Abstract
We present a high-coverage draft genome assembly of the aye-aye (Daubentonia madagascariensis), a highly unusual nocturnal primate from Madagascar. Our assembly totals ∼3.0 billion bp (3.0 Gb), roughly the size of the human genome, comprised of ∼2.6 million scaffolds (N50 scaffold size = 13,597 bp) based on short paired-end sequencing reads. We compared the aye-aye genome sequence data with four other published primate genomes (human, chimpanzee, orangutan, and rhesus macaque) as well as with the mouse and dog genomes as nonprimate outgroups. Unexpectedly, we observed strong evidence for a relatively slow substitution rate in the aye-aye lineage compared with these and other primates. In fact, the aye-aye branch length is estimated to be ∼10% shorter than that of the human lineage, which is known for its low substitution rate. This finding may be explained, in part, by the protracted aye-aye life-history pattern, including late weaning and age of first reproduction relative to other lemurs. Additionally, the availability of this draft lemur genome sequence allowed us to polarize nucleotide and protein sequence changes to the ancestral primate lineage—a critical period in primate evolution, for which the relevant fossil record is sparse. Finally, we identified 293,800 high-confidence single nucleotide polymorphisms in the donor individual for our aye-aye genome sequence, a captive-born individual from two wild-born parents. The resulting heterozygosity estimate of 0.051% is the lowest of any primate studied to date, which is understandable considering the aye-aye's extensive home-range size and relatively low population densities. Yet this level of genetic diversity also suggests that conservation efforts benefiting this unusual species should be prioritized, especially in the face of the accelerating degradation and fragmentation of Madagascar's forests.
Keywords: genome assembly, molecular clock, primate evolution, lemur
Introduction
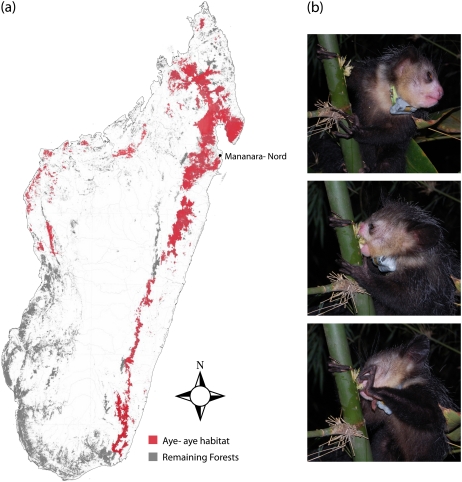
The only surviving representative of the primate family Daubentoniidae is the aye-aye (Daubentonia madagascariensis). The aye-aye is a nocturnal lemur with unusual derived traits including an elongated, thin, highly flexible third middle digit on the hand, a pair of relatively huge continuously growing incisors, the largest relative brain size of any strepsirrhine primate (lemurs and lorisoids), and a relatively slow life history including late weaning and a protracted learning period (Cartmill 1974; Martin 1990; Simons 1995; Fleagle 1999; Catlett et al. 2010). These features are likely adaptations that facilitate complex extractive foraging strategies to obtain seeds (endosperm) of hard-shelled ramy nuts or grubs from cavities gnawed in tree bark or bamboo stalks (Gibson 1986; Iwano and Iwakawa 1988; Erickson 1991; Sterling and Povinelli 1999). Aye-ayes have the largest species range of any extant lemur (fig. 1; Mittermeier et al. 2010). However, they are primarily solitary foragers, with extensive home ranges (Sterling 1993) and population densities that are likely very low (Mittermeier et al. 2010).
FIG. 1.—
Aye-aye species distribution in Madagascar and extractive foraging behavior (photo credit: Francois Randrianasolo). (a) Remaining forests (gray) and presumed current species distribution of aye-ayes (red) in Madagascar. The aye-aye species distribution is the most extensive of any lemur, but they have huge individual home ranges and low population densities. The two parents of the donor individual for our aye-aye genome sequence, Goblin, were caught near Mananara-Nord in Northeast Madagascar, indicated on the map. (b) Photos of a male aye-aye with a GPS satellite radio-collar being monitored near Kianjavato, Madagascar (top photo) using his large continuously growing incisors to gnaw through a bamboo stalk (middle photo) and extracting a grub with his long, thin, flexible left middle finger (bottom photo). Note the thin middle finger on the right hand wrapped around the bamboo stalk in all three photos. There are no woodpeckers on Madagascar to otherwise fill this extractive foraging niche (Cartmill 1974).
The slow life history, extensive home-range size, and low population densities of the aye-aye, when considered in the context of Madagascar's continued forest degradation and fragmentation, may have severe implications for the long-term survival of this important species. Presently, aye-ayes are considered “Near Threatened” (Red List of threatened species) but an elevation to “Endangered” status has been proposed (Mittermeier et al. 2010).
A publicly available resource of aye-aye genomic sequence data would facilitate future population studies designed to assess genetic diversity and characterize patterns of biogeographical variation to aid in aye-aye conservation efforts and could be compared with the published genomes of other primates for evolutionary analyses. With the advent of new sequencing technologies, obtaining complete genomic sequence of a new species should no longer be considered a prohibitive task. However, the full assembly and analysis of a finished or high-quality draft genome sequence, as typically produced by large collaborative consortiums, remain prohibitively challenging and would be slow for individual labs interested in advancing research with their own study organisms. In this paper, we demonstrate that readily attainable unfinished draft-quality genome sequences have high potential value and utility for many conservation, ecological, and evolutionary studies of nonmodel organisms.
Materials and Methods
DNA Isolation and Sequencing
Genomic DNA was isolated from a liver tissue sample taken at necropsy from an adult male aye-aye (Dm6514m; Goblin) at the Duke Lemur Center, Durham, North Carolina, with the Gentra PureGene purification kit (Qiagen). This captive-born individual was the offspring of two wild-born parents (Dm6451m and Dm6453f), each originally captured in Northeast Madagascar near Mananara-Nord (fig. 1). Genomic DNA libraries for paired-end Illumina Genome Analyzer IIx sequencing were prepared following manufacturer instructions. Three different libraries were constructed, with modes of insert sizes at 448, 580, and 957 bp as estimated with a DNA 1000 chip on the Agilent Bioanalyzer 2100 (supplementary fig. 1, Supplementary Material online).
The 580 bp insert library was pair-end (PE) sequenced on one flowcell (eight lanes) of the Illumina Genome Analyzer IIx for 108 cycles from each end (PE 108 × 108), generating a total of 263,743,085 PE reads (an average of 33.0 million PE reads per lane). On a second flowcell, we sequenced the 448 bp insert library on seven lanes (232,367,850 total PE reads; average 33.2 million per lane) and the 957 bp insert library on one lane (33,345,067 PE reads). As a control, one lane included a 1% PhiX genome spike-in. If the aye-aye genome is of similar size to that of humans (i.e., ∼3 Gb), then the total sequence data generated would equate to ∼38× average coverage (see “single nucleotide polymorphisms (SNP) identification” for observed coverage statistics from mapped reads to the final assembly). The sequence data have been deposited in the National Center for Biotechnology Information short read archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) as study no. SRA043766.1.
Genome Assembly
We first removed reads mapping to the PhiX genome (from the spike-in control), using the Burrows-Wheeler aligner (BWA; Li and Durbin 2009) and custom scripts. This resulted in the removal of 492,216 read pairs and 24,460 nonpaired reads. We performed all steps of the assembly on a single node of a Linux high-performance computing cluster with 24 cores and 128 GB of random access memory (RAM). We first used Quake (Kelley et al. 2010) to correct sequencing errors, using the q-mer counting option, a k-mer size of k = 19, and the “-I” flag. To reduce memory requirements associated with the storage of all k-mers during error correction, we modified Quake with a Map-Reduce algorithm developed on top of the MPI protocol (MR-MPI; Plimpton and Devine 2011), using the parameters “-k = 19 -c = 0.04 -p = 24 -q = 64.” The code is available upon request.
We used the command line CLC bio Assembly Cell (Aarhus, Denmark) to generate an initial assembly due to the small memory footprint of this assembler. Contig assembly was completed using a maximum of 56 GB RAM, assessed using the monitoring tool Memmon (https://software.sandia.gov/trac/utilib/wiki/Documentation/memmon). The parameters used for assembly were as follows: “-cpus=24 -v, -p fb ss 460 616 –q” for reads from the 448 bp insert size library, “-p fb ss 660 816 –q” for reads from the 580 bp insert size library, and “-p fb ss 1060 1216 –q” for reads from the 957 bp insert size library. This step produced 3,237,204 contigs with total length of 2,856,952,161 bp. The largest contig was 86,436 bp, and the N50 contig size was 3,650 bp (i.e., half of the total length of the contigs is contained within contigs of at least 3,650 bp).
The CLC bio Assemler does not include a scaffolding step. Therefore, we used SSPACE (Boetzer et al. 2011) to merge contigs into scaffolds, requiring a minimum of two paired reads between contigs for merging. The parameters used for SSPACE were “-x = 0 -k = 2 -v = 1,” with the reverse compliment option turned off and minimum allowed error of 0.125, 0.083, and 0.05 for reads from the 448, 580, and 957 bp libraries, respectively. We thus produced 2,564,533 scaffolds representing a total of 2,970,026,655 bp (maximum scaffold size = 252,467 bp; N50 scaffold size = 13,597 bp). The genome assembly data have been deposited in GenBank as a whole-genome shotgun sequencing project (Accession number AGTM000000000). In addition, because there may be differences in the deposited GenBank assembly sequence and the assembly sequence used for analyses in our paper due to GenBank trimming procedures, the specific scaffold sequences that were used for analyses in this paper are available at http://giladlab.uchicago.edu/data.html.
Quality Assessment
To evaluate the quality of our draft genome assembly, we compared our sequence data with aye-aye nuclear genome nucleotide sequence data previously deposited in GenBank that were generated by polymerase chain reaction (PCR) and Sanger sequencing methods (Perry et al. 2006, 2007; Horvath et al. 2008; Perelman et al. 2011). We used BLAST (Altschul et al. 1997) to identify 96 nonoverlapping best alignments >100 bp in size with ≥92% sequence identity (to exclude small and nonorthologous alignments) to 76 of the 77 aye-aye sequences that have been deposited in GenBank (supplementary fig. 2, Supplementary Material online). In total (including indels), 68,876 of 69,013 aligned base pairs (99.80%) were identical between the GenBank and genome sequences.
The proportion of nonidentical sites from the GenBank BLAST analysis is similar to a published estimate of aye-aye genetic diversity: π (average pairwise sequence divergence) = 0.08% (Perry et al. 2007) that was based on sequence data from 16 intergenic regions (>25 kb in total) and two opsin genes for eight wild-born aye-ayes. The eight individuals included the two wild-born parents of the individual whose genome we sequenced for this study. Therefore, we evaluated the sequence and polymorphism data from that study (34,187 bp) to further estimate our assembly error rate. Based on the BLAST analysis against the 18 regions from Perry et al. (2007), 31,871 of 31,936 aligned bp (99.80%) were identical in the aye-aye genome scaffolds. Of the 65 total differences, 37 were found to be SNPs (including 27 sites annotated with ambiguity codes in the GenBank sequence used in the BLAST analysis because that individual was heterozygous at those positions). Five differences were inferred sequence errors in the scaffold sequence (four of the five errors were clustered near the end of one scaffold sequence). Five differences were insertion/deletion errors in difficult-to-assemble regions (within each of the following repetitive regions: A(18), T(11), TACTAAAAAGA(13), T(14), GAAG tetranucleotide repeat). Two differences were due to N regions in the assembly (one within a GT dinucleotide repeat region, the other a run of seven Ns near the end of a scaffold sequence). Finally, 16 differences were due to multi-base pair indels or the runs of Ns because each nonidentical base pair is scored as a difference in the BLAST analysis rather than each event.
Based on this analysis and making the conservative assumption that any sequence errors occurred in this project and not in the Sanger sequencing, we estimate that the nucleotide sequence error rate (not including indels) in aligned nonrepetitive regions of the aye-aye genome assembly is approximately 0.00016 (5/31,913) or 1 error per 6,382 bp.
SNP Identification
We then aligned our sequence reads to the aye-aye genome scaffolds using the default parameters of the Burrows-Wheeler Aligner (BWA version 0.5.9rc1; Li and Durbin 2009). Low quality bases at the ends of the reads were trimmed using the “-q 20” option prior to mapping, and potential PCR duplicate reads were removed using Picard (http://www.picard.sourceforge.net) after alignment. The median coverage of reads mapped to the genome scaffolds was 17× (mean = 15.88; standard deviation [SD] = 11.63). After trimming 108 bp from the ends of each contig, where a reduction in mapped read coverage would be expected, the median coverage of mapped reads was 19× (mean = 18.94×; SD = 10.42). All sites with a minimum of 10× sequence coverage on each strand (20× total coverage) were analyzed using the default parameters of SAMtools version 0.1.12a (Li et al. 2009) to identify heterozygous positions. In a previous study (Perry et al. 2010), we found that this 20× coverage cutoff can facilitate the highly accurate identification of heterozygous sites. We also established a maximum total coverage cutoff of 50×, to limit potentially erroneous SNP calling in recently duplicated genomic regions. We used the SAMtools option “-C 50” to downgrade the mapping quality of reads with excessive mismatches to the reference sequence. Only positions where the root mean square mapping quality was greater than 20 were considered. The positions of the identified SNPs are available at http://giladlab.uchicago.edu/data.html.
Genome Alignment and Annotation of Exons
We focused on gene coding regions. For each nonoverlapping human exon from the ENSEMBL database (GRCh37), we used the lastz program (http://www.bx.psu.edu/∼rsharris/lastz/) to identify the sequence from our aye-aye scaffolds with the best match, requiring ≥70% nucleotide sequence identity. We also required alignment of the entire exon without insertions/deletions and ≥70% identity of the inferred amino acid sequences, generating a total of 120,241 exon alignments. If exons from multiple human genes had best matches to the same region of the aye-aye genome (within 50 bp of each other), then we removed those for which the nucleotide sequence identity was only 0–5% greater than all other alignments for that aye-aye genome region, resulting in the removal of 3,514 exon alignments. Using this approach, we identified putative aye-aye orthologs to 116,727 human exons from 21,494 human genes. The aye-aye exon sequences for each gene were concatenated, and ENSEMBL gene IDs were converted to RefSeq gene IDs.
The aye-aye gene sequences were then aligned against gene sequences predicted from the reference genomes of human, chimpanzee (Pan troglodytes), orangutan (Pongo abelii), rhesus macaque (Macaca mulatta), mouse (Mus musculus), and dog (Canis familiaris) (http://hgdownload-test.cse.ucsc.edu/goldenPath/hg19/multiz46way/alignments/) using the Fast Statistical Alignment (FSA) program (Bradley et al. 2009). These species represent four published primate genomes (International Human Genome Sequencing Consortium 2004; The Chimpanzee Sequencing and Analysis Consortium 2005; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007; Locke et al. 2011) and two nonprimate outgroups, which we included in the analysis so that nucleotide substitutions could be polarized to the ancestral primate lineage.
Evolutionary Analysis
For evolutionary analysis of gene coding region sequences, we removed start and stop codons from the multispecies alignments. To preserve the codon frame, we corrected any frame-shifting indels introduced in the alignment step by normalizing the alignments relative to the human RefSeq transcript coding sequences. That is, when the frame of the aligned sequences was shifted with respect to the human reference transcript, 1–2 bp gaps were introduced in all sequences as a correction. The resulting multispecies alignment files for each analyzed gene are available at http://giladlab.uchicago.edu/data.html. The MEGA4 program (Tamura et al. 2007) was used to estimate nucleotide sequence divergence and phylogeny and to perform relative rate tests.
We used PAML (Yang 2007) to estimate ancestral sequences and the numbers and rates of nonsynonymous and synonymous substitutions on each branch. The ratio of the rates of nonsynonymous to synonymous substitution (dN/dS) can be examined to make inferences about long-term selective pressures on amino acid sequences. We removed genes for which dS > 1 on any branch or dS > 0.5 on any branch excepting mouse, as these outlying values may reflect alignment or sequence artifacts. We also limited our analysis to genes with ≥100 synonymous sites that were aligned and analyzed across all species.
We calculated two dN/dS ratios for each remaining gene in each lineage. First, the conventional dN/dS value, where dS is based on the synonymous substitution rate for the individual gene on that branch. The second dN/dS value is based on a single genome-wide estimate of dS for each lineage; calculated as the total number of synonymous substitutions summed across all genes divided by the total number of synonymous sites summed across all genes. The second dN/dS value, which uses the genome-wide dS value in the denominator, may be valuable for interpreting results on shorter branches, where the number of gene-specific synonymous substitutions can be low or zero. We considered genes and branches that meet the following conditions as potential candidates to have evolved under positive selection: dN/dS > 1, dN/dS genome > 1, and ≥ 2 synonymous substitutions (on short branches, stochastic variation in the number of synonymous substitutions can lead to large numbers of genes with dN/dS > 1). Although it is unlikely that all genes and branches meeting these conditions were subjected to positive selection at the amino acid sequence level, the set of candidates we generated is likely enriched for such genes. Functional enrichment analyses using gene ontology annotations (Ashburner et al. 2000) were conducted using GeneTrail (Keller et al. 2008).
Results and Discussion
Our goal was not to produce a finished aye-aye genome assembly. With short read data only and currently available assembly algorithms, a high-quality “finished” assembly of a new mammalian genome requires truly extensive sequence coverage (on the order of 100-fold coverage) and considerable computational power that is not yet commonly available, extensive library preparation and sequencing efforts, or both (e.g., Gnerre et al. 2011). The complete annotation of the aye-aye genome (e.g., the annotation of genic regions, functional units, duplicated regions, etc.) was also beyond the scope of this project, as this would require considerable resources, typically achieved only through consortium-led projects.
Rather, we sought to efficiently generate a genomic resource that would be sufficient to facilitate most downstream applications relevant to an endangered nonmodel species. These applications might include evolutionary and phylogenetic analyses or the development of primers and markers for conservation- and behavior-related population genetic studies. The results of the initial analyses presented below illustrate this point.
Aye-Aye Genetic Diversity
We generated ∼19× sequence coverage of the assembled portions of the genome of one aye-aye individual. With these data, we estimated aye-aye genetic diversity using magnitudes more nucleotide sites than were previously available for such an analysis. Among the 581,674,772 sites with coverage sufficient for reliable SNP identification (see Materials and Methods), we identified 293,800 heterozygous sites (π = 0.051%). This genetic diversity estimate is slightly lower than those from each of the two previous aye-aye nuclear genome population genetic studies (π = 0.081% from 25,649 bp of intergenic region nucleotide sequence data generated by PCR and Sanger sequencing for eight individuals; Perry et al. 2007 and π = 0.073% from 197,784 synonymous sites in the coding regions of 1,175 genes based on RNA-seq data for two individuals; Perry et al. 2012). These previous studies included individuals with ancestry from multiple regions of Madagascar, whereas both parents of the donor individual for our genome reference sequence were captured in a single region. This difference may explain, at least in part, the slight variation in results. In addition, our estimate was computed from multiple magnitudes more sites over a broader representation of the genome but from one individual only.
Regardless, all three of these independently generated genetic diversity estimates place aye-ayes at the extreme low end of primate nuclear genome diversity, at least among the 14 other species for which comparable estimates are available (Yu et al. 2003, 2004; Fischer et al. 2004, 2006, 2011; The Chimpanzee Sequencing and Analysis Consortium 2005; Voight et al. 2005; Hernandez et al. 2007; Wall et al. 2008; Perry et al. 2010; Locke et al. 2011; Perry et al. 2012). The low genetic diversity estimate could be explained, at least in part, by aye-aye demography. Behavioral observational data have suggested that aye-ayes have both extensive home ranges and low population densities (Sterling 1993; Mittermeier et al. 2010). The low genetic diversity estimate could also be explained by an ancient bottleneck event. Because genetic diversity is an important variable to consider when assessing extinction risk (Frankham 2005; Palstra and Ruzzante 2008), when combined with our knowledge of aye-aye demography and in the face of habitat loss and forest fragmentation in Madagascar, our result supports the recommendation of Mittermeier et al. (2010) that the conservation status of this important Malagasy species be elevated from Near Threatened to Endangered.
Genomic Divergence and Aye-Aye Branch Length Analysis
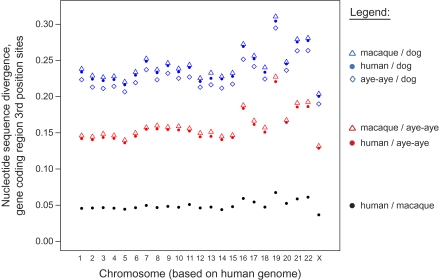
For third codon position sites in gene coding regions, the genome-wide aye-aye versus human nucleotide sequence divergence estimate = 0.152 (maximum composite likelihood model; Tamura et al. 2004). Estimated nucleotide sequence divergence varies by chromosome and is lowest on chromosome X (fig. 2), a finding consistent with those of previous primate comparative genomic studies (e.g., The Chimpanzee Sequencing and Analysis Consortium 2005).
FIG. 2.—
Nucleotide sequence divergence. Pairwise nucleotide sequence divergence estimates for third codon position sites in gene coding regions by chromosome. Chromosome definition is based on the human genome.
As depicted in figure 2, the human versus aye-aye sequence divergence estimate is relatively lower than that for rhesus macaque versus aye-aye. The same pattern is true for the comparisons with other species, including dog (fig. 2). This difference is likely explained by the “hominoid slowdown” effect, which describes a lower substitution rate in hominoids (including humans) relative to Old World monkeys (including macaques) and is hypothesized to result from variation in life-history patterns among lineages, for example, in generation time (Yi et al. 2002; Steiper et al. 2004; Kim et al. 2006).
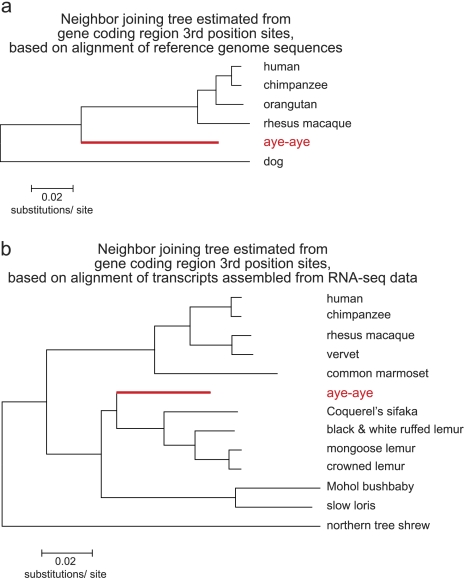
Unexpectedly, however, these data also suggest an even lower substitution rate in the aye-aye lineage relative to the human and macaque lineages. Specifically, the aye-aye versus dog nucleotide sequence divergence estimate is lower than the estimates for both human and macaque versus dog (fig. 2). Under the null model of equal substitution rates among branches, these estimates are expected to be similar (Tajima 1993). Yet, of the 4,261,562 total aligned third position sites among human, aye-aye, and dog, 275,396 are inferred to have substitutions that occurred along the human lineage, whereas 244,690 are inferred to have substitutions that occurred along the aye-aye lineage (relative-rate test; P < 0.00001). Thus, the nucleotide substitution rate in the lineage leading to humans from the primate common ancestor is more than 10% higher than that of the aye-aye lineage. This difference in substitution rates is readily visualized by a neighbor joining phylogeny estimated from the third position nucleotide sequence data (fig. 3).
FIG. 3.—
Estimated phylogenies highlighting the short aye-aye branch length. (a) Neighbor joining phylogeny estimated from third position sites for genes on human chromosome 1, based on the genome alignment analysis performed in this study. (b) Neighbor joining phylogeny estimated from gene coding region third position sites from 515 genes assembled from RNA-sequencing data in Perry et al. (2012). The aye-aye branch is highlighted in red in both panels.
We considered two potential explanations for the unexpected finding of a relatively low substitution rate in the aye-aye lineage. First, while the majority of third codon position substitutions are synonymous (having no effect on a translated protein's amino acid sequence) and therefore presumably neutral, a small proportion of third codon position substitutions are nonsynonymous. Therefore, theoretically, a markedly stronger strength of purifying selection in the aye-aye lineage could drive the pattern we observed. However, the maximum likelihood–estimated genome-wide rate of synonymous substitution (Yang 2007) in the aye-aye lineage (0.093; 260,589 substitutions at 2,811,311 sites) is also considerably lower than that in the human lineage following divergence from the primate common ancestor (0.140; 394,179 substitutions at 2,811,311 sites; substitutions summed across ancestral and extant lineages; summary in supplementary table 1, Supplementary Material online). This observation suggests that the estimated branch length difference based on the third codon position sites can be attributed principally to a lower neutral rate of substitution in the aye-aye lineage rather than to variation in selective constraint.
Second, we considered independently generated data sets to evaluate whether our finding could be explained by the presence of errors in the aye-aye genome assembly. Figure 3b depicts a neighbor joining phylogeny estimated from third codon position sites of 515 genes that were assembled de novo from independently collected RNA-sequencing data (Perry et al. 2012). Among the 12 primates studied, aye-ayes have the shortest aggregate (i.e., “root to tip”) branch length. In turn, Perelman et al. (2011) performed a phylogenetic analysis of 34,927 bp of nuclear genome nucleotide sequence data (comprised of 54 genes) in 186 primate species. Aye-ayes again have the shortest aggregate branch length of any species in their study, providing further support for our original finding. Finally, although based on only one nonrecombining locus, the aye-aye branch length based on third codon position sites of mitochondrial DNA genes was also the shortest among 18 primate genera (Yang and Yoder 2003).
The relatively short aye-aye branch length is consistent, in some respects, with the hypothesis that life-history traits such as generation time are inversely correlated with nucleotide substitution rate (for a recent review, see Tsantes and Steiper 2009). Specifically, aye-ayes have the most extended life-history profile of any extant lemur, with a weaning age of 1.5 years compared with a maximum of 0.68 years in any other studied species and an interbirth interval of 2.5 years versus a maximum of 2 years and the more typical 1 year in other lemurs (Catlett et al. 2010). The age at first reproduction of aye-aye females, 3.5 years (Ross 2003; males show behavioral evidence of sexual maturity at 2.5 years; Winn 1994), is obviously much earlier than that of modern humans. However, the modern human life-history pattern is likely very different from that of our longer term ancestors starting from the time that we last shared a common ancestor with aye-ayes ∼70 Ma.
Moreover, there is indirect evidence suggesting the possibility that ancestral aye-ayes had longer generation times than modern aye-ayes. Specifically, there were at least 17 now-extinct “subfossil” lemur species that survived on Madagascar until ∼2,300 to 1,000 years ago; the timing of extinctions is coincident with the arrival of humans to the island (Godfrey et al. 2006). The reconstructed body sizes of all subfossil lemurs are substantially larger than those of their extant relatives (Jungers et al. 2008), and dental histology patterns suggest that ages of weaning and interbirth intervals for many of these taxa were relatively protracted as well (Catlett et al. 2010). A giant aye-aye, Daubentonia robusta (Lamberton 1934; Macphee and Raholimavo 1988; Simons 1994), is believed to have had a body mass greater than five times that of the extant aye-aye, Daubentonia madagascariensis (Jungers et al. 2008). While life-history trait estimates are not yet available for the subfossil aye-aye, it is possible that the generation time of this species was also considerably greater than that of extant aye-ayes. If so, then it would be interesting to speculate and reconstruct the generation time of ancestral aye-ayes and consider the extent to which a protracted ancestral aye-aye generation time may have contributed to, at least in part, the shortest aggregate branch lengths of any extant primate studied to date.
Evolutionary Analysis of Gene Coding Regions
This aye-aye genome sequence resource is the first such high-coverage resource available for lemurs, thus providing an important step toward more comprehensive primate comparative genomic analyses. For our analysis, we first constructed a database of aye-aye gene sequences on the basis of homology to human exons (see Materials and Methods). We then generated a multispecies alignment of the aye-aye genes and the orthologous gene sequences of human, chimpanzee, orangutan, rhesus macaque, mouse, and dog (representing four other published primate genome sequences and two nonprimate outgroups). We used a maximum likelihood–based method (Yang 2007) to estimate ancestral gene sequences and the numbers and rates of nonsynonymous (amino acid changing) and synonymous (presumably neutral) substitutions on each internal and external branch in the tree. We then examined the ratio of the rates of nonsynonymous to synonymous substitution (dN/dS) to make inferences about long-term selective pressures on amino acid sequences. Specifically, in our analysis, we highlighted genes for which dN was substantially greater than dS. A dN/dS ratio significantly >1 may reflect a past long-term history of positive selection on amino acid substitutions.
Across all lineages, we identified 80 candidates that may have evolved under positive selection (see Materials and Methods; supplementary table 2, Supplementary Material online). Aye-ayes diverged from the other primates included in our analysis ∼70 Ma. On such a long branch, there is little power to identify positively selected genes, as the number of neutrally fixed synonymous substitutions may overpower any shorter term signal of a burst of nonsynonymous fixation. Accordingly, we identified only two candidate genes in the aye-aye lineage: AKIP1, involved in NF-κB signaling (Gao et al. 2010), and LRRC69, not yet studied in detail.
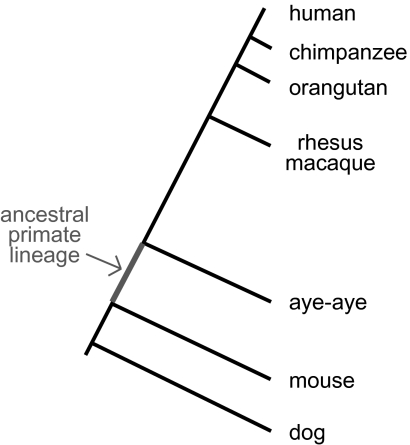
Importantly, the availability of the aye-aye genome sequence resource also provides us with the opportunity to infer nucleotide substitutions that occurred in ancestral primates (in our study, the lineage from the divergence of primates and mouse to the divergence of aye-aye from other primates; fig. 4). We identified 11 candidate genes that may have evolved under positive selection in the ancestral primate lineage (supplementary table 2, Supplementary Material online). Even with such a small number of genes, based on a functional enrichment analysis using gene ontology annotations, we found that three of these genes, IDO1, OSMR, and CD47, have roles in defense response and specifically in the positive regulation of inflammatory response, a significant enrichment over that expected by chance (expected number of genes = 0.03; false discovery rate [FDR] = 0.0006). IDO1, CD47, and another gene, IL18, are also involved in the regulation of T-cell activation (expected = 0.07; FDR = 0.0009). A fifth gene, IL1R1, is involved in cytokine-induced immune response. These results are unsurprising; scans for positive selection consistently identify an enrichment of immunity-related genes (Barreiro and Quintana-Murci 2010). Still, functional study of the specific amino acid substitutions identified in this analysis may help us to advance our understanding of primate-specific aspects of the immune system that may have played critical roles in our evolutionary history.
FIG. 4.—
Phylogenetic relationship of the species considered in this study. The ancestral primate lineage is highlighted.
Conclusion
We have produced a genome sequence resource of the aye-aye, Daubentonia madagascariensis. The assembly was generated in a cost-effective computationally efficient manner and cannot be considered finished quality (being comprised of ∼2.6 million scaffolds). However, the assembly is sufficient to facilitate subsequent comparative and population genetic studies that will benefit our understanding of lemur evolutionary biology in general and aye-aye conservation efforts in particular. For example, the sequence and SNP data that we produced will be useful for subsequent biogeographic studies of aye-aye genetic variation that use either PCR or DNA capture methods as well as for studies of gene expression profiles using RNA sequencing. In our analysis, we unexpectedly found that the substitution rate on the aye-aye lineage is relatively slow compared with other primates, which may reflect a protracted ancestral aye-aye life-history pattern. Additionally, our heterozygosity estimate for the aye-aye donor individual places this important species at the extreme low end of primate genetic diversity. Given the aye-aye's demographic profile of large home ranges and low population densities in the face of continuing deforestation and habitat fragmentation in Madagascar, this result leads us to emphasize the need to prioritize efforts that will benefit aye-aye conservation.
Supplementary Material
Supplementary figures 1 and 2 and tables 1 and 2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Sarah Zehr and the Duke Lemur Center for providing the aye-aye sample used for this study, Ying Wang for assistance with the PAML analysis, Lisa Kimmel for producing figure 1, Steve Salzberg and David Kelley for early access to the Quake sequence error correction program, and Walter Pirovano and Marten Boetzer for early access to the SSPACE scaffolding program. We are grateful to Anne Yoder and Nathaniel Dominy for discussions and comments on the manuscript. This work was funded by National Institutes of Health (NIH) grant GM084996 to Y.G., and G.H.P. was supported by NIH fellowship F32GM085998.
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11:17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- Bradley RK, et al. Fast statistical alignment. PLoS Comput Biol. 2009;5:e1000392. doi: 10.1371/journal.pcbi.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartmill M. Daubentonia, Dactylopsila, woodpeckers, and klinorhynchy. In: Martin RD, Doyle GA, Walker AC, editors. Prosimian biology. Pittsburgh (PA): University of Pittsburgh Press; 1974. pp. 655–670. [Google Scholar]
- Catlett KK, Schwartz GT, Godfrey LR, Jungers WL. “Life history space”: a multivariate analysis of life history variation in extant and extinct Malagasy lemurs. Am J Phys Anthropol. 2010;142:391–404. doi: 10.1002/ajpa.21236. [DOI] [PubMed] [Google Scholar]
- Erickson CJ. Percussive foraging in the aye-aye, Daubentonia madagascariensis. Anim Behav. 1991;41:793–801. [Google Scholar]
- Fischer A, Pollack J, Thalmann O, Nickel B, Paabo S. Demographic history and genetic differentiation in apes. Curr Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Fischer A, Wiebe V, Paabo S, Przeworski M. Evidence for a complex demographic history of chimpanzees. Mol Biol Evol. 2004;21:799–808. doi: 10.1093/molbev/msh083. [DOI] [PubMed] [Google Scholar]
- Fischer A, et al. Bonobos fall within the genomic variation of chimpanzees. PLoS One. 2011;6:e21605. doi: 10.1371/journal.pone.0021605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleagle JG. Primate adaptation and evolution. San Diego (CA): Academic Press; 1999. [Google Scholar]
- Frankham R. Genetics and extinction. Biol Conserv. 2005;126:131–140. [Google Scholar]
- Gao N, Hibi Y, Cueno M, Asamitsu K, Okamoto T. A-kinase-interacting protein 1 (AKIP1) acts as a molecular determinant of PKA in NF-kappaB signaling. J Biol Chem. 2010;285:28097–28104. doi: 10.1074/jbc.M110.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson KR. Cognition, brain size and the extraction of embedded food resources. In: Else JG, Lee PC, editors. Primate ontogeny, cognition and social behaviour. Cambridge: Cambridge University Press; 1986. pp. 93–103. [Google Scholar]
- Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey LR, Jungers WL, Schwartz GT. Ecology and extinction of Madagascar's subfossil lemurs. In: Gould L, Sauther ML, editors. Lemurs: ecology and adaptation. New York: Springer; 2006. pp. 41–64. [Google Scholar]
- Hernandez RD, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316:240–243. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- Horvath JE, et al. Development and application of a phylogenomic toolkit: resolving the evolutionary history of Madagascar's lemurs. Genome Res. 2008;18:489–499. doi: 10.1101/gr.7265208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Iwano T, Iwakawa C. Feeding behavior of the aye-aye (Daubentonia madagascariensis) on nuts of ramy (Canarium madagascariensis) Folia Primatol (Basel). 1988;50:136–142. [Google Scholar]
- Jungers WL, Demes B, Godfrey LR. How big were the “giant” extinct lemurs of Madagascar. In: Fleagle JG, Gilbert CC, editors. Elwyn Simons: a search for origins. New York: Springer; 2008. pp. 343–360. [Google Scholar]
- Keller A, et al. GeneTrailExpress: a web-based pipeline for the statistical evaluation of microarray experiments. BMC Bioinformatics. 2008;9:552. doi: 10.1186/1471-2105-9-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Schatz MC, Salzberg SL. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 2010;11:R116. doi: 10.1186/gb-2010-11-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Elango N, Warden C, Vigoda E, Yi SV. Heterogeneous genomic molecular clocks in primates. PLoS Genet. 2006;2:e163. doi: 10.1371/journal.pgen.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberton C. Contribution a la connaissance de la faune subfossile de Madagascar. Mem Acad Malgache. 1934;17:40–46. [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macphee RDE, Raholimavo EM. Modified subfossil aye-aye incisors from Southwestern Madagascar: species allocation and paleoecological significance. Folia Primatol (Basel). 1988;51:126–142. doi: 10.1159/000156363. [DOI] [PubMed] [Google Scholar]
- Martin RD. Primate origins and evolution: a phylogenetic reconstruction. Princeton (NJ): Princeton University Press; 1990. [Google Scholar]
- Mittermeier RA, et al. Lemurs of Madagascar. Arlington (VA): Conservation International; 2010. [Google Scholar]
- Palstra FP, Ruzzante DE. Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Mol Ecol. 2008;17:3428–3447. doi: 10.1111/j.1365-294x.2008.03842.x. [DOI] [PubMed] [Google Scholar]
- Perelman P, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Marioni JC, Melsted P, Gilad Y. Genomic-scale capture and sequencing of endogenous DNA from feces. Mol Ecol. 2010;19:5332–5344. doi: 10.1111/j.1365-294X.2010.04888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Martin RD, Verrelli BC. Signatures of functional constraint at aye-aye opsin genes: the potential of adaptive color vision in a nocturnal primate. Mol Biol Evol. 2007;24:1963–1970. doi: 10.1093/molbev/msm124. [DOI] [PubMed] [Google Scholar]
- Perry GH, Verrelli BC, Stone AC. Molecular evolution of the primate developmental genes MSX1 and PAX9. Mol Biol Evol. 2006;23:644–654. doi: 10.1093/molbev/msj072. [DOI] [PubMed] [Google Scholar]
- Perry GH, et al. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. Forthcoming 2012 doi: 10.1101/gr.130468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plimpton SJ, Devine KD. MapReduce in MPI for large-scale graph algorithms. Parallel Comput. 2011;37:610–632. [Google Scholar]
- Red List of Threatened Species [Internet]. 2010.4. Cambridge (UK): International Union for Conservation of Nature; 2010. [cited 2011 May 01]. Available from: www.iucnredlist.org [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Ross C. Life history, infant care strategies, and brain size in primates. In: Kappeler PM, Pereira ME, editors. Primate life histories and socioecology. Chicago (IL): Chicago University Press; 2003. pp. 266–284. [Google Scholar]
- Simons EL. The giant aye-aye Daubentonia robusta. Folia Primatol (Basel). 1994;62:14–21. doi: 10.1159/000156759. [DOI] [PubMed] [Google Scholar]
- Simons EL. History, anatomy, subfossil record and management of Daubentonia madagascariensis. In: Alterman L, Doyle GA, Izard MK, editors. Creatures of the dark: the nocturnal prosimians. New York: Plenum Press; 1995. pp. 133–140. [Google Scholar]
- Steiper ME, Young NM, Sukarna TY. Genomic data support the hominoid slowdown and an Early Oligocene estimate for the hominoid-cercopithecoid divergence. Proc Natl Acad Sci U S A. 2004;101:17021–17026. doi: 10.1073/pnas.0407270101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling EJ. The behavioral ecology of the aye-aye on Nosy Mangabe, Madagascar. [dissertation]. [New Haven (CT)]: Yale University; 1993. [Google Scholar]
- Sterling EJ, Povinelli DJ. Tool use, aye-ayes, and sensorimotor intelligence. Folia Primatol (Basel). 1999;70:8–16. doi: 10.1159/000021669. [DOI] [PubMed] [Google Scholar]
- Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Tsantes C, Steiper ME. Age at first reproduction explains rate variation in the strepsirrhine molecular clock. Proc Natl Acad Sci U S A. 2009;106:18165–18170. doi: 10.1073/pnas.0906686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, et al. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci U S A. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, et al. A novel DNA sequence database for analyzing human demographic history. Genome Res. 2008;18:1354–1361. doi: 10.1101/gr.075630.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn RM. Preliminary study of the sexual behaviour of three aye-ayes (Daubentonia madagascariensis) in captivity. Folia Primatol (Basel). 1994;62:63–74. doi: 10.1159/000156764. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yoder AD. Comparison of likelihood and Bayesian methods for estimating divergence times using multiple gene Loci and calibration points, with application to a radiation of cute-looking mouse lemur species. Syst Biol. 2003;52:705–716. doi: 10.1080/10635150390235557. [DOI] [PubMed] [Google Scholar]
- Yi S, Ellsworth DL, Li WH. Slow molecular clocks in Old World monkeys, apes, and humans. Mol Biol Evol. 2002;19:2191–2198. doi: 10.1093/oxfordjournals.molbev.a004043. [DOI] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, Ryder O, Li WH. Nucleotide diversity in gorillas. Genetics. 2004;166:1375–1383. doi: 10.1534/genetics.166.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, et al. Low nucleotide diversity in chimpanzees and bonobos. Genetics. 2003;164:1511–1518. doi: 10.1093/genetics/164.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]