Abstract
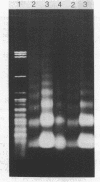
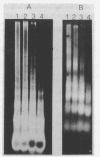
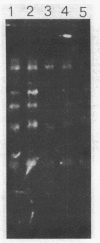
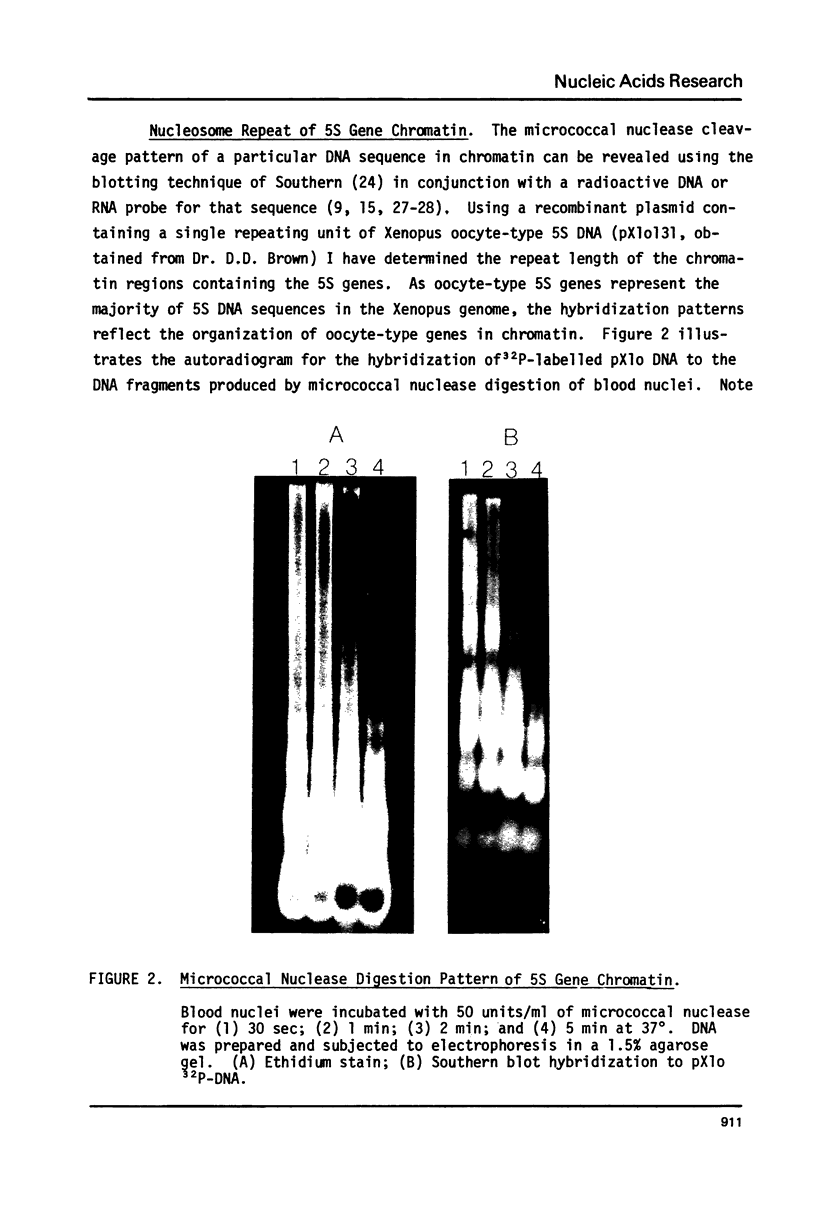
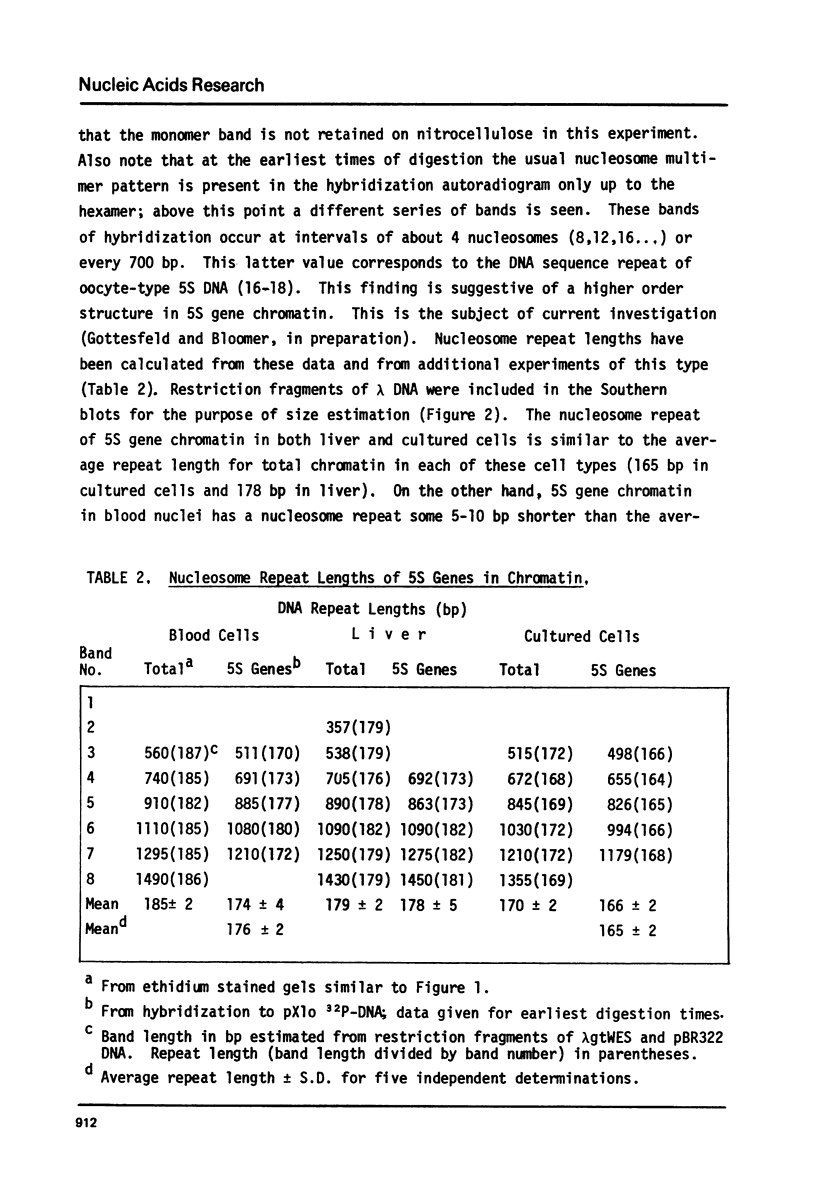
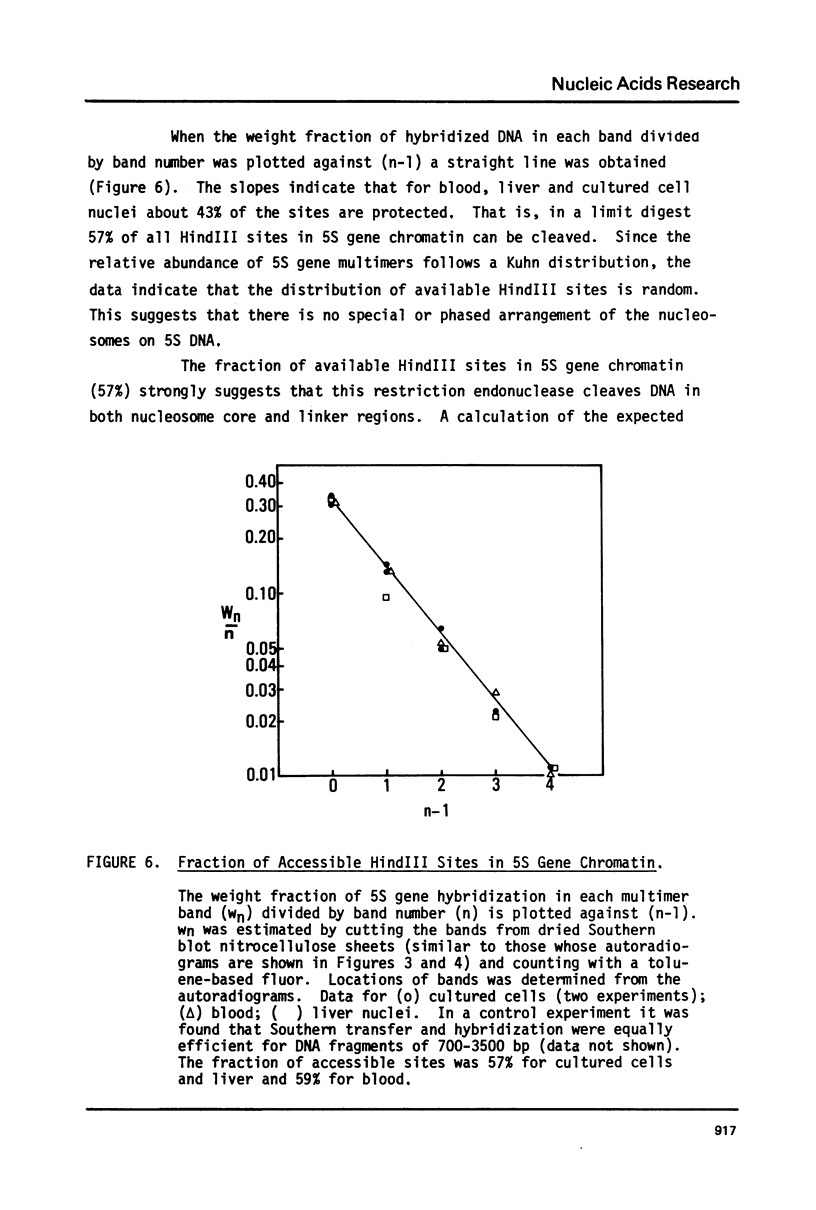
The chromatin organization of the genes coding for 5S RNA in Xenopus laevis has been investigated with restriction endonucleases and micrococcal nuclease. Digestion of nuclei from liver, kidney, blood and kidney cells maintained in culture with micrococcal nuclease reveals that these Xenopus cells and tissues have shorter nucleosome repeat lengths than the corresponding cells and tissues from other higher organisms. 5S genes are organized in nucleosomes with repeat lengths similar to those of the bulk chromatin in liver (178 bp) and cultured cells (165 bp); however, 5S gene chromatin in blood cells has a shorter nucleosome repeat (176 bp) than the bulk of the genome in these cells (184 bp). From an analysis of the 5S DNA fragments produced by extensive restriction endonuclease cleavage of chromatin in situ, no special arrangement of the nucleosomes with respect to the sequence of 5S DNA can be detected. The relative abundance of 5S gene multimers follows a Kuhn distribution, with about 57% of all HindIII sites cleaved. This suggests that HindIII sites can be cleaved both in the nucleosome core and linker regions.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biroc S. L., Reeder R. H. Iodination of Xenopus laevis histone F2a1 in chromatin. Biochemistry. 1976 Apr 6;15(7):1440–1448. doi: 10.1021/bi00652a014. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Fractionation of hen oviduct chromatin into transcriptionally active and inactive regions after selective micrococcal nuclease digestion. Cell. 1978 Sep;15(1):141–150. doi: 10.1016/0092-8674(78)90090-9. [DOI] [PubMed] [Google Scholar]
- Botchan M., McKenna G., Sharp P. A. Cleavage of mouse DNA by a restriction enzyme as a clue to the arrangement of genes. Cold Spring Harb Symp Quant Biol. 1974;38:383–395. doi: 10.1101/sqb.1974.038.01.041. [DOI] [PubMed] [Google Scholar]
- Brown F. L., Musich P. R., Maio J. J. The repetitive sequence structure of component alpha DNA and its relationship to the nucleosomes of the African green monkey. J Mol Biol. 1979 Jul 15;131(4):777–799. doi: 10.1016/0022-2836(79)90201-8. [DOI] [PubMed] [Google Scholar]
- Carroll D., Brown D. D. Repeating units of Xenopus laevis oocyte-type 5S DNA are heterogeneous in length. Cell. 1976 Apr;7(4):467–475. doi: 10.1016/0092-8674(76)90198-7. [DOI] [PubMed] [Google Scholar]
- Fedoroff N. V., Brown D. D. The nucleotide sequence of oocyte 5S DNA in Xenopus laevis. I. The AT-rich spacer. Cell. 1978 Apr;13(4):701–716. doi: 10.1016/0092-8674(78)90220-9. [DOI] [PubMed] [Google Scholar]
- Fittler F., Zachau H. G. Subunit structure of alpha-satellite DNA containing chromatin from African green monkey cells. Nucleic Acids Res. 1979 Sep 11;7(1):1–13. doi: 10.1093/nar/7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Axel R. The structure of the transcriptionally active ovalbumin genes in chromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):701–708. doi: 10.1101/sqb.1978.042.01.072. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Glover C., Johmann C. A., Keevert J. B., Mathis D. J., Samuelson M. Histones and chromatin structure in Tetrahymena macro- and micronuclei. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):493–503. doi: 10.1101/sqb.1978.042.01.052. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Butler P. J. Structure of transcriptionally-active chromatin subunits. Nucleic Acids Res. 1977 Sep;4(9):3155–3173. doi: 10.1093/nar/4.9.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Melton D. A. The length of nucleosome-associated DNA is the same in both transcribed and nontranscribed regions of chromatin. Nature. 1978 May 25;273(5660):317–319. doi: 10.1038/273317a0. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Humphries S. E., Young D., Carroll D. Chromatin structure of the 5S ribonucleic acid genes of Xenopus laevis. Biochemistry. 1979 Jul 24;18(15):3223–3231. doi: 10.1021/bi00582a006. [DOI] [PubMed] [Google Scholar]
- Hörz W., Igo-Kemenes T., Pfeiffer W., Zachau H. G. Specific cleavage of chromatin by restriction nucleases. Nucleic Acids Res. 1976 Nov;3(11):3213–3226. doi: 10.1093/nar/3.11.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Kumar A., Lindberg U. Characterization of messenger ribonucleoprotein and messenger RNA from KB cells. Proc Natl Acad Sci U S A. 1972 Mar;69(3):681–685. doi: 10.1073/pnas.69.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy E., Axel R. Analysis of DNA of isolated chromatin subunits. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3978–3982. doi: 10.1073/pnas.72.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy B., Dixon G. H. Partial purification of transcriptionally active nucleosomes from trout testis cells. Nucleic Acids Res. 1978 Nov;5(11):4155–4163. doi: 10.1093/nar/5.11.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipchitz L., Axel R. Restriction endonuclease cleavage of satellite DNA in intact bovine nuclei. Cell. 1976 Oct;9(2):355–364. doi: 10.1016/0092-8674(76)90125-2. [DOI] [PubMed] [Google Scholar]
- Lohr D., Tatchell K., Van Holde K. E. On the occurrence of nucleosome phasing in chromatin. Cell. 1977 Nov;12(3):829–836. doi: 10.1016/0092-8674(77)90281-1. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Characterization of DNase-I cleavage sites in the nucleosome. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):137–147. doi: 10.1101/sqb.1978.042.01.015. [DOI] [PubMed] [Google Scholar]
- Manteuil S., Hamer D. H., Thomas C. A., Jr Regular arrangement of restriction sites in Drosophila DNA. Cell. 1975 Aug;5(4):413–422. doi: 10.1016/0092-8674(75)90060-4. [DOI] [PubMed] [Google Scholar]
- Miller J. R., Cartwright E. M., Brownlee G. G., Fedoroff N. V., Brown D. D. The nucleotide sequence of oocyte 5S DNA in Xenopus laevis. II. The GC-rich region. Cell. 1978 Apr;13(4):717–725. doi: 10.1016/0092-8674(78)90221-0. [DOI] [PubMed] [Google Scholar]
- Morris N. R. A comparison of the structure of chicken erythrocyte and chicken liver chromatin. Cell. 1976 Dec;9(4 Pt 1):627–632. doi: 10.1016/0092-8674(76)90045-3. [DOI] [PubMed] [Google Scholar]
- Musich P. R., Brown F. L., Maio J. J. Subunit structure of chromatin and the organization of eukaryotic highly repetitive DNA: nucleosomal proteins associated with a highly repetitive mammalian DNA. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3297–3301. doi: 10.1073/pnas.74.8.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Ponder B. A., Crawford L. V. The arrangement of nucleosomes in nucleoprotein complexes from polyoma virus and SV40. Cell. 1977 May;11(1):35–49. doi: 10.1016/0092-8674(77)90315-4. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Relation of nucleosomes to nucleotide sequences in the rat. Philos Trans R Soc Lond B Biol Sci. 1978 May 11;283(997):269–273. doi: 10.1098/rstb.1978.0023. [DOI] [PubMed] [Google Scholar]
- Reeves R., Jones A. Genomic transcriptional activity and the structure of chromatin. Nature. 1976 Apr 8;260(5551):495–500. doi: 10.1038/260495a0. [DOI] [PubMed] [Google Scholar]
- Singer D. S. Arrangement of a highly repeated DNA sequence in the genome and chromatin of the African green monkey. J Biol Chem. 1979 Jun 25;254(12):5506–5514. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Thompson R. J. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977 Apr;10(4):633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- Van Lente F., Jackson J. F., Weintraub H. Identification of specific crosslinked histones after treatment of chromatin with formaldehyde. Cell. 1975 May;5(1):45–50. doi: 10.1016/0092-8674(75)90090-2. [DOI] [PubMed] [Google Scholar]
- Weinstock R., Sweet R., Weiss M., Cedar H., Axel R. Intragenic DNA spacers interrupt the ovalbumin gene. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1299–1303. doi: 10.1073/pnas.75.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]