Abstract
Plasmids pRSB113 and pRSB115 were recovered from an activated sludge bacterial community of a municipal wastewater treatment plant in Germany. Both plasmids carry the same blaGES-5 carbapenemase gene, located within two distinct class 1 integrons. These plasmids have different backbones, belong to different incompatibility groups, and could replicate in both Pseudomonas aeruginosa and Escherichia coli.
TEXT
The aquatic environment may be considered a reservoir for dissemination of antibiotic resistance determinants, since resistant bacteria, resistance genes, and mobile genetic elements carrying resistance determinants have been isolated frequently from bacterial communities residing in lakes, rivers, or wastewater treatment plants (WWTP) (10, 11, 12, 13, 22, 29, 31). Genes encoding carbapenem-hydrolyzing β-lactamases have been identified in several water samples, such as those encoding the Ambler class A β-lactamases IMI-2 from Enterobacter asburiae (1, 2) or BIC-1 from Pseudomonas fluorescens (11), the class B β-lactamase VIM-2 from Pseudomonas pseudoalcaligenes (27), or the class D β-lactamase OXA-23 from Acinetobacter baumannii (10). Ambler class A GES-type carbapenemases (GES-2, GES-4, GES-5, and GES-8) are inhibited by clavulanic acid, and the corresponding genes, located on plasmids, have been identified in the Enterobacteriaceae and in Pseudomonas aeruginosa (23, 24, 26). GES-5 was originally found in a clinical isolate of Escherichia coli collected in Athens, Greece, in 2004 (33). GES-5 has also been identified in Klebsiella pneumoniae from Korea and Brazil (3, 17, 24), in Enterobacter cloacae from Canada (19), and in P. aeruginosa from different countries, such as China (34, 35), Spain (32), Republic of South Africa (20), and Brazil (23). The blaGES-5 gene was always plasmid located and usually is part of class 1 integrons. Therefore, the carbapenemase blaGES-5 gene seems to be quite widespread worldwide.
The mobilization of antibiotic resistance genes in Gram-negative bacteria occurs through several mechanisms. The transfer of plasmids (through natural transformation or conjugation) is one of the most frequent factors for the horizontal transfer of resistance genes. Additionally, their acquisition is related mainly to different mechanisms, including recombination, integron-mediated mobilization of gene cassettes, transposition, or mobilization via integron-mobilization units, as recently described for the blaGES-5 gene (IMU) (25).
This work was initiated by the selection of carbapenem-resistant Pseudomonas knackmussii B13 (30), formerly Pseudomonas sp. B13 (7), which had been transformed with plasmids isolated from bacterial communities residing in the activated sludge compartment of a wastewater treatment plant located in Bielefeld-Heepen, Germany. Transformants were selected on spectinomycin-containing agar (70 μg/ml). Unexpectedly, two resulting recombinants, Pseudomonas sp. B13(pRSB113) and Pseudomonas sp. B13(pRSB115), exhibited resistance or reduced susceptibility to ceftazidime, aztreonam, and carbapenems. MICs were interpreted according to the CLSI breakpoints (6). MIC values of imipenem were increased from 0.064 μg/ml to 2 and 1.5 μg/ml in Pseudomonas sp. B13(pRSB113) and Pseudomonas sp. B13(pRSB115), respectively. MIC values of ertapenem were increased from 0.38 μg/ml to >32 μg/ml in both Pseudomonas sp. B13 (pRSB113) and Pseudomonas sp. B13 (pRSB115). MIC values of meropenem were increased from 0.023 μg/ml to 4 and 6 μg/ml in Pseudomonas sp. B13(pRSB113) and Pseudomonas sp. B13(pRSB115), respectively. In addition, they showed resistance to chloramphenicol, trimethoprim-sulfamethoxazole, tobramycin, and gentamicin, and Pseudomonas sp. B13(pRSB115) was also resistant to netilmicin. Preliminary PCR detection of the most common carbapenemase genes failed (26). However, PCR amplification gave positive results with primers specific for the blaGES-like gene (26). Sequencing of the amplicon showed 100% identity with blaGES-5. Since blaGES-1-like genes are known to be integron encoded, the consensus primers 5′-CS and 3′-CS for class 1 integrons were used (26), and PCR amplification resulted in a ca. 3.2-kb DNA fragment using both pRSB113 and pRSB115 plasmid DNAs as templates.
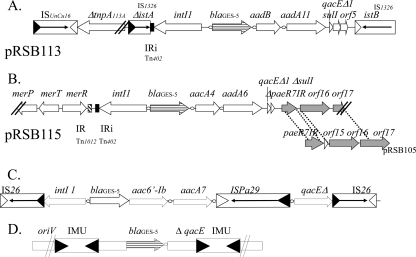
The genetic environment of the blaGES-5 gene was determined by sequencing (Applied Biosystems sequencer ABI 3130) of the class 1 integron and surrounding sequences directly from natural plasmid DNA, using a primer-walking approach (Maxiprep, Qiagen, France). However, for plasmid pRSB113, this primer-walking approach failed. Therefore, a 10,000-bp fragment containing the blaGES-5 gene was subcloned into plasmid pBKCMV by using EcoRI-restricted DNA. The resulting recombinant plasmid, p113Eco, was introduced into E. coli DH10B by electroporation as described previously (28). E. coli DH10B (p113Eco) was selected on ticarcillin (50 μg/ml) and kanamycin (30 μg/ml)-containing Trypticase soy agar (TSA) plates. Plasmid p113Eco was sequenced by using combinations of universal T3 and T7 primers and specific walking primers designed based on the obtained sequences. The sequences of 10,047 bp- and 7,235-bp fragments, respectively, containing the blaGES-5 gene were determined for plasmids pRSB113 (after subcloning) and pRSB115 (directly from natural plasmid extract). On both plasmids, the blaGES-5 gene represents a gene cassette, located in the first position of a class 1 integron, as described previously (4). On plasmid pRSB113, the blaGES-5 gene is followed by the aadB and aadA11 genes, encoding resistance to aminoglycosides, whereas it is followed by the aacA4 and aadA6 genes, also encoding aminoglycoside resistance, on plasmid pRSB115. The blaGES-5-containing class 1 integrons were inserted into two distinct structures: between the two open reading frames (ORFs) istA and istB of the IS21 family insertion sequences (IS1326) on plasmid pRSB113 and between genes (paeR7IR, orf16, and orf17) of the previously described plasmid pRSB105 (29) and a mercuric resistance operon on pRSB115 (Fig. 1). Notably, the 3′ end of the pRSB115-located class 1 integron was deleted, since the sul1 gene is partially missing, interrupting the paeR7IR gene, encoding a type II restriction enzyme on pRSB115. In contrast to pRSB105, plasmid pRSB115 therefore encodes a nonfunctional restriction/modification system (ΔpaeR7IR) (Fig. 1). Notably, the 25-bp inverted repeat (IR) element of a Tn402-like transposon was identified on the left extremity of the intI1 integrase-encoding gene on both the pRSB113 and pRSB115 plasmids (Fig. 1). Moreover, the terminal inverted repeat of Tn1012, a Tn21-related terminal inverted repeat frequently associated with merR, was identified on the left-hand extremity of the class 1 integron on pRSB115 (Fig. 1). On pRSB113, two ORFs that encode two transposase-like proteins, termed TnpA113A (truncated) and TnpAISUncu16, were identified on the left-hand extremity of the class 1 integron. Interestingly, TnpAISUncu16 shares 59% amino-acid identity with a TnpA-like protein from Alteromonas macleodii, found in the deep water column of the Mediterranean (16). This result strongly suggests genetic exchanges between waterborne bacteria and indicates again that water habitats are important reservoirs of antibiotic resistance genes, as previously shown by the diversity of these genes found in Aeromonas spp. in the Seine River, France (13). The capture of the blaGES-5 gene by integrons containing aminoglycoside resistance gene cassettes, which are overrepresented in environmental plasmids of the Enterobacteriaceae or of Pseudomonas, may enhance the dissemination of this carbapenemase gene.
Fig 1.
Genetic environment of the blaGES-5 gene on plasmid pRSB113 (A), plasmid pRSB115 (B), chromosomes of P. aeruginosa isolates from Brazil (23) (C), or plasmid pCHE-A from E. cloacae (25) (D). Arrows indicate the directions of transcription of the coding regions, the left and right inverted repeats of IS elements are shown by filled and empty triangles, respectively, and core sites are indicated as circles. Tn1012 and IRi of the Tn402-like transposon are shown as small hatched and black rectangles. IS1326 is a IS21 family member and encodes two consecutive open reading frames: a long upstream frame, designated istA, which is truncated on pRSB113, and a shorter downstream frame, istB, which is separated from istA in this plasmid. ISUnCu16 is an IS66 family member, ISBstXII subgroup, and tnpAISUnCu16 encodes a protein sharing similarity with a transposase-like protein from Alteromonas macleolii (16). Open reading frame ΔtnpAIS113A encodes a truncated transposase-like protein sharing similarity with the product of the IS408-like insertion sequence. paeR7IR, orf16, and orf17, previously identified in plasmid pRSB105 (29), are indicated as gray arrows.
Plasmids pRSB113 and pRSB115, extracted by using the Kieser method, were of ca. 30 and 40 kb in size, respectively (18). Direct transfer of the blaGES-5 determinant by electroporation (28) to E. coli DH10B (Invitrogen, Life Technologies, Cergy-Pontoise, France) and selection on ticarcillin-containing agar (50 μg/ml) resulted in recombinant E. coli DH10B (pRSB113) and E. coli DH10B (pRSB115) derivatives. Conjugation experiments were attempted between Pseudomonas sp. B13 (pRSB113) or Pseudomonas sp. B13 (pRSB115) and azide-resistant E. coli J53 or between E. coli DH10B (pRSB113) or E. coli DH10B (pRSB115) and E. coli J53, as previously described (21, 22). Mating-out transfer of the plasmids harboring the blaGES-5 gene to E. coli remained unsuccessful, suggesting that these plasmids were not self-transferable.
Both plasmids replicated in Pseudomonas sp. and E. coli DH10B, suggesting a possible horizontal transfer between members of the Enterobacteriaceae and nonfermenting rods. Incompatibility (Inc)-group typing of resistance plasmids carried out by PCR with primer pairs specific for the Inc groups IncP, IncQ, IncN, IncW, and IncA/C (5) showed that these plasmids did not belong to any of the tested Inc groups. Plasmids pRSB113 and pRSB115 were isolated from an unknown activated sludge bacterium, such as the previously identified erythromycin resistance plasmids pRSB101 and pRSB105 (29, 31). Plasmid pRSB105 possesses two replicase genes: Rep1 clearly derives from plasmids of Pseudomonas syringae, while Rep2 is more divergent and might derive from plasmids previously found in P. aeruginosa (Rep protein CAI46990). Plasmid pRSB101 possessed a Rep identical to Rep1 from pRSB105. This suggests that they likely derive from the same ancestor; however, the mobA gene on those two plasmids was only 61% identical, suggesting a mosaicism or a fusion of the two replicons in a multireplicon status. Although a backbone similarity was observed only between pRSB105 and pRSB115 in the vicinity of the blaGES-5 gene (Fig. 1), further comparisons were performed in order to elucidate the typing, replication, and mobilization features. PCR mapping of the repA and mobA genes was performed using primers listed in Table 1. The two repA genes (rep1 and rep2) from pRSB105 were detected by PCR in pRSB113, whereas only the repA gene from pRSB101 was detected in pRSB115. Sequencing of the resulting amplicons containing the repA genes from pRSB113 showed that the deduced RepA proteins showed 80% and 94% amino acid identity with the corresponding Rep1 and Rep2 proteins from pRSB105 (IncP-6 group) (31). Sequencing of the resulting amplicons containing the repA gene from pRSB115 showed that the deduced RepA protein showed 92% amino acid identity with RepA from pRSB101 (unknown Inc group, pRSB101-like) (29). Rep1 is part of a narrow-host-range replicon that is able to replicate in the gammaproteobacteria, and Rep2 extends the host range of the plasmid to the betaproteobacteria. As previously described, the Rep1 replicon may originate with a plant-associated bacterium. pRSB101, pRSB105, and pRSB115 might have evolved from a common ancestor.
Table 1.
Sequences of primers used in this study for replication/mobilization modules characterizationa
| Primer | Sequence (5′–3′) |
|---|---|
| pRSB105 rep1 F | ATGGCTCGACAATCCACCGTC |
| pRSB105 rep1 R | TACGAAGAATCGAGAGGCTCC |
| pRSB105 rep2 F | AAGCACAGCTACGACTTTTCAGCC |
| pRSB105 rep2 R | TTTGAACACCTCACGATCAGCC |
| pRSB101MobA-F | ATGATCTCAAAGCACATTCG |
| pRSB101MobA-R | TATATAGAGCAGAGTCCAGCG |
| pRSB105MobA-F | GTGATCGTTAAGAAGGTGCC |
| pRSB105MobA-R | AAGTCTTATCGACTCATGCCGGGTG |
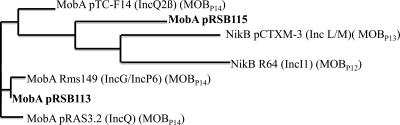
As shown by Garcillan-Barcia et al. (9), most classical Inc groups of plasmids are located in the relaxase (or MobA) phylogenies. The MobA proteins encoded by plasmids pRSB113 and pRSB115 were therefore investigated as evolutionary markers. The whole mobA gene from pRSB113 was successfully amplified with primers specific for the mobA gene from pRSB105, and that from pRSB115 was amplified with primers specific for the mobA gene from pRSB101 (Table 1). Corresponding proteins showed, respectively, 93 and 90% amino acid identity with the corresponding fragments, and the MobA proteins from pRSB113 and pRSB115 shared only 43% amino acid identity. Dendrograms were derived from the multiple relaxase protein sequence alignment by a parsimony method using the ClustalW2 software program (Fig. 2). MobA from pRSB113 shared the highest amino acid identity with that from plasmids Rms149 from P. aeruginosa (15) and pRAS3.2 from Aeromonas salmonicida (19), with 98% and 92% identities, respectively. According to classification by Garcillan-Barcia et al., pRSB113 belongs to clade P14 of the MOBp family, a clade composed of a series of mobilizable plasmids (9). MobA from pRSB115 shared low amino acid identity with other relaxase proteins, and the highest amino acid identities were with those from plasmids R64 from Salmonella enterica serovar Typhimurium (8) and pCTXM-3 from Citrobacter freundii (14) (44% and 39% identities, respectively). The latter could then belong to a clade related to MOBP12 and MOBP13. This result suggests that the pRSB113 and pRSB115 plasmids belong to two distinct clades of the MOBP plasmid family, and it might be speculated that these plasmids are mobilizable (Fig. 2).
Fig 2.
Phylogeny tree of relaxase of plasmids pRSB113 and pRSB115 and MobA of pTC-F14 from Acidithiobacillus caldus (GenBank accession no. AAP04747), NikB of pCTXM-3 from Citrobacter freundii (GenBank accession no. AAN87675), NikB of R64 from Salmonella enterica serovar Typhimurium (GenBank accession no. BAK64476), MobA of Rms146 from Pseudomonas aeruginosa (GenBank accession no. CAI46961), and MobA of pRAS3.2 from Aeromonas salmonicida (GenBank accession no. AAK97758). Known incompatibility groups and MOB families are indicated.
Conclusions.
Comparative genomic analysis of plasmids pRSB113 and pRSB115 in the vicinity of the blaGES-5 gene showed that they are related. They may have evolved by integration of distinct modules from different plasmid sources and thus represent mosaic plasmids. No hot spot of integration of the blaGES-5-gene-containing class 1 integron could be evidenced, suggesting a high ability of integration in distinct genetic loci in plasmids found in the environment. This study emphasizes that aquatic environments may be the reservoir of carbapenemase genes of clinical relevance.
Nucleotide sequence accession numbers.
The sequences corresponding to the genetic elements described in this work were assigned to GenBank accession numbers JN849689 (pRSB113) and JN849690 (pRSB115).
ACKNOWLEDGMENTS
This work was funded by a grant from the INSERM (U914), Paris, France, the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France, and grants from the European Community (TEMPOtest-QC, HEALTH-2009-241742, and TROCAR [HEALTH-F3-2008-223031]). A.S. gratefully acknowledges the METAEXPLORE grant from the European Commission (KBBE-222625).
Footnotes
Published ahead of print 9 December 2011
REFERENCES
- 1. Ambler RP, et al. 1991. A standard numbering scheme for class A β-lactamases. Biochem. J. 276:269–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aubron C, Poirel L, Ash RJ, Nordmann P. 2005. Carbapenemase-producing Enterobacteriaceae, U.S. rivers. Emerg. Infect. Dis. 11:260–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bae IK, et al. 2007. Genetic and biochemical characterization of GES-5, an extended-spectrum class A β-lactamase from Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 58:465–468 [DOI] [PubMed] [Google Scholar]
- 4. Bonnin R, et al. 2011. Carbapenem-hydrolyzing GES-type extended-spectrum β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 19th informational supplement M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Dröge M, Pühler A, Selbitschka W. 2000. Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol. Gen. Genet. 263:471–482 [DOI] [PubMed] [Google Scholar]
- 8. Furuya N, Nisioka T, Komano T. 1991. Nucleotide sequence and functions of the oriT operon in IncI1 plasmid R64. J. Bacteriol. 173:2231–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcillan-Barcia MP, Francia MV, de la Cruz F. 2009. The diversity of conjugative relaxases and its application classification. FEMS Microbiol. Rev. 33:657–687 [DOI] [PubMed] [Google Scholar]
- 10. Girlich D, Poirel L, Nordmann P. 2010. First isolation of the blaOXA-23 carbapenemase gene from an environmental Acinetobacter baumannii isolate. Antimicrob. Agents Chemother. 54:578–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Girlich D, Poirel L, Nordmann P. 2010. Novel Ambler class A carbapenem-hydrolyzing β-lactamase from a Pseudomonas fluorescens isolate from the Seine River, Paris, France. Antimicrob. Agents Chemother. 54:328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Girlich D, Poirel L, Nordmann P. 2010. PER-6, an extended-spectrum β-lactamase from Aeromonas allosaccharophila. Antimicrob. Agents Chemother. 54:1619–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Girlich D, Poirel L, Nordmann P. 2011. Diversity of clavulanic-inhibited extended-spectrum β-lactamases in Aeromonas spp. from the Seine River, Paris, France. Antimicrob. Agents Chemother. 55:1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Golebiewski M, et al. 2007. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum beta-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51:3789–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haines AS, Jones K, Cheung M, Thomas CM. 2005. The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J. Bacteriol. 187:4728–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ivars-Martinez E, et al. 2008. Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J. 2:1194–1212 [DOI] [PubMed] [Google Scholar]
- 17. Jeong SH, et al. 2005. First outbreak of Klebsiella pneumoniae clinical isolates producing GES-5 and SHV-12 extended-spectrum β-lactamases in Korea. Antimicrob. Agents Chemother. 49:4809–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kieser T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19–36 [DOI] [PubMed] [Google Scholar]
- 19. L'Abée-Lund TM, Sørum H. 2002. A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid 47:172–181 [DOI] [PubMed] [Google Scholar]
- 20. Labuschagne CDJ, Weldhagen GF, Ehlers MM, Dove MG. 2008. Emergence of class 1 integron-associated GES-5 and GES-5-like extended-spectrum β-lactamases in clinical isolates of Pseudomonas aeruginosa in South Africa. Int. J. Antimicrob. Agents 31:527–530 [DOI] [PubMed] [Google Scholar]
- 21. Nordmann P, Lartigue M-F, Poirel L. 2008. Beta-lactam induction of ISEcp1B-mediated mobilization of the naturally occurring blaCTX-M β-lactamase gene of Kluyvera ascorbata. FEMS Microbiol. Lett. 288:247–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Picão CR, et al. 2008. Expanded-spectrum β-lactamase PER-1 in an environmental Aeromonas media isolate from Switzerland. Antimicrob. Agents Chemother. 52:3461–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Picão CR, Poirel L, Gales AC, Nordmann P. 2009. Diversity of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolates causing bloodstream infections in Brazil. Antimicrob. Agents Chemother. 53:3908–3913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Picão CR, Santos AF, Nicoletti AG, Furtado GH, Gales AC. 2010. Detection of GES-5-producing Klebsiella pneumoniae in Brazil. J. Antimicrob. Chemother. 65:796–807 [DOI] [PubMed] [Google Scholar]
- 25. Poirel L, Carrër A, Pitout JD, Nordmann P. 2009. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob. Agents Chemother. 53:2492–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poirel L, et al. 2001. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 45:2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quinteira S, Peixe L. 2006. Multiniche screening reveals the clinically relevant metallo-β-lactamase VIM-2 in Pseudomonas aeruginosa far from the hospital setting: an ongoing dispersion process? Appl. Environ. Microbiol. 72:3743–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sambrook J, Russell DW. 2001Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Schlüter A, et al. 2007. Erythromycin resistance-conferring plasmid pRSB105, isolated from a sewage treatment plant, harbors a new macrolide resistance determinant, an integron-containing Tn402-like element, and a large region of unknown function. Appl. Environ. Microbiol. 73:1952–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stolz A, Busse HJ, Kämpfer P. 2007. Pseudomonas knackmussii sp. nov. Int. J. Syst. Evol. Microb. 57:572–576 [DOI] [PubMed] [Google Scholar]
- 31. Szczepanowski R, et al. 2004. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613–3630 [DOI] [PubMed] [Google Scholar]
- 32. Viedma E, et al. 2009. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum β-lactamases GES-1 and GES-5 in Spain. Antimicrob. Agents Chemother. 53:4930–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vourli S, et al. 2004. Novel GES/IBC extended-spectrum beta-lactamase variants with carbapenemase activity in clinical enterobacteria. FEMS Microbiol. Lett. 234:209–213 [DOI] [PubMed] [Google Scholar]
- 34. Wang C, Cai P, Chang D, Mi Z. 2006. A Pseudomonas aeruginosa isolate producing the GES-5 extended-spectrum β-lactamase. J. Antimicrob. Chemother. 57:1261–1262 [DOI] [PubMed] [Google Scholar]
- 35. Wang J, et al. 2010. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Chinese hospitals. Int. J. Antimicrob. Agents 35:486–491 [DOI] [PubMed] [Google Scholar]