Abstract
Crp/Fnr-type global transcriptional regulators regulate various metabolic pathways in bacteria and typically function in response to environmental changes. However, little is known about the function of four annotated Crp/Fnr homologs (DVU0379, DVU2097, DVU2547, and DVU3111) in Desulfovibrio vulgaris Hildenborough. A systematic study using bioinformatic, transcriptomic, genetic, and physiological approaches was conducted to characterize their roles in stress responses. Similar growth phenotypes were observed for the crp/fnr deletion mutants under multiple stress conditions. Nevertheless, the idea of distinct functions of Crp/Fnr-type regulators in stress responses was supported by phylogeny, gene transcription changes, fitness changes, and physiological differences. The four D. vulgaris Crp/Fnr homologs are localized in three subfamilies (HcpR, CooA, and cc). The crp/fnr knockout mutants were well separated by transcriptional profiling using detrended correspondence analysis (DCA), and more genes significantly changed in expression in a ΔDVU3111 mutant (JW9013) than in the other three paralogs. In fitness studies, strain JW9013 showed the lowest fitness under standard growth conditions (i.e., sulfate reduction) and the highest fitness under NaCl or chromate stress conditions; better fitness was observed for a ΔDVU2547 mutant (JW9011) under nitrite stress conditions and a ΔDVU2097 mutant (JW9009) under air stress conditions. A higher Cr(VI) reduction rate was observed for strain JW9013 in experiments with washed cells. These results suggested that the four Crp/Fnr-type global regulators play distinct roles in stress responses of D. vulgaris. DVU3111 is implicated in responses to NaCl and chromate stresses, DVU2547 in nitrite stress responses, and DVU2097 in air stress responses.
INTRODUCTION
Transcription factors, promoter sequences, and regulatory circuit architecture are often referred as “the regulatory genome” (27, 33), and transcription factors are central to the function of regulatory networks (5). Organisms rely on regulatory networks to orchestrate the transcription of genes in response to internal or external environmental changes in order to optimize metabolism and enhance survival. Crp/Fnr regulators are global transcriptional regulators widely distributed in bacteria. The characteristic structure of Crp/Fnr is a C-terminal helix-turn-helix (HTH) motif that fits the DNA major groove and an N-terminal nucleotide binding domain (34). This class of transcriptional factors is named after the first two representatives discovered in Escherichia coli, i.e., the Crp (cyclic AMP [cAMP] receptor protein) and Fnr (fumarate and nitrate reductase regulator protein) (4, 21, 24, 27). Crp/Fnr regulators are generally classified into different subfamilies (e.g., CooA, Crp, Dnr, FixK, Fnr, HbaR, NnrR, etc.) according to phylogenetic affiliations (50). The increasing number of sequenced whole genomes has added to a growing list of Crp/Fnr family regulators identified in bacteria (9, 26); however, functions for most are not well defined.
Members of the Crp/Fnr family of global transcription regulators typically function as transcriptional activators. Previous studies on several bacterial species suggest that Crp/Fnr regulators are involved in responses to a variety of intracellular or extracellular signals, such as anoxia, carbon monoxide, temperature, nitric oxide, and oxidative and nitrosative stress (1, 13, 35, 42) and control metabolic pathways such as photosynthesis, nitrogen fixation, aromatic compound degradation, and respiration (11, 25, 31, 39). The E. coli Crp, also known as catabolite activator protein (CAP), regulates expression of over 100 genes, the majority of which are involved in energy metabolism (5). Fnr, the other Crp/Fnr-type transcriptional regulator in E. coli, positively or negatively regulates the expression of approximately 40 genes under anaerobic conditions, including the fumarate reductase, nitrate and nitrite reductase, and cytochrome d oxidase genes (29). It does so by its direct interactions with molecular oxygen to regulate the metabolic transition between aerobic and anaerobic growth (23, 41). In contrast, Rhodopseudomonas palustris HbaR, another Crp/Fnr-type transcriptional factor, regulates gene expression of the 4-hydroxybenzoate coenzyme A ligase involved in the initial enzymatic step of aromatic compound degradation (8, 11, 14). CooA, the only known heme protein of the Crp/Fnr-type regulators, functions as a CO-sensing transcriptional regulator and positively regulates the coo operon encoding enzymes for the oxidation of CO to CO2 coupled with proton reduction in Rhodospirillum rubrum (17, 38), highlighting the functional diversity of the Crp/Fnr-type regulators.
Analysis of the genome of the model sulfate-reducing bacterium (SRB) Desulfovibrio vulgaris Hildenborough revealed four annotated Crp/Fnr-type regulators (DVU0379, DVU2097, DVU2547, and DVU3111), suggesting potential functional differentiations among the multiple Crp/Fnr-type regulators, which exhibit a remarkable ability to survive and adapt to environmental perturbations and stress conditions (32, 40, 46, 52). Indeed, functional genomics studies of the response of D. vulgaris to various stress conditions have shown that the expression of the four Crp/Fnr-type transcriptional regulator genes significantly changed when D. vulgaris cells were exposed to nitrate (15), nitrite (16), oxygen (37), air (37), H2O2 (51), ethanol (unpublished data [http://www.microbesonline.org]), heat (7), or acetone (unpublished [http://www.microbesonline.org]) stress, suggesting that Crp/Fnr-type global regulators may play important roles in regulating a variety of stress responses in D. vulgaris. However, the functions and specificity of the Crp/Fnr-type regulators in D. vulgaris remain unclear.
Therefore, the objectives of this study were to (i) examine the activities of the four fnr/crp-type regulatory genes in the control of stress responses and (ii) identify the potential specificity of each Crp/Fnr-type regulator for various stress conditions in D. vulgaris. To address these objectives, knockout mutants for each Crp/Fnr regulator were generated and functionally characterized by their response to various environmental stress conditions that included air, NaCl, nitrite, and Cr(VI) by growth phenotype, transcriptional profiling, and competition analysis. Our results indicate that these four Crp/Fnr-type global transcriptional regulators have distinct functions in D. vulgaris in response to various environmental conditions.
MATERIALS AND METHODS
Phylogenetic analysis of Crp/Fnr superfamily members.
An initial data set of all annotated Crp/Fnr genes in all currently available SRB genomes was compiled by extracting the genes from the Joint Genome Institute Integrated Microbial Genomes database (33). Additional reference sequences were also extracted based on homology to SRB sequences to obtain a representative data set of the major Crp/Fnr subfamilies. The final data set containing 1,588 Crp/Fnr sequences was aligned using MUSCLE (10). The evolutionary history was inferred by the neighbor-joining method (44). The percentages of replicate trees in which the associated taxa clustered together were analyzed with the bootstrap test (500 replicates) (12). The evolutionary distances were computed by the Poisson correction method (53), and units represent the number of amino acid substitutions per site. The rate of variation among sites was modeled with a gamma distribution (shape parameter = 0.7241). All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons. Phylogenetic analyses were conducted in MEGA4 (47). The final tree was prepared for publication with Interactive Tree of Life (IToL) software (28).
Bacterial strains, growth conditions, and biomass production.
D. vulgaris was cultured in defined lactate/sulfate medium (LS4D) (36) at 37°C. A knockout mutant for each crp/fnr pair was generated by a marker replacement approach (Table 1) (2). For mutant complementation, the entire operons of DVU2097, DVU2547, and DVU3111 or a partial operon of DVU0379 as well as the upstream and downstream sequences were included in the complementation plasmids (Table 1). The DNA fragments were amplified from wild-type (WT) D. vulgaris genomic DNA (gDNA) by PCR with Phusion high-fidelity DNA polymerase (Finnzymes Oy, Finland) and the primers listed in Table S1 in the supplemental material and were cloned into pMO719, which is stably replicated in D. vulgaris (22). The plasmids were propagated in Escherichia coli DH5α, and the correctness of the construct was confirmed with Sanger sequencing. The complementation plasmids carrying the right inserts were electroporated (1,750 V, 1-mm gap) into competent cells of each crp/fnr knockout mutant. The transformants were confirmed by plasmid isolation and digestion of plasmid DNA.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description and relevant feature(s)a | Source or reference |
|---|---|---|
| Strains | ||
| D. vulgaris Hildenborough | Wild type; NCIMB 8303 | 19 |
| D. vulgaris Hildenborough JW9007 | ΔDVU0379, Kmr | This study |
| D. vulgaris Hildenborough JW9009 | ΔDVU2097, Kmr | This study |
| D. vulgaris Hildenborough JW9011 | ΔDVU2547, Kmr | This study |
| D. vulgaris Hildenborough JW9013 | ΔDVU3111, Kmr | This study |
| Plasmids | ||
| pMO719 | Desulfovibrio shuttle vector; SRB replicon pBG1 (EcoRI-EcoRI from pSC27) ligated to the EcoRI-bounded pCR8/GW/TOPO fragment; Specr | 22 |
| pMO719-DVU0379 | Complementation plasmid for JW9007; DNA fragment 425654-429479 (3,826 bp) cloned into the EcoRV site in pMO719; chromosome position of DVU0379, 428780-429457; Specr | This study |
| pMO719-DVU2097 | Complementation plasmid for JW9009; DNA fragment 2185067-2186208 (1,142 bp) cloned into the EcoRV site in pMO719; chromosome position of DVU2097, 2185336-2186007; Specr | This study |
| pMO719-DVU2547 | Complementation plasmid for JW9011; DNA fragment 2659090-2660860 (1,771 bp) cloned into the EcoRV site in pMO719; chromosome position of DVU2547, 2659978-2660664; Specr | This study |
| pMO719-DVU3111 | Complementation plasmid for JW9013; DNA fragment 3260525-3261583 (1,059 bp) cloned into the EcoRV site in pMO719; chromosome position of DVU3111, 3260648-3261382; Specr | This study |
Kmr, kanamycin resistance; Specr, spectinomycin resistance.
To test the effects of nitrite, NaCl, and chromate on D. vulgaris growth, the wild-type (WT) strain and four knockout mutants were inoculated into LS4D medium containing different concentrations of nitrite (0 mM, 0.5 mM, 1.0 mM, and 1.5 mM), NaCl (0 mM, 200 mM, 300 mM, and 400 mM), or chromate (0 mM, 0.5 mM, 0.7 mM, and 0.9 mM). An anaerobic titanium solution [titanium (III) chloride (20%) (wt/vol), sodium citrate (0.2 M), and sodium carbonate (8% wt/vol)] was used as a reductant for the LS4D medium before inoculation. To avoid an abiotic reaction, cysteine HCl (0.025%) prepared in NaHCO3 (8 mM) was used as a reductant in the chromate-related stress response test. For air stress conditions, 10 ml of air was aseptically injected into the mid-exponential-phase cell culture (10 ml of culture with 20 ml of headspace with N2 gas) and 10 ml of the headspace gas was removed before air injection. Cell growth was monitored at 600 nm with a spectrophotometer.
For the transcriptomic analysis, mid-exponential-phase (optical density at 600 nm [OD600] of about 0.4) cell cultures of WT D. vulgaris and the four Crp/Fnr mutants were harvested and flash-frozen in liquid nitrogen. Three biological replicates were prepared for each sample.
Microarray database search.
A survey of the gene transcription level changes of four crp/fnr genes as well as recA in D. vulgaris under different growth conditions or stress conditions was carried out by searching the microarray database (http://www.microbesonline.org/cgi-bin/microarray/viewExp.cgi). Most of the data sets have been published and deposited in the NCBI GEO database as follows: nitrate stress (accession no. GSE20079), nitrite stress (16), air stress (GSE8196), H2O2 stress (GSE21441), ethanol stress (GSE7489), heat shock stress (7), and salt adaptation stress (GSE14343).
Reverse transcription-PCR.
Expression patterns of four crp/fnr genes in cultures of cells at the mid-exponential-growth phase and the stationary-growth phase and of cells in the mid-exponential-growth phase harvested at 60 min after the application of different stressors such as 300 mM NaCl, 2.0 mM NaNO2, 0.5 mM potassium chromate [Cr(VI)], and 20 ml of air were determined. The application of air stress was performed as described above. Isolation of total RNA and on-column DNase digestion were performed as described previously (7). Total RNAs (1.5 μg) were reverse transcribed with SuperScript reverse transcriptase (Qiagen, Valencia, CA) in a final reaction volume of 20 μl following the manufacturer's manual. A 1-μl volume of the reverse transcription product was used for PCR to amplify the full-length open reading frame (ORF) of crp/fnr genes (28 cycles) and the recA control gene (25 cycles) with primers listed in Table S1 in the supplemental material.
Transcriptional profiling assay.
Isolation, fluorophore dye labeling, and handling of total RNA or gDNA were performed as described previously (48). DNA microarrays used in this study covered 3,482 of the 3,531 annotated protein-coding sequences of the D. vulgaris genome (16). Each array was hybridized with Cy3-labeled gDNA (as a control for normalization) and Cy5-labeled cDNA on a Tecan HS4800 hybridization station (Tecan Group, Durham, NC). Hybridization conditions as well as washing conditions and performance of image scanning were as described previously (7, 16, 36, 51). The microarray raw data processing was performed as described previously (36).
The similarities of the transcription profiles of the crp knockout mutants were analyzed with detrended correspondence analysis (DCA). The transcript level of each ORF was calculated as the ratio of signal intensity of Cy5 to that of Cy3, and the genes detected in at least two out of three replicates were kept for statistical analysis. The ratio of (Cy5/Cy3mutant)/(Cy5/Cy3wildtype) with three biological replicates for each mutant was used for DCA with PC-ORD (version 5; MjM Software Design). The heat map of gene expression data was rendered as vector graphics in Encapsulated PostScript format with JColorGrid (20).
Cr(VI) reduction assay with washed cell.
Experiments with washed cells were conducted to determine the chromate reduction capability of the WT strain and the four crp/fnr knockout mutants. Mid-exponential-phase cells (100 ml each; OD600 of approximately 0.4) were anoxically harvested by centrifugation. After being washed twice with anoxic bicarbonate buffer (30 mM NaHCO3, 62 mM sodium lactate, 20 mM Na2SO4, 0.025% cysteine HCl), the cells were resuspended in 1 ml of anoxic bicarbonate buffer. Then, 0.2-ml cells were transferred to 9.8 ml of anoxic bicarbonate buffer containing 0.5 mM K2CrO4 and the culture was incubated at 37°C. Samples taken at different time points were centrifuged, and the supernatants were used for quantification of Cr(VI) (30). Cr(VI) concentrations were determined colorimetrically (OD540) using the diphenylcarbazide method with a Hach ChromaVer 3 chromium reagent powder pillow (Hach Company, Loveland, CO) (49).
Fitness assay.
WT D. vulgaris and the four crp/fnr knockout mutants were subjected to competition under standard growth conditions (LS4D) or stress conditions with extra NaCl (200 mM), nitrite (0.1 mM), or chromate (0.2 mM) in LS4D. Low concentrations of stressors were chosen to obtain enough biomass at the sampling time point. As mentioned above, cysteine HCl (0.025%) was used as a reductant for chromate stress. For competition under air stress conditions, 10 ml of air was injected into the medium about 1 h after the reduction of the medium. All strains were revived, and approximately equal numbers of cells were mixed and inoculated into the competition media and allowed to compete for 2 days. Samples were taken at day 0 and day 2. Genomic DNA was isolated from each sample and quantified with PicoGreen (Quant-iT PicoGreen dsDNA assay kit; Invitrogen). Equal amounts of template DNA (20 ng) were used for real-time PCR with strain-specific PCR primers (see Table S1 in the supplemental material). Copy numbers were used to represent the cell numbers of each strain in the population. Fitness (w) was calculated as follows:
Microarray data accession number.
Microarray data for this study have been deposited in the NCBI GEO database under accession number GSE29380.
RESULTS
Phylogeny of D. vulgaris Crp/Fnr homologs.
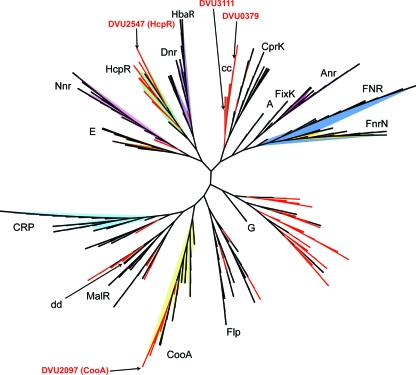
A total of 1,588 protein sequences, including all annotated Crp/Fnr proteins from sequenced SRB genomes, were used for phylogenetic analysis. As shown in Fig. 1, the four D. vulgaris Crp/Fnr homologs are localized in three subfamilies. DVU0379 and DVU3111 are both located in the cc subfamily, which is constituted entirely of SRB sequences. DVU2547 is located in the HcpR branch and is highly conserved in most SRB species analyzed. In addition, DVU2097 is located in the CooA subfamily with other SRB genes and its localization in the genome with the coo operon suggests that it is the likely regulator of carbon monoxide dehydrogenase operon expression. While it contains no D. vulgaris genes, the phylogeny also revealed a novel clade (related to subfamily G) containing numerous SRB genes. The phylogenetic positioning of the SRB genes suggests that the Crp/Fnr homologs in D. vulgaris and other SRB species may have different functions in gene regulation and that the diversity of crp/fnr genes in SRB species is much greater than originally envisioned.
Fig 1.
Phylogenetic analysis of Crp/Fnr superfamily enzymes (1,588 proteins) with an emphasis on SRB sequences. Red indicates genes from SRB species. Established subfamily names are marked for each clade. Multiple sequence alignment was conducted using MUSCLE. The evolutionary history was inferred using the neighbor-joining method. The optimal tree with the sum of branch lengths = 394.5 is shown. The percentage of replicate trees in which the associated taxa clustered together was assessed using the bootstrap test (500 replicates). Branch lengths are ignored for clarity. The evolutionary distances were computed using the Poisson correction method, and units represent the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.7241). All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons. Phylogenetic analyses were conducted in MEGA4. The final tree was prepared using the Interactive Tree of Life (IToL) software.
Expression changes of crp/fnr under different stress or growth conditions.
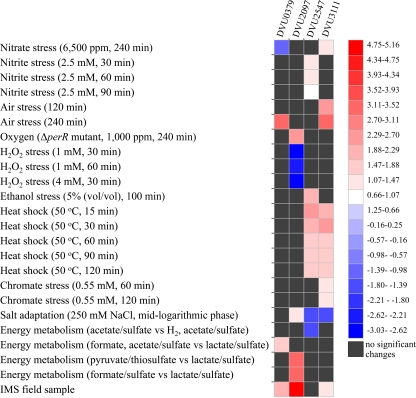
To examine whether these four crp/fnr homologs are actively expressed, gene expression changes of the four genes were examined with a currently available microarray database. As shown in Fig. 2, the four genes were differentially transcribed in stress responses or growth conditions with different electron donors or acceptors. For instance, the DVU2547 expression increased in the nitrite stress response and DVU3111 in the chromate stress response; the expression of both DVU2547 and DVU3111 significantly increased in the heat shock response; DVU2097, DVU2547, and DVU3111 altered their expression under conditions of salt (NaCl) adaptation; expression of DVU2097 and DVU0379 increased when D. vulgaris was cultured with different electron donors or acceptors; and the expression of DVU0379, DVU2097, and DVU3111 increased under conditions of oxygen stress, but expression of DVU2097 decreased in the H2O2 stress response. Similar expression changes of four crp/fnr genes under stress conditions, in addition to their expression at both the mid-exponential phase and stationary phase, were detected with reverse transcription-PCR (see Fig. S2 in the supplemental material). It is likely that these regulators are involved in various stress responses but play different roles to some extent.
Fig 2.
Expression changes of the four crp/fnr genes under different stress conditions or with different electron donors or acceptors. JColorgrid was used to generate the heat map.
Characterization of knockout mutants for crp/fnr.
To further investigate the function of the four Crp/Fnr-type regulator genes, single knockout mutants for each crp/fnr gene were generated (Table 1) and confirmed by PCR amplification of the full-length open reading frames (see Fig. S1 in the supplemental material). The growth phenotypes of crp/fnr knockout mutants under different environmental stress conditions, including air, NaCl, nitrite, and chromate stress, were examined.
(i) Air stress.
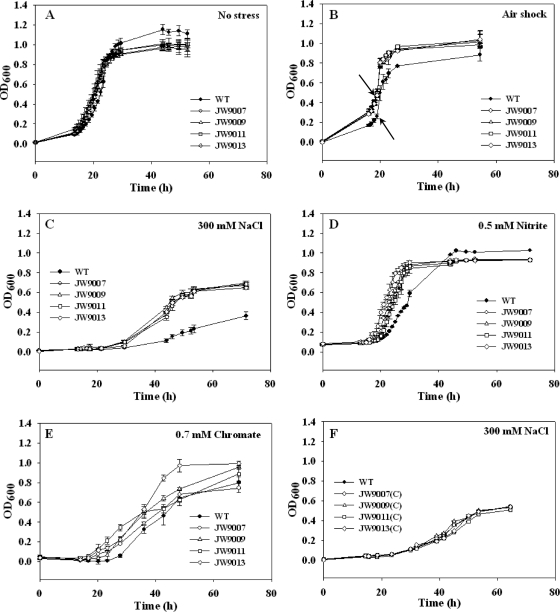
To find out whether these crp/fnr ortholog regulators are involved in defense against increased oxygen levels, the growth phenotypes of each crp/fnr knockout mutant in response to air shock were examined. The growth of WT D. vulgaris and the growth of crp/fnr knockout mutants under standard growth conditions were quite similar except for a higher final optical density in the WT culture (Fig. 3A). Although the growth of WT and knockout mutants was inhibited by air shock, the four knockout mutants were more resistant to air stress conditions, with quicker recovery from the stress and higher biomass yield (Fig. 3B). The results suggest the involvement of these crp/fnr regulators in response to altered oxygen levels in D. vulgaris.
Fig 3.
Growth phenotypes of the four crp/fnr knockout mutants. (A) no stress; (B) air shock; (C) 300 mM NaCl; (D) 0.5 mM nitrite; (E) 0.7 mM chromate (VI); (F) growth phenotype of complemented mutants with extra 300 mM NaCl in the medium. The standard deviations of the results from four biological replicate experiments are shown as error bars. The arrows in panel B indicate the addition of air (10 ml).
(ii) Salt stress.
Growth inhibition by extra NaCl (200 mM or 300 mM) in the medium was observed in this study. Very limited growth was observed with the medium supplemented with 400 mM NaCl. Interestingly, all crp/fnr mutants had increased resistance to NaCl at all concentrations tested. As an example, growth phenotypes of the WT and mutants with extra 300 mM NaCl in the medium are shown in Fig. 3C. Higher growth rates, shorter lag phases, and higher biomass yields were observed for mutants. These data indicate that crp/fnr regulators are involved in the response of D. vulgaris to NaCl stress.
(iii) Nitrite stress.
Growth of D. vulgaris was inhibited by nitrite in the medium. With 0.5 mM nitrite in the medium, longer lag phase was observed for both WT and all crp/fnr knockout mutants. A lower final biomass yield was obtained from mutants, although they grew faster than WT D. vulgaris (Fig. 3D). Growth of all strains was further delayed with 1.0 mM nitrite in the medium, and complete growth arrest was observed for all strains with 1.5 mM nitrite in the medium (data not shown). These results provide evidence for the involvement of crp/fnr genes in nitrite stress response.
(iv) Chromate stress.
Growth was slightly stimulated for both WT and four mutants with a low concentration (0.5 mM) of Cr(VI) in the medium (data not shown). However, growth was inhibited by an increase in the Cr(VI) concentration (e.g., 0.7 mM) in the medium, with strain JW9013 containing a deletion of DVU3111 showing the highest growth rate (Fig. 3E). With 0.9 mM chromate in the medium, the growth of all crp/fnr mutants was completely inhibited; WT D. vulgaris was able to survive but produced only half of the final biomass seen with WT in the absence of stress (data not shown). These results indicate that these four regulators are involved in chromate response and that DVU3111may play more important roles in the response of D. vulgaris to chromate.
To further confirm whether the phenotypes observed in knockout mutants resulted from the deletion of each specific crp/fnr gene, the phenotypes of the complemented knockout mutants carrying the WT copy of crp/fnr and the flanking upstream and downstream sequences were analyzed under conditions of NaCl stress, which resulted in the most conspicuous changes in growth between the knockout mutants and WT strain. As expected, WT-like phenotypes were observed for all complemented strains (Fig. 3F), indicating that these crp/fnr genes were indeed responsible for the observed phenotypes under these conditions.
Altered global gene transcription profiling in crp/fnr knockout mutants.
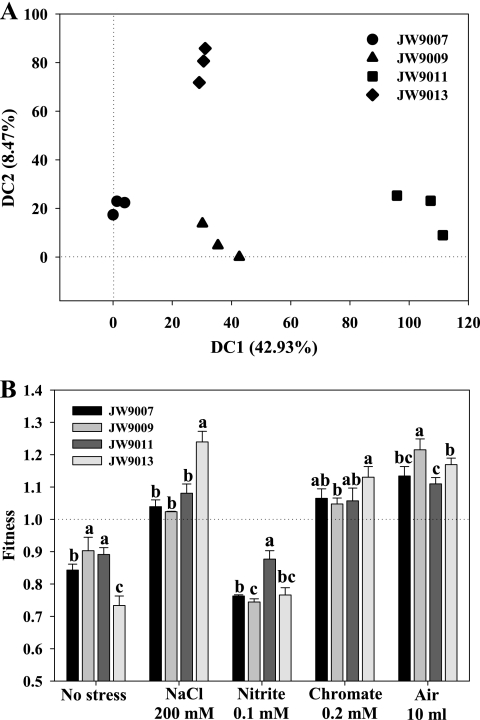
In order to better understand the roles of Crp/Fnr-type regulators in D. vulgaris, the global transcriptional profiling of the four knockout mutants at the mid-exponential phase was analyzed. Compared with the WT strain, a small percentage of D. vulgaris genes with a significant change in expression (|log2 R| > 1.5; Z score > 1.5) was detected in each knockout mutant, with 136 in JW9013, 33 in JW9011, 31 in JW9009, and 23 in JW9007. Nevertheless, gene transcription profiling showed that the four crp/fnr mutants were well separated in a DCA plot (Fig. 4A). The gene transcription profiling of JW9011 was quite different from that of other three mutants, and the profile was separated by from the others by axis 1, which explains 42.93% of the total variance among the four mutants. The transcriptional profile of JW9013 was separated from those of JW9007 and JW9011 by axis 1 and that of JW9009 by axis 2, which explains 8.47% of the total variance. In addition, the transcriptional profile of JW9009 was separated from that of JW9007 by axis 1 (Fig. 4A). The greater number of transcriptionally changed genes detected in JW9013 suggested that DVU3111might be involved in more cellular pathways than other crp/fnr gene products in D. vulgaris. A further analysis of transcription profiling demonstrated that the four crp/fnr products regulate both common and distinct genes (Table 2). A few categories of clusters of orthologous groups (COG) consisting of genes involved in stress responses, energy metabolism, and signal transduction are described below, further supporting the distinct functions of the four crp/fnr regulators in stress responses of D. vulgaris.
Fig 4.
Transcriptional profiling of crp/fnr mutants analyzed by DCA (A) and the fitness changes of the crp/fnr mutants compared to the WT under the standard growth and stress conditions (B). Each symbol in panel A indicates microarray data from one biological replicate experiment. Fitness data represent averages of the results of three biological replicate experiments, with the standard deviations shown as error bars. A pairwise t test was performed to assess the significance of fitness among four mutants, with a, b, and c indicating significant differences at the P < 0.05 level.
Table 2.
Selected genes with significant expression level changes in crp/fnr knockout mutants
| Regulator and category | Gene name | Annotation | Log2R (mutant/wild type)a |
|||
|---|---|---|---|---|---|---|
| JW9007 | JW9009 | JW9011 | JW9013 | |||
| Amino acid transport and metabolism | ||||||
| DVU0465 | trpE | Anthranilate synthase, component I | 1.33 (1.78) | 0.23 (0.32) | N/A | N/A |
| DVU0466 | trpG | Anthranilate synthase, glutamine amidotransferase | 1.52 (2.35) | 0.08 (0.12) | −1.33 (−1.66) | −0.81 (−0.60) |
| DVU0467 | trpD | Anthranilate phosphoribosyltransferase | 1.44 (2.24) | −0.17 (−0.18) | −0.32 (−0.49) | −0.08 (−0.12) |
| DVU0468 | trpC | Indole-3-glycerol phosphate synthase | 1.23 (2.08) | 0.12 (0.21) | −0.90 (−1.41) | −0.19 (−0.28) |
| DVU0469 | trpF-1 | N-(5-phosphoribosyl)anthranilate isomerase | 0.88 (1.30) | −0.17 (−0.24) | −1.00 (−1.45) | −0.13 (−0.16) |
| DVU0470 | trpB-2 | Tryptophan synthase, beta subunit | 1.43 (2.52) | 0.34 (0.56) | −0.56 (−0.95) | 0.15 (0.25) |
| DVU0471 | trpA | Tryptophan synthase, alpha subunit | 1.48 (2.66) | 0.42 (0.70) | −0.44 (−0.74) | 0.32 (0.54) |
| DVU0386 | glnH | Amino acid ABC transporter, periplasmic amino acid binding protein | 0.55 (1.01) | 1.83 (3.32) | 1.64 (2.89) | 1.51 (2.76) |
| Inorganic ion transport and metabolism | ||||||
| DVU2571 | feoB | Ferrous iron transport protein B | −2.11 (−2.53) | −1.81 (−2.16) | −1.32 (−2.14) | −1.78 (−1.96) |
| DVU2572 | feoA | Ferrous iron transport protein A | −1.69 (−2.51) | −1.73 (−2.17) | −1.50 (−2.25) | −1.75 (−1.97) |
| DVU2573 | N/A | Hypothetical protein | −0.62 (−0.96) | −0.52 (−0.85) | −0.64 (−1.13) | −0.46 (−0.62) |
| DVU2574 | feoA | Ferrous iron transporter component feoA | −2.17 (−2.79) | −1.34 (−1.30) | −1.70 (−2.51) | −2.00 (−2.98) |
| Energy production and conversion | ||||||
| DVU3171 | N/A | Cytochrome c3 | 0.63 (0.64) | 1.26 (1.30) | 0.88 (0.93) | 1.45 (1.52) |
| DVU2784 | lldD | Dehydrogenase, flavin mononucleotide -dependent family | 0.63 (0.97) | 1.36 (2.32) | 1.07 (1.90) | 1.51 (2.15) |
| DVU3143 | N/A | Iron-sulfur cluster-binding protein | 1.30 (1.68) | 0.97 (1.32) | 1.21 (1.72) | 1.74 (2.08) |
| DVU1165 | ndh | NADH respiratory dehydrogenase | −0.54 (−0.55) | −0.80 (−0.66) | −0.75 (−1.06) | −1.42 (−2.27) |
| DVU2543 | b0873 | Hybrid cluster protein | −0.72 (−1.07) | −1.22 (−1.81) | −2.79 (−3.61) | −1.46 (−1.96) |
| DVU2544 | N/A | Iron-sulfur cluster binding protein | −0.45 (−0.59) | −0.82 (−0.95) | −1.26 (−1.32) | −1.06 (−1.33) |
| DVU2680 | fld | Flavodoxin, iron repressed | −2.66 (−2.64) | −2.64 (−2.44) | −2.91 (−3.67) | −2.78 (−3.26) |
| DVU2247 | ahpC | Alkyl hydroperoxide reductase C | −1.41 (−2.47) | −0.29 (−0.47) | −0.34 (−0.61) | −0.74 (−1.33) |
| Transcription | ||||||
| DVU2577 | N/A | DNA-binding response regulator, LuxR family | 1.60 (2.53) | 1.15 (1.76) | 0.71 (1.09) | 1.01 (1.57) |
| DVU2785 | pdhR | Transcriptional regulator, GntR family | 1.28 (2.00) | 1.46 (2.14) | 1.16 (1.82) | 1.94 (2.15) |
| DVU2827 | N/A | Sigma-54 dependent transcriptional regulator | −0.76 (−0.91) | −2.20 (−1.75) | −0.63 (−1.12) | −1.24 (−1.64) |
| Signal transduction mechanisms | ||||||
| DVU0878 | N/A | dnaK suppressor protein, putative | 2.13 (3.75) | 0.68 (1.13) | 0.19 (0.31) | 1.00 (1.83) |
| DVU0888 | N/A | Response regulator | 0.62 (0.67) | 0.72 (0.82) | 0.21 (0.24) | 1.81 (1.57) |
| DVU1169 | N/A | Methyl-accepting chemotaxis protein | 0.72 (0.77) | 1.57 (2.01) | 1.97 (2.83) | 1.94 (2.86) |
| DVU1594 | cheA-1 | Chemotaxis protein CheA | −0.98 (−1.33) | −1.07 (−1.27) | −1.40 (−2.25) | −1.71 (−2.56) |
| DVU1595 | cheR-1 | Chemotaxis protein methyltransferase | −0.54 (−0.66) | −1.33 (−1.04) | −1.55 (−2.03) | −1.76 (−2.47) |
| DVU3182 | dcrA | Methyl-accepting chemotaxis protein DcrA | −0.38 (−0.49) | −1.15 (−0.75) | −0.21 (−0.35) | −1.69 (−2.14) |
| DVU3155 | dcrH | Methyl-accepting chemotaxis protein DcrH | −1.28 (−1.36) | −1.83 (−1.62) | −1.29 (−1.53) | −1.75 (−2.20) |
| DVU2577 | N/A | DNA-binding response regulator, LuxR family | 1.60 (2.53) | 1.15 (1.76) | 0.71 (1.09) | 1.01 (1.57) |
| DVU0039 | N/A | C4-type zinc finger protein, DksA/TraR family | 0.58 (0.62) | 0.78 (0.77) | 0.82 (0.92) | 2.14 (1.80) |
| DVU0598 | N/A | Carbon starvation protein A, putative | −2.76 (−2.15) | −1.79 (−2.04) | −1.69 (−2.91) | −3.07 (−4.02) |
| DVU0599 | N/A | Carbon starvation protein A, putative | −1.38 (−1.23) | −1.44 (−1.32) | −1.05 (−1.09) | −1.69 (−1.87) |
| DVU3023 | atoC | Sigma-54-dependent DNA-binding response | −1.49 (−1.33) | −1.50 (−1.02) | −0.08 (−0.11) | −2.01 (−1.65) |
N/A, no gene name has been assigned to the locus yet or no data for the gene. Z scores are shown in parentheses.
(i) Amino acid transport and metabolism.
glnH (DVU0386) encoding an amino acid ABC transporter was found significantly upregulated in three mutants, JW9009, JW9011, and JW9013 (Table 2). In contrast, trp operon genes (DVU0465 to DVU0471 for tryptophan synthase) were upregulated in JW9007 only (Table 2). Increased expression of trp operon genes was observed when mid-exponential-phase D. vulgaris cells encountered NaCl shock (36), and glutamine was accumulated in salt-adapted D. vulgaris (18). Therefore, these gene expression changes may contribute to the enhanced salt resistance phenotype of the four crp/fnr mutants.
(ii) Inorganic ion transport and metabolism.
Changes in the expression of the genes involved in inorganic ion transport, particularly iron homeostasis, were found in all crp/fnr mutants. The expression of the primary iron uptake system genes, feoAB (DVU2571 to DVU2574), decreased significantly in all crp/fnr mutants (Table 2). The decreased expression of the feo system in the knockout mutants provided evidence for the involvement of these crp/fnr regulators as activators for the acquisition of Fe(III).
(iii) Energy production and conversion.
The expression of a hybrid cluster protein-encoding gene, hcp2 (DVU2543), a predicted target gene for DVU2547 (43), was significantly downregulated in JW9011 (Table 2). Also, gene DVU2544, encoding an iron-sulfur cluster binding protein in the same operon, was concurrently downregulated in JW9011 (Table 2). Downregulation of these two genes was observed in JW9013 as well (Table 2). Another gene, fld (DVU2680), predicted to be an iron-repressed flavodoxin, was significantly downregulated in all crp/fnr mutants (Table 2). Moderately increased expression of DVU3171 (a cytochrome c3-encoding gene) was detected in JW9013 and JW9009 (Table 2). Cytochrome c3 was previously reported to be the major electron carrier for metal ion reduction (19), and the upregulation of this gene may be related to an increased chromate reduction capacity in crp/fnr mutants. The alkyl hydroperoxide reductase C gene ahpC (DVU2247), known for reducing hydrogen peroxide (H2O2) into H2O (6), was downregulated in JW9007 (Table 2), suggesting the possible role of DVU0379 in oxidative stress response.
(iv) Transcription.
The same trend of changes was found in all crp/fnr mutants. For example, increased expression of genes for a GntR family transcription regulator, pdhR (DVU2785), and a LuxR family transcriptional regulator (DVU2577) was detected in all mutants (Table 2). Also, expression of DVU2827, a sigma-54-dependent transcriptional regulator, decreased in all mutants. Future work is needed to reveal the relationship between Crp/Fnr and other transcription regulators, which may form complex networks in D. vulgaris.
(v) Signal transduction mechanisms.
Many genes involved in signal transduction mechanisms were found to be shared by some or all crp/fnr mutants (Table 2). Expression of two “carbon starvation protein A” genes in the operon DVU0598-DVU0599 was significantly downregulated in all crp/fnr mutants. Also, expression of a predicted methyl-accepting chemotaxis protein (MCP) encoded by DVU1169 increased significantly in three crp/fnr mutants (but not the JW9007 mutant), and expression of chemotaxis protein-encoding gene cheA-1(DVU1594) and chemotaxis protein methyltransferase-encoding gene cheR-1 (DVU1595) decreased more than 2-fold in three crp/fnr mutants (but not the JW9007 mutant). Decreased expression of MCP gene dcrA (DVU3182) in JW9009 and JW9013 and dcrH (DVU3155) in all mutants was detected. dcrA and dcrH were previously reported to be involved in locating SRB in the gradients of oxic and anoxic interfaces in pipelines or sewers (3). Therefore, these results suggest the roles of the four crp/fnr regulators in regulation of alternative energy pathways during carbon starvation and DVU2097, DVU2547, and DVU3111 in cell chemotaxis.
Functions of crp/fnr regulators confirmed by competition assay.
The assay of competition between the WT strain and the four knockout mutants provided further evidence of the specific contributions of each crp/fnr gene in response to different stresses (Fig. 4B). A significant (P ≤ 0.05) decrease of fitness was observed for all crp/fnr mutants compared to the WT strain under the standard growth conditions, and significant (P ≤ 0.05) differences were observed among mutants, with the lowest fitness exhibited by JW9013, medium fitness exhibited by JW9007, and the best fitness exhibited by JW9009 and JW9011 (but with no significant difference between these two mutants). Consistent with the growth phenotype, a fitness decrease was also observed for all knockout mutants under nitrite stress conditions, and significant (P ≤ 0.05) differences occurred among four mutants, with the highest fitness for JW9011, the lowest for JW9009, and medium fitness for JW9007, but no significant difference between JW9009 and JW9013 or between JW9007 and JW9013. In contrast, all four mutants generally showed improved fitness under air, NaCl, or chromate stress conditions compared to the WT. Under NaCl stress conditions, the highest fitness was seen in JW9013, but no significant difference was observed for the three other mutants. Under chromate stress conditions, the highest fitness was found in JW9013, and significant differences appeared only between JW9009 and JW9013. Under air shock conditions, the highest fitness was observed in JW9009, and significant differences were observed between JW9009 and other three mutants and also between JW9011and JW9013 (Fig. 4B). These results indicate that each crp/fnr gene product performs a different role(s) in response to different stresses. For example, more important roles were seen with DVU3111 in response to NaCl or chromate stresses, DVU2547 in response to nitrite stress, and DVU2097 in response to air stress.
Increased chromate reduction in crp/fnr knockout mutants.
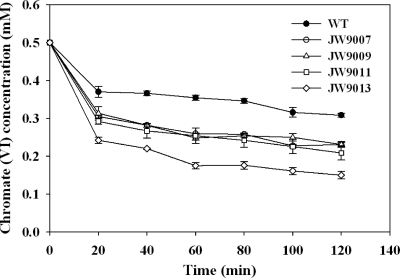
To further confirm the function of Crp/Fnr regulators in hexavalent chromate Cr(VI) reduction, experiments with washed cells were conducted to examine the Cr(VI) reduction capability of each strain. Increased Cr(VI) reduction was observed in four crp/fnr mutants with 0.5 mM Cr(VI) present in the medium (Fig. 5). Consistent with the fitness data, the highest chromate reduction rate was observed in JW9013.
Fig 5.
Cr(VI) reduction in experiments with washed cells. The error bars represent the standard deviations.
DISCUSSION
Regulation of gene expression is critical for bacterial response and survival in an ever-changing environment. Crp/Fnr regulators are global transcriptional factors present in many bacteria and serve key roles in response to environmental stresses. In this study, taking advantage of bioinformatics, mutagenesis, high-throughput transcriptomics, and competition analysis, functional characterization of four annotated Crp/Fnr-type regulators in D. vulgaris was performed to better understand the physiological responses associated with the regulators.
Each of the crp/fnr regulators may have distinct functions in response to different environmental stresses in D. vulgaris. First, the overall similarities of the transcriptional profiles of each mutant are low (Fig. 4A). The differences were also obvious in terms of significantly changed gene numbers as well as the genes involved in different functional processes (Table 2). Second, fitness analysis of four crp/fnr mutants indicated that each crp/fnr mutant had different contributions in response to different stresses. The highest fitness was observed for JW9013 under NaCl stress or chromate stress conditions. JW9011 had the highest fitness under conditions of nitrite stress, and highest fitness was observed for JW9009 under air stress conditions. Third, an analysis of chromate reduction provided further evidence for more important functions of DVU3111 in chromate reduction. Besides this experimental evidence, the phylogenetic position of the four Crp/Fnr homologs in three different branches and subfamilies also suggests different functions for the crp/fnr genes in D. vulgaris.
Results from this study suggest that four crp/fnr regulators D. vulgaris may function differently from E. coli fnr but similarly to E. coli crp to some extent. In E. coli, nitrate and nitrite reductases have been shown to be regulated by Fnr (29); however, no significant changes in expression of nitrite reduction genes (e.g., the nrfHA operon encoding cytochrome c nitrite reductase) were observed in crp/fnr mutants of D. vulgaris. Increased resistance to air stress in crp/fnr knockout mutants suggested that crp/fnr regulators may act as transcriptional repressors in D. vulgaris, which is different from E. coli Fnr functioning as a global regulator under anaerobic growth conditions (21, 41). Decreased expression of two carbon starvation protein A genes (DVU0598 and DVU0599) in all crp/fnr mutants suggested the possible similarity between crp/fnr regulators in D. vulgaris and E. coli crp, which positively regulates about two-thirds of carbon starvation genes induced at the onset of carbon starvation (45).
Cooperation among crp/fnr and other transcriptional regulators in stress responses was also suggested by results from this study. Fur regulon genes such as the iron uptake system feoAB genes are repressed by Fur, and trp operon is repressed by iron-free Fur (2). A significant decrease in expression of feoAB and the iron-repressed flavodoxin gene fld (DVU2680) in the four crp/fnr mutants and upregulation of the trp operon in JW9007 (Table 2) suggested the possibility of cross-talk between Crp/Fnr and Fur. On the other hand, expression of DVU2543, one of the predicted target genes of DVU2547 (43), was found to be significantly downregulated in JW9011. However, no significant expression changes of other predicted target genes such as apsAB, sat, DVU1080-DVU1081, and genes in the coo operon (43) were found in this study. It is possible that additional cooperating factors contribute to the regulation of these genes. If such cooperation exists in D. vulgaris, complicated gene regulatory networks are expected.
In summary, the systematic study provided evidence for distinct functions of the crp/fnr regulators in D. vulgaris: for example, the roles of DVU3111 in responses to NaCl or chromate stresses, DVU2547 in nitrite stress response, and DVU2097 in air stress response. Future in-depth studies with double- or triple-knockout mutants and chromatin immunoprecipitation-microarray technology (ChIP-chip) should provide more and higher-resolution information about the functional redundancy and gene regulatory networks of Crp/Fnr regulators in D. vulgaris.
Supplementary Material
ACKNOWLEDGMENT
This work conducted by ENIGMA (Ecosystems and Networks Integrated with Genes and Molecular Assemblies) was supported by the Office of Science, Office of Biological and Environmental Research, U.S. Department of Energy, under contract DE-AC02-05CH11231.
Footnotes
Published ahead of print 9 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Aono S, Nakajima H, Saito K, Okada M. 1996. A novel heme protein that acts as a carbon monoxide-dependent transcriptional activator in Rhodospirillum rubrum. Biochem. Biophys. Res. Commun. 228:752–756 [DOI] [PubMed] [Google Scholar]
- 2. Bender KS, et al. 2007. Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 73:5389–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borenstein SB. 1994. Microbiologically influenced corrosion handbook. Industrial Press Inc, New York, NY [Google Scholar]
- 4. Botsford JL, Harman JG. 1992. Cyclic AMP in prokaryotes. Microbiol. Mol. Biol. Rev. 56:100–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199–213 [DOI] [PubMed] [Google Scholar]
- 6. Chae HZ, et al. 1994. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. U. S. A. 91:7017–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chhabra SR, et al. 2006. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188:1817–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dispensa M, et al. 1992. Anaerobic growth of Rhodopseudomonas palustris on 4-hydroxybenzoate is dependent on AadR, a member of the cyclic AMP receptor protein family of transcriptional regulators. J. Bacteriol. 174:5803–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dufour YS, Kiley PJ, Donohue TJ. 2010. Reconstruction of the core and extended regulons of global transcription factors. PLoS Genet. 6:e1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Egland PG, Harwood CS. 2000. HbaR, a 4-hydroxybenzoate sensor and FNR-CRP superfamily member, regulates anaerobic 4-hydroxybenzoate degradation by Rhodopseudomonas palustris. J. Bacteriol. 182:100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 13. Galimand M, Gamper M, Zimmermann A, Haas D. 1991. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J. Bacteriol. 173:1598–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gibson J, Dispensa M, Fogg GC, Evans DT, Harwood CS. 1994. 4-Hydroxybenzoate-coenzyme A ligase from Rhodopseudomonas palustris: purification, gene sequence, and role in anaerobic degradation. J. Bacteriol. 176:634–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He Q, et al. 2010. Impact of elevated nitrate on sulfate-reducing bacteria: a comparative study of Desulfovibrio vulgaris. ISME J. 4:1386–1397 [DOI] [PubMed] [Google Scholar]
- 16. He Q, et al. 2006. Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough, inferred from global transcriptional analysis. Appl. Environ. Microbiol. 72:4370–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He Y, Shelver D, Kerby RL, Roberts GP. 1996. Characterization of a CO-responsive transcriptional activator from Rhodospirillum rubrum. J. Biol. Chem. 271:120–123 [DOI] [PubMed] [Google Scholar]
- 18. He Z, et al. 2010. Global transcriptional, physiological, and metabolite analyses of the responses of Desulfovibrio vulgaris Hildenborough to salt adaptation. Appl. Environ. Microbiol. 76:1574–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heidelberg JF, et al. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22:554–559 [DOI] [PubMed] [Google Scholar]
- 20. Joachimiak M, Weisman J, May B. 2006. JColorGrid: software for the visualization of biological measurements. BMC Bioinformatics 7:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 187:1135–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keller KL, Bender KS, Wall JD. 2009. Development of a markerless genetic exchange system for Desulfovibrio vulgaris Hildenborough and its use in generating a strain with increased transformation efficiency. Appl. Environ. Microbiol. 75:7682–7691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kiley PJ, Beinert H. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341–352 [DOI] [PubMed] [Google Scholar]
- 24. Kiley PJ, Reznikoff WS. 1991. Fnr mutants that activate gene expression in the presence of oxygen. J. Bacteriol. 173:16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kolb A, Busby S, Buc H, Garges S, Adhya S. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62:749–797 [DOI] [PubMed] [Google Scholar]
- 26. Körner H, Sofia HJ, Zumft WG. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27:559–592 [DOI] [PubMed] [Google Scholar]
- 27. Lawson CL, et al. 2004. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 14:10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Letunic I, Bork P. 2007. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128 [DOI] [PubMed] [Google Scholar]
- 29. Li B, Wing H, Lee D, Wu H-C, Busby S. 1998. Transcription activation by Escherichia coli FNR protein: similarities to, and differences from, the CRP paradigm. Nucleic Acids Res. 26:2075–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X, Krumholz LR. 2009. Thioredoxin is involved in U(VI) and Cr(VI) reduction in Desulfovibrio desulfuricans G20. J. Bacteriol. 191:4924–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lopez O, et al. 2001. Regulation of gene expression in response to oxygen in Rhizobium etli: role of FnrN in fixNOQP expression and in symbiotic nitrogen fixation. J. Bacteriol. 183:6999–7006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lovley DR, Phillips EJP. 1994. Reduction of chromate by Desulfovibrio vulgaris and its c3 cytochrome. Appl. Environ. Microbiol. 60:726–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Markowitz VM, et al. 2010. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38:D382–D390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McKay DB, Steitz TA. 1981. Structure of catabolite gene activator protein at 2.9 Å resolution suggests binding to left-handed B-DNA. Nature 290:744–749 [DOI] [PubMed] [Google Scholar]
- 35. Mesa S, Hennecke H, Fischer HM. 2006. A multitude of CRP/FNR-like transcription proteins in Bradyrhizobium japonicum. Biochem. Soc. Trans. 34:156–159 [DOI] [PubMed] [Google Scholar]
- 36. Mukhopadhyay A, et al. 2006. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol. 188:4068–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mukhopadhyay A, et al. 2007. Cell-wide responses to low-oxygen exposure in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 189:5996–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakajima H, et al. 2001. Redox properties and coordination structure of the heme in the CO-sensing transcriptional activator CooA. J. Biol. Chem. 276:7055–7061 [DOI] [PubMed] [Google Scholar]
- 39. Oh J-I, Eraso JM, Kaplan S. 2000. Interacting regulatory circuits involved in orderly control of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 182:3081–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ouattara AS, Jacq VA. 1992. Characterization of sulfate-reducing bacteria isolated from Senegal ricefields. FEMS Microbiol. Ecol. 101:217–228 [Google Scholar]
- 41. Poole RK, et al. 1996. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 178:5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poole RK, Hughes MN. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36:775–783 [DOI] [PubMed] [Google Scholar]
- 43. Rodionov D, Dubchak I, Arkin A, Alm E, Gelfand M. 2004. Reconstruction of regulatory and metabolic pathways in metal-reducing delta-proteobacteria. Genome Biol. 5:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 45. Schultz JE, Matin A. 1991. Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J. Mol. Biol. 218:129–140 [DOI] [PubMed] [Google Scholar]
- 46. Spring S, et al. 2009. Complete genome sequence of Desulfotomaculum acetoxidans type strain (5575T). Stand. Genomic Sci. 1:242–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 48. Thompson DK, et al. 2002. Transcriptional and proteomic analysis of a ferric uptake regulator (Fur) mutant of Shewanella oneidensis: possible involvement of Fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl. Environ. Microbiol. 68:881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Viamajala S, Peyton BM, Apel WA, Petersen JN. 2002. Chromate/nitrite interactions in Shewanella oneidensis MR-1: evidence for multiple hexavalent chromium [Cr(VI)] reduction mechanisms dependent on physiological growth conditions. Biotechnol. Bioeng. 78:770–778 [DOI] [PubMed] [Google Scholar]
- 50. Vollack K-U, Härtig E, Körner H, Zumft WG. 1999. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol. Microbiol. 31:1681–1694 [DOI] [PubMed] [Google Scholar]
- 51. Zhou A, et al. 2010. Hydrogen peroxide-induced oxidative stress responses in Desulfovibrio vulgaris Hildenborough. Environ. Microbiol. 12:2645–2657 [DOI] [PubMed] [Google Scholar]
- 52. Zhou J, et al. 2011. How sulfate reducing microorganisms cope with stress: lessons from systems biology. Nat. Rev. Microbiol. 9:452–466 [DOI] [PubMed] [Google Scholar]
- 53. Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins, p 97–166 In Bryson V, Vogel HJ. (ed), Evolving genes and proteins. Academic Press, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.