Abstract
Imatinib mesylate (Imatinib), clinically employed for chronic myeloid leukemia and gastrointestinal stromal tumors, is a selective inhibitor of the tyrosine kinases, c-abl, c-kit and PDGFRs. Due to the frequent expression of these genes in breast cancer cells, the clinical efficacy of Imatinib has recently been investigated in patients with advanced and metastatic breast cancer. Here, we have studied the effects of Imatinib on human MA-11 breast carcinoma cells, expressing both c-abl and PDGFRbeta, in vitro and in mouse xenografts. Daily intraperitoneal treatment with 60 mg/kg Imatinib for 9 days of athymic nude mice pre-implanted subcutaneously with MA-11 cells did not result in an antitumor effect, but rather increased the take rate of 3×104 cells from 30.8% to 84.6% and caused the appearance of large abdominal masses in 30% of mice. To investigate the mechanism(s) of the observed effects of Imatinib on MA-11 tumors, we exposed the cells in vitro to Imatinib for 9 days. The surviving population, expanded in culture, showed increased motility and over-expressed a set of genes associated with aggressive behavior. Also, several genes belonging to the Wnt and the MAPK pathway were differentially expressed. In promoter activation assays, Imatinib increased the promoter activity driven by both Wnt and MAPK/ERK-1/2. Our data suggest caution in the clinical use of Imatinib in breast cancer patients; the comparison of Imatinib-surviving breast cancer cells with parental cells may help define the regulatory pathways involved in the increased malignancy of residual tumor cells that survive therapy, ultimately providing important therapeutic targets.
Keywords: Breast cancer, Imatinib, cancer stem cells, CD24, microarray
INTRODUCTION
Every year, more than one million women are newly diagnosed with breast cancer worldwide (1). One of the most exciting areas of improvement in the treatment of breast cancer is the development of novel targeted and biological therapeutic agents, such as Imatinib mesylate (Imatinib), a 2-phenylaminopyrimidine derivative that specifically inhibits the tyrosine kinase enzymes, c-Abl, PDGFR, and c-Kit (2,3). Imatinib was first developed to specifically inhibit Bcr–Abl, a fusion protein with kinase activity present in chronic myeloid leukemia (CML) patients. It has been widely used for treatment of patients with CML and gastrointestinal stromal tumors, and is under investigation in several malignancies, including breast cancer (4,5). Also, potentiation of chemotherapy by Imatinib in breast carcinoma cell lines has recently been reported (6).
Recent evidence suggests that breast cancer originates from and is maintained by a sub-population of stem cell-like cells, named cancer-initiating cells or cancer stem cells (CSC), able to differentiate along different pathways. The identification and characterization of breast CSC has important diagnostic, prognostic and therapeutic implications [4–7]. Originally, breast CSC were described as CD44+CD24−/low cells. Employing as breast cancer model the human MA-11 breast carcinoma cell line, established from bone marrow micrometastases of a breast cancer patient, we have previously reported (7) that (i) MA-11 cells grown as mammospheres under stem cell-like conditions acquire increased tumorigenicity and lose CD24 expression compared with the parental adherent cell line; (ii) the level of CD24 expression is neither a stable feature of mammosphere-forming cells nor confers tumorigenic potential to MA-11 cells; (iii) cancer-initiating cells-enriched MA-11 mammospheres have activated specific signal transduction pathways, including MAPK and Wnt, potential targets for anti-breast cancer therapy. Here, we have studied the effects of Imatinib on MA-11 cells in vitro and in vivo, and report that treatment with Imatinib results in increased tumorigenicity and malignancy of MA-11 cells.
MATERIALS AND METHODS
Cell culture
MA-11 cells were cultured in RPMI-1640 (Mediatech Inc. Herndon, VA) additioned with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) at 37 °C in a 5% CO2 humidified incubator. Cells were used between passages 10 and 20 and tested routinely for mycoplasma contamination.
Tumor implantation
Animal studies were done under a protocol approved by the Institutional Animal Care and Use Committee. For s.c. injection, cells were resuspended in 100 μl PBS, and injected subcutaneously into the right flank of the animals.
Wound healing assay
To perform the wound healing assay, a wound in a confluent MA-11 cell monolayer was introduced using a pipette tip. The monolayers were observed over a time course of 3–24 hrs.
Flow cytometric analysis
Mouse anti-human CD24 (clone ML5) and anti-human CD44 (clone G44-26) monoclonal antibodies were from BD Biosciences (San Jose, CA). Cell surface CD24 expression was quantitated using the Quantum Simply Cellular System (Bangs Laboratories, Fishers, IN), as previously described (8). 5×105 cells/sample were incubated with saturating concentrations (10 μg/ml) of antibodies for 30 min at 4°C. Standard curves of beads with fixed antibody-binding capacity and samples were analyzed on a FACSVantage flow cytometer (BD Biosciences, San Jose, CA). Antibody binding capacity for each cell population was calculated using the QuickCal v.2.3. software (Bangs Laboratories), employing median histogram values and linear regression analyses.
Microarray analysis
Total RNA was isolated from Imatinib- and mock-treated MA-11 cells using Trizol phenol-based extraction. The quality of the RNA was assessed by gel electrophoresis and A260/A280 ratio. Microarray analyses were performed by Ocean Ridge Biosciences (ORB, Palm Beach Gardens, FL) using human exonic evidence-based oligonucleotide (HEEBO) microarrays (http://alizadehlab.stanford.edu/) containing approximately 50,000 70-mer probes complementary to constitutive exons of most human genes, as well as alternatively spliced exons, ESTs, and control sequences. Biotinylated UTP complementary RNA (cRNA) probes were prepared, fragmented and hybridized to the microarrays for 16–18 h with constant rotation. The microarray slides were washed under stringent conditions, stained with Streptavidin-Alexa-647 (Invitrogen), and scanned using an Axon GenePix 4000B scanner. For data analyses, the local background was subtracted and the spot intensities were log2-transformed. The spot intensities were then normalized by subtracting the 70th percentile of spot intensity of the probes against human constitutive exons and adding back a scaling factor (grand mean of 70th percentiles). After removing data for low quality spots, control sequences, and non-human probes, 41,213 human probe intensities remained. The human probes intensities were filtered to identify all probes with intensity above a normalized threshold (log2 (3* standard deviation of raw local background) + mean of log2-transformed negative controls), to arrive at 23,291 probes above threshold in both samples from at least one group. For statistical analysis, samples were binned in two groups (Imatinib and control). The log2-transformed and normalized spot intensities for the detectable probes were examined for differences between the treatment groups by 1-way ANOVA using National Institute of Aging (NIA) Array Analysis software. This ANOVA was conducted using the Bayesian Error Model and 20 degrees of freedom. Gene ontology analysis was performed using GenMAPP software (Gladstone Institute, San Francisco, CA) for all detectable probes with current Entrez Gene IDs. Specifically, the MAPPfinder module of GenMAPP was first used to map all detectable probes, based on their gene targets, to GO and Local MAPP categories. Then MAPPfinder compared the relative representation in each functional group of genes associated with probes meeting one of 5 differential expression criteria to the relative representation of genes associated with the full set of detectable probes. Significance was determined by permutation of Z scores with correction for multiple comparisons as described in the GenMAPP software manual. To shed light on the Imatinib-specific signaling pathways, all differentially expressed genes linked to GO terms identified in the microarray were subjected to pathway analysis using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
Transduction of MA-11 cells with lentiviral reporter gene constructs
MA-11 cells were transduced with VSV-pseudotyped lentiviral particles expressing inducible reporter constructs encoding the luciferase gene under the control of a basal promoter element, joined to tandem repeats of a specific transcriptional response element (Cignal Finder Lenti Cancer 10-Pathway Reporter Array kit, from SABiosciences, Frederick, MD). For transduction of MA-11 cells, lentiviral supernatants were preloaded onto recombinant fibronectin (Retronectin, Takara Shuzo, Japan)-coated plates and centrifuged at 950×g for 30 min at 4 °C. The operation was repeated a second time with fresh supernatant. The supernatant was then removed and the plates washed with PBS before addition of MA-11 cells in their complete culture medium. After transduction, stable cell lines expressing the specific transcriptional response elements were isolated via selection with 2 μg/ml puromycin. Pathway-specific MA-11 cells were seeded at a density of 2 × 104 per well in 24-well plates and grown to 50% to 60% confluence. Cells were then treated with Imatinib or with the solvent for 24 h or for 9 days. The preparation of cell extracts and measurement of luciferase activities were carried out using the Steady-Glo Luciferase Reporter Assay System according to recommendations by the manufacturer (Promega, Madison, WI). The assays for luciferase activity were done in a 20/20 luminometer (Turner Biosystems, Sunnyvale, CA). The ratio of luciferase activity between Imatinib- and mock-treated samples were then calculated.
RESULTS
Effects of Imatinib on MA-11 xenografts
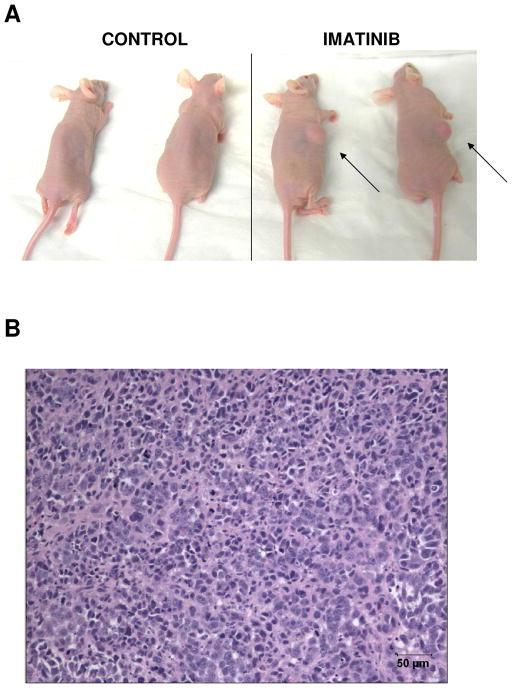
To investigate the activity of Imatinib on breast cancer xenografts, we implanted 30,000 MA-11 cells s.c. in athymic nude mice and treated the animals i.p. with 60 mg/Kg Imatinib for 9 consecutive days, starting the day after the implantation of the tumor cells. Imatinib did not have an antitumor effect, rather increased the tumorigenic potential of MA-11 cells and in 30% of animals caused the formation of large tumor masses in the intraperitoneal cavity. Table 1 shows the take rates in control and Imatinib-treated animals (30.8 and. 84.6%, respectively). The average time to reach 1 cm3 was 83.5 days for control animals and 41.1 days for animals exposed to Imatinib. A representative experiment with two control and two Imatinib-treated mice is shown in Fig. 1A. An hematoxylin-eosin-stained section from an abdominal MA-11 mass, showing a very cellular tumor, is represented in Fig. 1B.
Table 1.
In vivo effects of i.p. Imatinib on the formation of MA-11 tumors in athymic s.c. implanted nude mice
| Parameter | Control | Imatinib |
|---|---|---|
| Take rate (%) | 30.8 (n=13) | 84.6 (n=13)* |
| Time to 1 cm3 tumor (days) | 83.5 ± 24.5+ | 41.1 ± 4.7+* |
| Presence of an abdominal tumor mass (%) | 0 (n=13) | 30(n=13)* |
Mice were implanted with 3×104 MA-11 cells s.c. and treated daily i.p. with 60 mg/Kg Imatinib for 9 days starting the day after the implantation.
p<0.05, Wilcoxon two-tailed test;
SD.
Figure 1. In vivo effects of Imatinib treatment on MA-11 tumors.
A. Athymic nude mice implanted s.c. with MA-11 cells and treated with 9 daily doses of 60 mg/Kg Imatinib or with the solvent alone. B. Hematoxylin-eosin staining of an abdominal tumor mass from a nude mouse injected s.c. with Imatinib-surviving cells.
Reversible changes in the CD44+CD24−/low phenotype of MA-11 cells by Imatinib
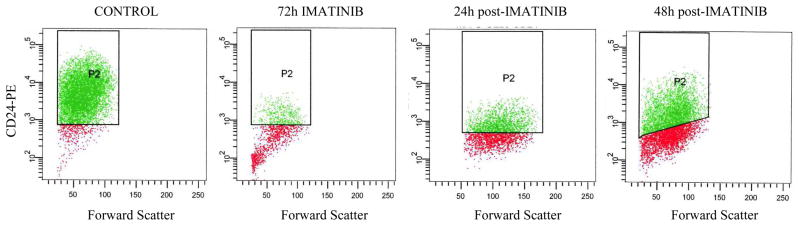
Originally, breast CSC were described as CD44+CD24−/low cells. We have previously reported (7) that in MA-11 cells the level of CD24 expression is neither a stable feature of CSC-enriched cells grown as mammospheres nor confers tumorigenic potential to the cells. We now found that upon treatment of MA-11 cells with Imatinib, CD24 expression on the plasma membrane was almost completely lost in 3 days (Fig. 2). The average number of CD24 molecules on the surface of MA-11 cells, measured by flow cytometry, employing phycoerythrin-conjugated anti-human CD24 monoclonal antibodies and the Quantum Simply Cellular System, decreased from 214,000 to 7,000 .molecules/cell. Instead, CD44 did not change significantly with Imatinib treatment, with the average number of CD44 molecules/cell going from 4,100,000 to 4,000,000 after 72h exposure to Imatinib. 24 and 48 h after removal of Imatinib from the culture medium, the average number of CD24 molecules/cell increased very rapidly to 21,000 and 45,000, respectively (Fig. 2). Microarray analysis (data not shown) confirmed that four days after removal of Imatinib from the culture medium, CD24 expression was similar to that of control mock-treated cells.
Figure 2. Imatinib-induced changes in CD24 expression on the surface of MA-11 cells.
MA-11 cells were exposed to 25 μM Imatinib for 72 h, followed by incubation in drug-free medium for up to 48 h. After labeling of MA-11 cells with an anti-CD24-PE monoclonal antibody, cells were analyzed by flow cyotmetry for CD24 expression. P2 gates include only the CD24+ cells based on their respective isotype controls.
In vitro study of the Imatinib-surviving fraction of MA-11 cells
To investigate the mechanism of the effects of Imatinib on MA-11 cells, we simulated the in vivo treatment by exposing MA-11 cells in vitro for 9 days to 25 μM Imatinib. After 9 days, the great majority of cells died. After growth and expansion in culture of the surviving cells for four additional days in the absence of Imatinib, we compared Imatinib-treated cells and the parental cell line for biological properties, chemosensitivity and gene expression pattern. First, we noticed that the motility of the cells increased, as measured by a conventional wound healing assay (9). After 24 hours, MA-11 cells pre-treated with Imatinib polarized toward the wound, initiated protrusion, migrated, and closed the wound faster than mock-treated cells (Suppl. Fig. 1).
We then investigated the pattern of chemosensitivity of Imatinib-surviving cells by an MTS/PMS microtiter plate assay. Cells were exposed to the investigational drugs for 72 h. The level of resistance to Imatinib was 2-fold, while no changes in sensitivity to the cytotoxic activity of etoposide, paclitaxel, doxorubicin, antimony potassium tartrate and chlorambucil were observed (Table 2).
Table 2.
Drug sensitivity profile of Imatinib-surviving MA-11 cells (Imatinib)Cancer Chemotherapy and Pharmacology
| Drug | IC50 Control (mean ± SD) | IC50 Imatinib (mean ± SD) |
|---|---|---|
| Imatinib | 20 ± 4 | 40 ± 5* |
| Etoposide | 1.2 ± 0.3 | 1.3 ± 0.2 |
| Paclitaxel | 0.05 ± 0.01 | 0.06 ± 0.02 |
| Doxorubicin | 0.2 ± 0.07 | 0.15 ± 0.06 |
| Antimony | 6 ± 1 | 5 ± 1.5 |
| Chlorambucil | 500 ± 150 | 600 ± 100 |
MA-11 cells were exposed to 25 μM Imatinib for 9 days. After 4 days in drug-free medium, their sensitivity to a panel of drugs was compared with that of mock-treated cells by an MTT assay. IC50 values are expressed in μM concentrations, except for Imatinib (μg/ml). SD, standard deviation;
, p>0.05, ANOVA test.
To investigate whether specific genes and/or signaling pathways were involved in the increased tumorigenicity and invasiveness of Imatinib-surviving MA-11 cells, we analyzed Imatinib-induced changes in gene expression profile. By HEEBO microarray analysis of the gene expression profiles of two independent cultures each for control and Imatinib-surviving MA-11 cells, we found 877 probes showing significant differences with P<0.05 and FDR (false discovery rate) <5% (Suppl. Table 1). Excluding the non-annotated genes and multiple probes, only 301 and 241 genes were found to be significantly up- and down-regulated, respectively (Suppl. Table 2). In all cases where multiple probes were available for the 542 genes, the changes in expression were very similar. First, we searched for Imatinib target genes, whose over-expression could cause resistance to Imatinib. We found that c-kit and pdgfr-alpha were not expressed in MA-11 cells, abl and pdgfr-beta were equally expressed between parental and Imatinib-treated cells, while CACNA1G, a T-type calcium channel, recently identified as an additional target of Imatinib, unrelated to the inhibition of tyrosine kinases (10), was 3-fold over-expressed in Imatinib-treated cells (Suppl. Table 3).
Although no difference in the cytotoxic activity of several chemotherapeutic agents was observed, 35 transporters were found differentially regulated in Imatinib-surviving versus control cells, including ABCC3 (MRP3), ABCC6 (MRP6), and several SLC (solute carrier) genes. The over-expression of SLC22A3 and ABCC3 was confirmed by RT-PCR (Table 3).
Table 3.
Fold-increase in expression for 10 genes over-expressed in Imatinib-surviving MA-11 cells by microarray and RT-PCR.
| Entrez Gene ID | Entrez Symbol | Microarray Fold Change | RT-PCR Fold Change |
|---|---|---|---|
| 22943 | NTS | 6.9 | 10.0 |
| 4804 | DACT1 | 4.0 | 5.0 |
| 4897 | FAIM2 | 2.5 | 10.0 |
| 7045 | SLC22A3 | 4.9 | 2.5 |
| 28513 | ABCC3 | 2.2 | 3.3 |
| 1000 | COP1 | 2.2 | 2.5 |
| 558 | CASP9 | 1.9 | 2.0 |
| 3725 | PCDHGA11 | 5.2 | 5.0 |
| 4035 | CACNA1G | 2.2 | 1.7 |
| 51339 | MMP9 | 1.6 | 1.3 |
To shed light on Imatinib-induced or repressed signaling pathways, all genes differentially expressed in the microarray were subjected to pathway analysis and assigned to pathway networks. In particular, 8 genes of the Wnt pathway were up-regulated and 5 and 6 genes of the MAPK pathway were respectively up- and down-regulated in Imatinib-surviving cells (Suppl. Tables 4 and 5). We then looked for genes associated with tumorigenicity, invasiveness, and/or the breast cancer stem cell phenotype, and we found that Imatinib-treated cells overexpress many genes associated with malignancy of breast cancer cells, including the mitochondrial benzodiazepine receptor TSPO, neurotensin, MMP-9, the inhibitor of cell death, COP1, and the aldehyde dehydrogenase gene, ALDH3A1, associated with the breast cancer stem cell phenotype (11) (Suppl. Table 6). We then employed RT-PCR to confirm the differential expression of ten representative over-expressed genes. A good correlation between microarray and RT-PCR data was observed for all ten genes (Table 3).
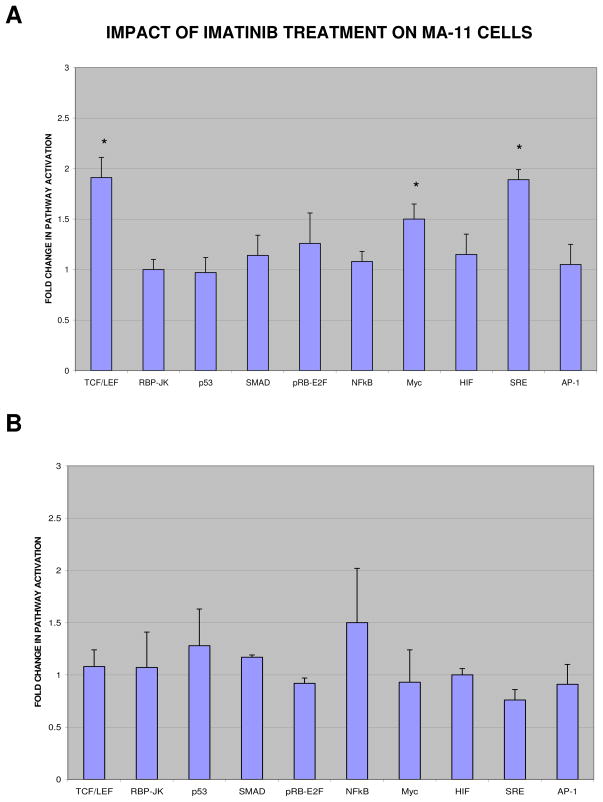
To further investigate the effects of Imatinib on the WNT/β-catenin and the MAPK pathways, we analyzed the transcriptional activation of promoters driven by Wnt and MAPK proteins, using luciferase as reporter. To this aim, we transduced MA-11 cells with TCF/LEF and SRE lentiviral reporters, as well as with lentiviral reporters for eight additional cancer-related pathways, namely Notch, p53, TGFβ, pRB-E2F, NFkB, Myc, Hypoxia, and JNK. After puromycin selection, we exposed each reporter cell line to a short treatment (24h) or a long treatment (9 days + 4 days recovery) with 25 μM Imatinib and analyzed the activity of the different pathways by a luciferase assay. Fig. 3 shows that in Imatinib-surviving cells only the Wnt/β-catenin, the MAPK/Erk1-2, and the Myc pathways were activated (Fig. 3A), while after 24h-exposure to Imatinib no significant changes could be observed for any of the ten pathways investigated (Fig. 3B).
Figure 3. Imatinib enhances promoter activation driven by Wnt and MAPK proteins.
MA-11 cells were transduced with ten different lentiviral reporters (Cignal Finder Lenti Cancer 10-Pathway Reporter Array kit, from SABiosciences). After puromycin selection, each reporter cell line was exposed to 25 μM Imatinib for 9 days, followed by a 4-day recovery period (A) or for 24 h (B). The activity of the different pathways was analyzed by a luciferase assay, and expressed as the ratio between the luciferase activity of Imatinib-treated and mock-treated MA-11 cells. Data points are the mean of three separate experiments; bars, SD; *, P<0.05 (ANOVA test).
DISCUSSION
Despite novel therapeutic strategies for advanced and metastatic breast cancer, many patients relapse after an initial response to conventional and/or experimental drug treatments. In most cases, local and distant recurrences are associated with a more aggressive phenotype and resistance to therapy. It has been suggested that a small subpopulation of “tumor-initiating cells” or “cancer stem cells” (CSC) are resistant to therapy and hence may reinitiate tumor growth after treatment (1). Chemotherapy-induced selection of CSC has been reported in several studies (12,13). However, the CSC that have survived first-line therapy should re-establish a recurrent cancer that is similar to the original cancer both in biological behavior and susceptibility to first-line therapy.
We have found that treatment of MA-11 cells in vitro with Imatinib results in a surviving population of cells with increased motility, that over-express a set of genes associated with tumorigenic and invasive behavior. In vivo, nude mice subcutaneously implanted with MA-11 cells and treated with Imatinib developed more tumors at low levels of inoculum and developed with high frequency abdominal tumor masses. There are three possible non mutually exclusive explanations for our findings: (i) Imatinib selects a sub-population of breast CSC; (ii) Imatinib induces a more malignant gene expression pattern; (iii) Imatinib exerts an immunosuppressive activity in vivo, thus facilitating tumor engraftment.
While selection of a CSC population could explain our in vitro data, it is not sufficient to justify our data of increased tumorigenicity and malignancy in immunodeficient mice. Also, microarray analysis showed that stem cell markers or cancer stem cell markers were either not expressed in MA-11 cells or not differentially expressed: in particular, ALDH1, Notch3, CD133, Rex-1, Oct-4, Nanog were not expressed, and Sox-2 was equally expressed in Imatinib-treated and control MA-11 cells. In vitro treatment with Imatinib resulted in decreased expression of CD24, a marker negatively associated with the breast CSC phenotype. This is in agreement with observations by other groups of a decrease in CD24 expression in patients treated with neoadjuvant chemotherapy (14), and that the CD44+/CD24−/low phenotype can be induced by genetic events or epigenetic events, such as exposure to TGF-beta, a factor secreted by tumor-associated stroma (15). However, the rapidity of changes in CD24 expression observed in the present study was consistent with changes in the expression level of the antigen, not with selection of a CD24−/low sub-population. This is in agreement with our previous findings that the CD44+CD24−/low phenotype of MA-11 cells is reversible and unrelated to tumorigenicity and metastatic potential (7).
Our in vivo findings may be attributed to epigenetic changes induced by the drug treatment, i.e. Imatinib-surviving cells activate some cell machinery that results in expression of a set of pro-malignant genes and consequently become endowed with pro-malignant properties. Alternatively, the in vivo effects of Imatinib on MA-11 xenografts could be mediated, at least in part, by an immunosuppressive effect of the drug can not be excluded. Imatinib can adversely affect the immune system by blocking immune surveillance of the malignancy and permitting disease recurrence. In fact, Imatinib reportedly inhibits activation, differentiation, and proliferation of normal T lymphocytes (16), antigen presentation by dendritic cells (17), and TNFα secretion (18). Also, Cristofanilli et al. (4) recently reported Imatinib-mediated inhibition of interferon-gamma production by TCR-activated CD4+ T cells in patients with PDGFR-beta-expressing advanced breast cancer.
By microarray analysis, we found that resistance to Imatinib of the surviving MA-11 cell population could be attributed to increased expression of the CACNA1G gene, a T-type calcium inhibitor recently identified as an additional target of Imatinib, and/or over-expression of the MRP3 transporter (19,20). The observed changes in expression for 35 SLC genes, that typically mediate uptake of nutrients (glucose, amino acids, organic anions, and peptides), may be related to increased energy and nutrition needs of cells with increased malignancy.
Interestingly, several genes potentially associated with the malignant phenotype of Imatinib-surviving cells, were found to be over=expressed. Among these, TSPO, a mitochondrial permeability transition-pore protein involved in the regulation of apoptosis and cell proliferation, that potentiates proliferation, motility, and transmigration capabilities of cancer cells (21); COP1, inhibitor of cell death, that binds caspase 1 and caspase 4, blocking apoptosis (22); MMP-9, involved in the breakdown of the extracellular matrix and in the metastatic process; and Neurotensin, a trophic and anti-apoptotic factor associated with breast cancer progression, that interacts with the neurotensin receptor on the cell membrane and stimulates Rho GTPases (23,24). Interestingly, many RhoGTPases were also over-expressed in Imatinib-surviving cells, and a link between neurotensin and MMP-9 has recently been demonstrated in breast cancer cells (23). We have also found several signal transduction pathways, potential targets for anti-breast cancer therapy, differentially expressed in the Imatinib-surviving population vs. the parental cell line. Among these, the Wnt/β-Catenin and the MAPK pathway, both recently found by our group to be altered in CSC-enriched MA-11 mammospheres compared with parental MA-11 cells (7). The association of significant changes in the Wnt/β-Catenin and the MAPK pathway with increased malignancy of breast cancer cells is supported by recent studies showing that aberrant activation of Wnt signaling induces mammary tumors from stem/progenitor cells (25), and that activation of the classical MAPK/ERK pathway, leading to c-ras activation, is associated with breast cancer stem cells and breast tumor epithelial-mesenchymal transition (26,27). Our data suggest caution in the clinical use of Imatinib in breast cancer patients; the comparison of Imatinib-surviving and parental breast cancer cells may help understand the role of differentially expressed genes in tumorigenicity and invasiveness and define the regulatory pathways involved in the increased malignancy of residual tumor cells that survive therapy, ultimately providing important therapeutic targets.
Supplementary Material
Acknowledgments
Financial Support: The project described was supported by Award Number R01CA133797 (G. R.) from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 3.Joensuu H, Dimitrijevic S. Ann Med. 2001;33:451–455. doi: 10.3109/07853890109002093. [DOI] [PubMed] [Google Scholar]
- 4.Cristofanilli M, Morandi P, Krishnamurthy S, Reuben JM, Lee BN, Francis D, Booser DJ, Green MC, Arun BK, Pusztai L, Lopez A, Islam R, Valero V, Hortobagyi GN. Ann Oncol. 2008;19:1713–1719. doi: 10.1093/annonc/mdn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chew HK, Barlow WE, Albain K, Lew D, Gown A, Hayes DF, Gralow J, Hortobagyi GN, Livingston R. Clin Breast Cancer. 2008;8:511–515. doi: 10.3816/CBC.2008.n.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sims JT, Ganguly S, Fiore LS, Holler CJ, Park ES, Plattner R. Biochem Pharmacol. 2009;78:249–260. doi: 10.1016/j.bcp.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rappa G, Lorico A. Exp Cell Res. doi: 10.1016/j.yexcr.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappa G, Fodstad O, Lorico A. Stem Cells. 2008;26:3008–3017. doi: 10.1634/stemcells.2008-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lampugnani MG. Methods Mol Biol. 1999;96:177–182. doi: 10.1385/1-59259-258-9:177. [DOI] [PubMed] [Google Scholar]
- 10.Cataldi M, Gaudino A, Lariccia V, Russo M, Amoroso S, di Renzo G, Annunziato L. J Pharmacol Exp Ther. 2004;309:208–215. doi: 10.1124/jpet.103.061184. [DOI] [PubMed] [Google Scholar]
- 11.Moreb JS. Curr Stem Cell Res Ther. 2008;3:237–246. doi: 10.2174/157488808786734006. [DOI] [PubMed] [Google Scholar]
- 12.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 13.Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. PLoS One. 2008;3:e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, Wong H, Rosen J, Chang JC. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 15.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cwynarski K, Laylor R, Macchiarulo E, Goldman J, Lombardi G, Melo JV, Dazzi F. Leukemia. 2004;18:1332–1339. doi: 10.1038/sj.leu.2403401. [DOI] [PubMed] [Google Scholar]
- 17.Seggewiss R, Price DA, Purbhoo MA. Cytotherapy. 2008;10:633–641. doi: 10.1080/14653240802317639. [DOI] [PubMed] [Google Scholar]
- 18.Wolf AM, Wolf D, Rumpold H, Ludwiczek S, Enrich B, Gastl G, Weiss G, Tilg H. Proc Natl Acad Sci U S A. 2005;102:13622–13627. doi: 10.1073/pnas.0501758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, Franke RM, Filipski KK, Hu C, Orwick SJ, de Bruijn EA, Burger H, Baker SD, Sparreboom A. Clin Cancer Res. 2008;14:3141–3148. doi: 10.1158/1078-0432.CCR-07-4913. [DOI] [PubMed] [Google Scholar]
- 20.Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B, Sawyers C, Shah N, Stock W, Willman CL, Friend S, Linsley PS. Proc Natl Acad Sci U S A. 2006;103:2794–2799. doi: 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rechichi M, Salvetti A, Chelli B, Costa B, Da Pozzo E, Spinetti F, Lena A, Evangelista M, Rainaldi G, Martini C, Gremigni V, Rossi L. Biochim Biophys Acta. 2008;1782:118–125. doi: 10.1016/j.bbadis.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Narayanan M, Bruey JM, Rigamonti D, Cattaneo E, Reed JC, Friedlander RM. Biochim Biophys Acta. 2006;1762:742–754. doi: 10.1016/j.bbadis.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Souaze F, Dupouy S, Viardot-Foucault V, Bruyneel E, Attoub S, Gespach C, Gompel A, Forgez P. Cancer Res. 2006;66:6243–6249. doi: 10.1158/0008-5472.CAN-06-0450. [DOI] [PubMed] [Google Scholar]
- 24.Dupouy S, Viardot-Foucault V, Alifano M, Souaze F, Plu-Bureau G, Chaouat M, Lavaur A, Hugol D, Gespach C, Gompel A, Forgez P. PLoS One. 2009;4:e4223. doi: 10.1371/journal.pone.0004223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindvall C, Bu W, Williams BO, Li Y. Stem Cell Rev. 2007;3:157–168. doi: 10.1007/s12015-007-0025-3. [DOI] [PubMed] [Google Scholar]
- 26.Dreesen O, Brivanlou AH. Stem Cell Rev. 2007;3:7–17. doi: 10.1007/s12015-007-0004-8. [DOI] [PubMed] [Google Scholar]
- 27.Liu M, Casimiro MC, Wang C, Shirley LA, Jiao X, Katiyar S, Ju X, Li Z, Yu Z, Zhou J, Johnson M, Fortina P, Hyslop T, Windle JJ, Pestell RG. Proc Natl Acad Sci U S A. 2009;106:19035–19039. doi: 10.1073/pnas.0910009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.