DCKB1_14310 to DCKB1_14860 on contig DCKB1_c338 and is phylogenetically related to Siphoviridae-type phage, characterized by long, noncontractile tails (3). The tape measure protein responsible for determination of phage tail length in the KB-1 phage genome (DCKB1_14440) is about half as long as λ's tape measure protein, indicating that this phage likely has a relatively short tail. Several genes upstream of the predicted prophage genome have some similarity to phage genes (DCKB1_14270 to DCKB1_14300) but share much stronger homology with known Dehalococcoides genes and appear distinct from this predicted phage genome. This suggests that similar proteins are transduced or encoded in other phages and that the prophage insertion may have been targeted to a feature in this region rather than being a random insertion event.
Fig 4.
Nucleotide alignments of Dehalococcoides ethenogenes strain 195 genome segments with the tceA- and prophage-containing Dehalococcoides contig from the KB-1 metagenome. In both alignments, tceA and tceB are highlighted in red, annotated phage genes are highlighted in blue, and genes associated with transposable elements are highlighted in purple. Letters A to F correspond to genes examined using PCR (Fig. 5). (Upper alignment) Alignment of two noncontiguous segments of the DET195 genome with the first ∼70 kbp of the Dehalococcoides KB-1 contig. Numbers correspond to the nucleotide positions on the KB-1 Dehalococcoides contig and the DET195 genome coordinates (32). The origin and terminus of the predicted phage genome are marked with blue boundaries. There was no homologous region in the DET195 genome to align to the prophage portion of the KB-1 contig. The bar graph above the alignment depicts identity between the aligned sequences, where a green bar indicates 100% identity and yellow indicates disagreement and where the height of a yellow bar gives the percent identity across a 50-bp window. (Lower alignment) Zoomed-in view of the first 15 kbp of the upper alignment (dashed box), depicting the highly conserved transposable element containing the tceA and tceB genes. Alignments were created using the Geneious alignment algorithm (9), and annotations for the KB-1 contig were determined by the RAST server (2).
There are two other sequenced Dehalococcoides prophages: one in the type strain, DET195 (30), and one we identified from the metagenome of the Dehalococcoides-containing ANAS consortia (IMG/M website; sample 196) (28). A comparison of the KB-1 prophage to the DET195 and ANAS prophages indicates relatively low homology within the phage structural proteins, with some segments of high similarity between the DET195 and ANAS prophages not shared with the KB-1 prophage (see Fig. S4 in the supplemental material). These results are in agreement with the mosaic nature of phage genomes (4).
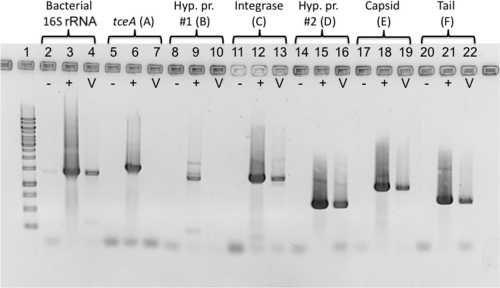
The identification of tceA-like reductive dehalogenase genes (97% nucleotide [nt] identity to DET0079) located in close proximity to the predicted prophage genomes in ANAS and KB-1 (see Fig. S4 in the supplemental material) prompted further experimentation to investigate the possibility that the tceA gene is contained within or associated with the prophage. To determine whether the tceA gene was present within the prophage genome and to confirm the activity of the putative phage, the viral fraction of a starved KB-1 culture was purified and DNA was extracted. The viral DNA fraction was tested using PCR for the presence of prophage marker genes (capsid protein, tail protein, and integrase genes), the tceA gene, hypothetical protein genes at either side of the boundary of the predicted phage genome, and the bacterial 16S rRNA gene (see Table S8 in the supplemental material for primer sequences). The PCRs showed clear presence of the phage marker genes in the viral DNA fraction, a significant reduction in the bacterial 16S rRNA gene signal compared to KB-1 community DNA, and an absence of the tceA gene (Fig. 5). From the higher transcription levels of the prophage genes on the array and the presence of Dehalococcoides prophage-specific genes within the viral metagenomic DNA, it seems reasonable to conclude that the prophage contained in the Dehalococcoides genome is activated by electron acceptor-limiting conditions. A similar phenomenon was reported in Dehalococcoides DET195, where many genes in a putative prophage were upregulated in stationary phase compared to exponential phase (14). In addition, the PCR assays confirmed the integrase gene as the terminal gene in the phage genome, as all examined genes downstream of the integrase gene (tceA and one gene encoding a hypothetical protein) were not detected in the viral fraction, while all examined genes upstream of the integrase gene (second hypothetical protein, capsid, and tail protein genes) were detected (Fig. 5 shows PCR results; gene locations are in Fig. 4).
Fig 5.
Agarose gel image of PCR amplification designed to examine the KB-1 viral metagenomic DNA sample for purity. In-house tceA primers were used (see Table S8 in the supplemental material). Lane numbers correspond to the following primer sets and samples: 1, Fermentas 1-kb ladder; 2 to 4, general bacterial 16S rRNA genes; 5 to 7, tceA gene (A); 8 to 10, hypothetical protein (hyp. pr.) gene outside putative phage genome boundary (B); 11 to 13, phage integrase gene (C); 14 to 16, hypothetical protein gene within putative phage genome boundary (D); 17 to 19, bacteriophage capsid protein gene (E); 20 to 22, bacteriophage tail protein gene (F). For all primer sets, a no-template control (−), a KB-1 community gDNA sample (+), and the KB-1 viral metagenomic DNA (V) were assayed. Letters A to F correspond to the letters identifying these genes in Fig. 4.
Based on phylogenetic affiliation with known phage terminases, the KB-1 Dehalococcoides terminase gene (DCKB1_14560), responsible for determination of genome boundaries for phage DNA packaging, most likely utilizes a 3′-extended cos-dependent packaging mechanism (10). The high variability of cos sites across species means we were unable to define a specific 3′ cos boundary for the prophage genome, but the PCR-based examination of the viral fraction metagenomic DNA suggests that tceA is not permanently contained within the phage genome. However, the proximity of the tceA gene to the phage lysogen suggests that it could be packaged into the viral particle and transferred to a new host by specialized transduction. In this process, the phage genome is aberrantly excised from the host genome upon lytic growth and adjacent host genomic DNA is packaged into the capsid along with the phage genome. Thus, we cannot rule out the possibility that the tceA gene is occasionally packaged with the prophage DNA into a mobile viral particle, despite no evidence of this phenomenon in our experiments.
An alternate explanation for the presence of the tceA gene can be offered based on the gene neighborhood surrounding the reductive dehalogenase gene. In the strain DET195 genome, the tceA gene is located near a transposase gene within insertion element I (IE_I). In KB-1_c338 and ANAS_c818 prophages, the tceA-like gene is also located near a transposase gene. A more in-depth examination of the tceA region in the KB-1 metagenome indicates that the transposase gene is located between tceA and the predicted prophage genome terminus (Fig. 4). Based on the sequence conservation between the DET195 and KB-1 tceA-containing regions and the indications of the presence of a mobile element in the form of a transposase gene upstream of and two resolvase genes flanking the tceAB operon, it seems more likely that, if tceA has been acquired through some form of lateral gene transfer (LGT), this acquisition was via a transposable element rather than by the insertion of the prophage. A study of environmental tceA distribution identified the resolvase gene downstream of tceAB as a conserved marker for a putative transposable element (14).
Notably, many RDH genes are associated with transposable element markers or otherwise altered genomic signatures (20, 21, 26, 27), and there is one reported identification of a circularized transposable element carrying a reductive dehalogenase gene, the Tn-Dha1 of Desulfitobacterium hafniense strain TCE1, which carries the pceABC operon (18). However, a clear understanding of the origins of dehalogenase genes and their frequency of LGT remains elusive. Prophages often constitute main sources of variation between strains and often confer beneficial environment-specific capacities to the host strain through the genes that they contain (2, 4). While the prophage within Dehalococcoides in KB-1 does not appear to be currently functioning as an LGT vehicle for the functionally important tceA, this mode of transfer of reductive dehalogenases cannot be eliminated as a possibility.
Conclusions.
We used a shotgun metagenome microarray to compare levels of gene expression in the Dehalococcoides-containing microbial community KB-1 in the presence and absence of the electron acceptor VC. Subsequent sequencing of spotted genomic fragments identified genes from the culture's metagenome that are transcriptionally active and provided key sequences of interest for further molecular studies and for protein expression and characterization. Sequences from Spirochaetes, Geobacter, and methanogens indicate the importance of these organisms to the KB-1 culture. Techniques to interrogate transcription in mixed communities continue to evolve at a rapid pace; a shotgun metagenome microarray is perhaps not the easiest approach currently available but proved to be an effective screening tool for identification of genes of interest. Our microarray results prompted experiments that confirmed the existence of a Siphoviridae-like Dehalococcoides prophage that is activated in response to starvation conditions. While the tceA gene is in very close genomic proximity to this prophage, we determined that its acquisition was more likely via a transposable element than via the prophage. Starvation and other stressful conditions (high salt concentrations, heavy metals, low nutrient concentrations) are common at contaminated sites and may cause activation of Dehalococcoides prophages, thus allowing increased genetic variation through phage-mediated gene transfer in Dehalococcoides.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Natural Sciences and Engineering Research Council of Canada and the U.S. Department of Defense Strategic Environmental Research Defense Program for funding. We also acknowledge support from the Government of Canada through Genome Canada and the Ontario Genomics Institute (2009-OGI-ABC-1405) and a Canadian Institutes of Health Research Operating Grant to K.L.M. (fund number MOP-6279). A.S.W. was supported by an Ontario Graduate Scholarship (OGS). L.A.H. was supported by an NSERC CGS-D scholarship. D.R.R. was supported by OGS and NSERC CGS-D scholarships over the course of this work.
We acknowledge the University Health Network microarray center (Toronto) for printing the arrays and for invaluable technical guidance. Dingling Zhang (Toronto) assisted in preparing DNA for the arrays. Limin Chen and Jing Sun (Toronto) offered advice for performing the array experiments. Sequencing was performed at The Atlantic Genome Center (Halifax, NS, Canada) with the help of Sharen Bowman, Catherine Kozera, and Bruce Curtis and at the DoE Joint Genome Institute with the help of Kerrie Barry and Susannah Green Tringe. Help with the prophage DNA extraction was generously provided by Mostafa Fatehi and Diane Bona (University of Toronto). Heterologous expression clones were generated by Alexei Savchenko and Alexander Yakunin (University of Toronto).We thank Frank Löffler (University of Tennessee) and Krishna Mahadevan (University of Toronto) for valuable discussions.
Footnotes
Published ahead of print 16 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Brussow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brussow H, Desiere F. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213–222 [DOI] [PubMed] [Google Scholar]
- 4. Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeSantis TZ, Jr., et al. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394–W399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duhamel M, Edwards EA. 2007. Growth and yields of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2-dichloroethane. Environ. Sci. Technol. 41:2303–2310 [DOI] [PubMed] [Google Scholar]
- 8. Duhamel M, et al. 2002. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-dichloroethene and vinyl chloride. Water Res. 36:4193–4202 [DOI] [PubMed] [Google Scholar]
- 9. Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763 [DOI] [PubMed] [Google Scholar]
- 10. Feiss M, Siegele DA. 1979. Packaging of the bacteriophage-lambda chromosome-dependence of cos cleavage on chromosome length. Virology 92:190–200 [DOI] [PubMed] [Google Scholar]
- 11. He S, et al. 2010. Metatranscriptomic array analysis of Candidatus Accumulibacter phosphatis'-enriched enhanced biological phosphorus removal sludge. Environ. Microbiol. 12:1205–1217 [DOI] [PubMed] [Google Scholar]
- 12. Hug LA, Salehi M, Nuin P, Tillier ER, Edwards EA. 2011. Design and verification of a pangenome microarray oligonucleotide probe set for Dehalococcoides spp. Appl. Environ. Microbiol. 77:5361–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jensen LJ, et al. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37:D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson DR, et al. 2008. Temporal transcriptomic microarray analysis of “Dehalococcoides ethenogenes” strain 195 during the transition into stationary phase. Appl. Environ. Microbiol. 74:2864–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kube M, et al. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269–1273 [DOI] [PubMed] [Google Scholar]
- 16. Lee PK, et al. 2011. Comparative genomics of two newly isolated Dehalococcoides strains and an enrichment using a genus microarray. ISME J. 5:1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mahadevan R, et al. 2008. Characterizing regulation of metabolism in Geobacter sulfurreducens through genome-wide expression data and sequence analysis. OMICS 12:33–59 [DOI] [PubMed] [Google Scholar]
- 18. Maillard J, Regeard C, Holliger C. 2005. Isolation and characterization of Tn-Dha1, a transposon containing the tetrachloroethene reductive dehalogenase of Desulfitobacterium hafniense strain TCE1. Environ. Microbiol. 7:107–117 [DOI] [PubMed] [Google Scholar]
- 19. Markowitz VM, et al. 2008. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 36:D534–D538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McMurdie PJ, et al. 2009. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 5:e1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McMurdie PJ, Hug LA, Edwards EA, Holmes S, Spormann AM. 2011. Site-specific mobilization of vinyl chloride respiration islands by a mechanism common in Dehalococcoides. BMC Genomics 12:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muller JA, et al. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parro V, Moreno-Paz M. 2003. Gene function analysis in environmental isolates: the nif regulon of the strict iron oxidizing bacterium Leptospirillum ferrooxidans. Proc. Natl. Acad. Sci. U. S. A. 100:7883–7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rahm BG, Morris RM, Richardson RE. 2006. Temporal expression of respiratory genes in an enrichment culture containing Dehalococcoides ethenogenes. Appl. Environ. Microbiol. 72:5486–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rahm BG, Richardson RE. 2008. Dehalococcoides' gene transcripts as quantitative bioindicators of tetrachloroethene, trichloroethene, and cis-1,2-dichloroethene dehalorespiration rates. Environ. Sci. Technol. 42:5099–5105 [DOI] [PubMed] [Google Scholar]
- 26. Regeard C, Maillard J, Dufraigne C, Deschavanne P, Holliger C. 2005. Indications for acquisition of reductive dehalogenase genes through horizontal gene transfer by Dehalococcoides ethenogenes strain 195. Appl. Environ. Microbiol. 71:2955–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rhee SK, Fennell DE, Haggblom MM, Kerkhof LJ. 2003. Detection by PCR of reductive dehalogenase motifs in a sulfidogenic 2-bromophenol-degrading consortium enriched from estuarine sediment. FEMS Microbiol. Ecol. 43:317–324 [DOI] [PubMed] [Google Scholar]
- 28. Richardson RE, Bhupathiraju VK, Song DL, Goulet TA, Alvarez-Cohen L. 2002. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ. Sci. Technol. 36:2652–2662 [DOI] [PubMed] [Google Scholar]
- 29. Rowe AR, Lazar BJ, Morris RM, Richardson RE. 2008. Characterization of the community structure of a dechlorinating mixed culture and comparisons of gene expression in planktonic and biofloc-associated “Dehalococcoides” and Methanospirillum species. Appl. Environ. Microbiol. 74:6709–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seshadri R, et al. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105–108 [DOI] [PubMed] [Google Scholar]
- 31. Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. [DOI] [PubMed] [Google Scholar]
- 32. Smyth GK, Michaud J, Scott HS. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067–2075 [DOI] [PubMed] [Google Scholar]
- 33. Smyth GK, Speed T. 2003. Normalization of cDNA microarray data. Methods 31:265–273 [DOI] [PubMed] [Google Scholar]
- 34. Soding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. (Erratum, 21:2144.) [DOI] [PubMed] [Google Scholar]
- 35. Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22:281–285 [DOI] [PubMed] [Google Scholar]
- 36. Ulstrup JC, Jeansson S, Wiker HG, Harboe M. 1995. Relationship of secretion pattern and MPB70 homology with osteoblast-specific factor 2 to osteitis following Mycobacterium bovis BCG vaccination. Infect. Immun. 63:672–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Waller AS. 2010. Molecular investigation of chloroethene reductive dehalogenation by the mixed microbial community KB-1. Ph.D. ; thesis. University of Toronto, Toronto, Canada. [Google Scholar]
- 38. Waller AS, Krajmalnik-Brown R, Loffler FE, Edwards EA. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71:8257–8264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. West KA, et al. 2008. Comparative genomics of “Dehalococcoides ethenogenes” 195 and an enrichment culture containing unsequenced “Dehalococcoides” strains. Appl. Environ. Microbiol. 74:3533–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. White DC, et al. 2005. Phospholipid furan fatty acids and ubiquinone-8: lipid biomarkers that may protect dehalococcoides strains from free radicals. Appl. Environ. Microbiol. 71:8426–8433 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.