Abstract
This work presents simultaneous imaging and detection of three different cell receptors using three types of plasmonic nanoparticles (NPs). The size, shape, and composition-dependent scattering profiles of these NPs allow for a system of multiple distinct molecular markers using a single optical source. With this goal in mind, tags consisting of anti-epidermal growth factor receptor gold nanorods, anti-insulin-like growth factor 1-R silver nanospheres, and human epidermal growth factor receptor 2Ab gold nanospheres were developed to monitor the expression of receptors commonly overexpressed by cancer cells. These labels were chosen because they scatter strongly in distinct spectral windows. A hyperspectral darkfield microspectroscopy system was developed to record the scattering spectra of cells labeled with these molecular tags. Simultaneous monitoring of multiple tags may lead to applications such as profiling of cell line immunophenotype and investigation of receptor signaling pathways. Single, dual, and triple tag experiments were performed to analyze NP tag specificity as well as their interactions. Distinct resonance peaks were observed in these studies, showing the ability to characterize cell lines using conjugated NPs. However, interpreting shifts in these peaks due to changes in a cellular dielectric environment may be complicated by plasmon coupling between NPs bound to proximal receptors and other coupling mechanisms due to the receptors themselves.
Keywords: molecular imaging, multiplexing, plasmonics, nanoparticles, immunophenotype, cancer cells
Introduction
A wide range of imaging modalities have been used for multiplexed molecular imaging, motivated by the molecular level information these imaging modalities can provide about relevant biological systems. When used for in vitro studies, molecular imaging can help identify receptor expression and facilitate the understanding of cellular signaling pathways by identifying the binding of labels to a cell and observing subsequent interactions. Improving the understanding of cell receptor pathways can lead to the development of new treatments for various diseases. When used in vivo, molecular imaging may be used for both diagnosis and as a tool to determine optimal therapeutic treatment. For example, detecting cancer cells that overexpress certain receptors can enable early diagnosis, perhaps before other phenotypic changes in cellular structure are apparent. By identifying cells with different molecular phenotypes, multiplexed molecular imaging may be used to guide therapy, as particular phenotypes may be resistant to certain drugs or therapies.1 Molecular imaging can also be an important tool for guiding surgical resection of tumors. Many tumors have poorly defined margins preventing surgeons from removing the entire tumor without damaging normal tissues. This is especially important for brain surgery, where the tumor may reside near vital brain regions. However, molecular imaging of receptors that are overexpressed by these tumors can allow the surgeon to see exactly where the tumor remains and remove it with decreased risk for the surrounding tissues. For these reasons, molecular imaging with various labels has been studied extensively.
While fluorescent molecules are the most commonly used molecular labels, they are mainly limited to visible wavelengths. This makes multiplexing more than seven fluorophores quite difficult. While the emission spectra of quantum dots may be extended into the near-infrared (NIR) range, cytotoxicity issues prevent their widespread use for in vivo applications.2, 3 Currently there are only a few fluorophores approved for clinical use in the United States, due to their potentially harmful effects. Plasmonic nanoparticles (NPs) provide an alternative labeling system that circumvents these limitations.
Noble metal NPs exhibit localized surface plasmon resonance, which is manifested by enhanced absorption and scattering at a specific optical frequency, termed the peak resonant wavelength. The peak resonance wavelength for a given particle is dependent on many factors including its composition, size, shape, and the surrounding dielectric medium.4 The first three factors can be easily controlled and tuned by the choice of metal and the synthesis procedure. NPs are now readily available that exhibit their plasmon resonant peaks over a range of wavelengths in the visible and near-infrared regions of the spectrum. For example, the scattering peaks of gold nanorods extend from 600 to 2200 nm as a function of their aspect ratio. Introducing other NP geometries and compositions broadens the potential peak scattering range throughout the visible spectrum. Gold nanospheres scatter from 500 to 600 nm while silver nanospheres scatter from 400 to 500 nm. This wide range of peak resonance wavelengths potentially allows for a much larger number of distinct labels in a multiplexed molecular imaging system than is possible by other labels such as quantum dots or organic fluorophores.
In this study, three plasmonic NP labels were developed with distinct, nonoverlapping plasmon resonant peaks that are specific to three different cellular receptors: epidermal growth factor receptor (EGFR), human epidermal growth factor receptor 2 (HER-2), and insulin like growth factor receptor (IGF-1R). These receptors were chosen because they are commonly overexpressed in cancer cells. EGFR (ErbB-1) and HER-2 (ErbB-2) are two structurally similar receptors classified in the ErbB receptor family. They are among the many receptors that are responsible for regulating signaling pathways which control cell differentiation, proliferation, survival, and adhesion.5, 6 While the overexpression of one of these receptors is common in many forms of cancer cells, detecting high levels of both EGFR and HER-2 is a much more accurate measure for diagnosing malignant cells.7 This is one example where multiplexed molecular imaging would provide important information regarding cellular receptors relevant to diagnosing potentially cancerous tissues. IGF-1R plays a role in preventing apoptosis, the mechanism which causes programmed cell death.8 IGF-1R is overexpressed in many, but not all, cancer types, including breast cancer, pancreatic cancer, colon cancer, and melanomas.9 Access to this third receptor could provide unique insight into cancer phenotype, and potentially guide therapeutic action. As many potential therapies involve reducing or blocking these cellular receptors,10 determining the tumors immunophenotype should help to determine which receptors to target and improve the therapy's effectiveness. For the experiments described in this paper, cells were chosen with different expression levels of these three receptors to test both the molecular specificity of the labels as well as the ability to determine cell immunophenotype through multiplexed molecular imaging. A complete list of the cells used in these studies, along with their ELISA measured receptor expression levels, are presented below in Table 1.
Table 1.
Cell lines used in the imaging experiments and their receptor expression levels measured by ELISA. N/A = “not applicable” refers to expression levels that were not measured. Given uncertainties are the standard deviation of the measurements.
| Cell line | IGF-1R protein (ng/ml) | HER-2 protein (ng/ml) | EGFR protein (ng/ml) |
|---|---|---|---|
| A549 | N/A | N/A | 16.09±1.94 |
| A549/HER-2 | 2.75 ± 3.10 | 38.45 ± 7.40 | 22.97 ± 3.90 |
| A549/IGF-1R | 3.53 ± 0.68 | N/A | N/A |
| BT-474 | 6.32 ± 0.40 | 2027.85 ± 75.94 | N/A |
| COLO-320DM | 3.70 ± 0.55 | 34.6 ± 4.74 | 12.56 ± 0.62 |
| MCF-7 | 6.55 ± 1.22 | 45.88 ± 9.97 | N/A |
| MDA-MB-453 | 2.40 ± 0.26 | 51.44 ± 10.81 | 10.98 ± 0.82 |
| MDA-MB-468 | 0.90 ± 0.25 | 21.95 ± 9.66 | 55.96 ± 3.68 |
| SK-BR-3 | N/A | 439.30 ± 14.98 | N/A |
Spectrally distinct plasmonic tags were developed for each of the three targeted receptors. Gold nanorods (∼600 nm peak) were conjugated to anti-EGFR Abs, 60 nm gold nanospheres (∼560 nm peak), were conjugated to HER-2 monoclonal antibody (Ab), and 100 nm silver nanospheres (∼530 nm peak) were conjugated to anti-IGF-1R Ab. These three labels provided distinct spectral signatures, thereby allowing simultaneous imaging of all three receptors. Although the results presented in this study are a proof of principle of multiplexed molecular imaging using plasmonic NPs, more than three receptors could be labeled with NPs with nonoverlapping plasmonic scattering peaks, by synthesis of particles of varied geometries, and by controlling their size and shape variation to minimize their peak width.11
In this study, the molecular specificity of each tag was confirmed with in vitro imaging and microspectroscopy analysis. Hyperspectral images of cells incubated with conjugated NPs were acquired using a custom-built hyperspectral darkfield imaging microspectroscopy system. The recorded data were analyzed to determine the peak wavelength of scattering from ligand-functionalized NPs bound to individual cells. The distributions of these peaks within a population of cells were fit to Gaussian curves in order to develop a statistical measure of the scattering properties of each type of bound NP label. Experiments with two and three simultaneous tags show the ability of this multiplex system to determine aspects of cell immunophenotype. Single- and multilabel experiments were also performed to analyze how interaction of different NP tags can influence their scattering properties when bound to cells. Changes in the observed scattering peaks in the multiple tag experiments as compared to the single tag experiments suggest either a change in the dielectric environment due to the crosstalk between receptors, or plasmon coupling between particles bound to proximal receptors. The impact of these findings is discussed and future directions are suggested for separating these two effects.
Methods
NP-Antibody Conjugation
Gold nanorods were synthesized using established seed-mediated methods.12 To an aqueous mixture of 7.5 ml of 0.1 M hexadecyltrimethylammonium bromide (CTAB, Sigma-Aldrich, St. Louis, Missouri) and 0.250 ml of 0.01 M hydrogen tetrachloroaurate trihydrate (HAuCl4, Sigma-Aldrich, St. Louis, Missouri), 0.6 ml of ice cold 0.01 M sodium borohydride (Sigma-Aldrich, St. Louis, Missouri) was added under vigorous stirring to produce the gold seed, which was then gently heated and stirred for a few minutes. To a separate bottle containing 95 ml of 0.1 M CTAB in water at 29°C (precision microprocessor-controlled 280 series water bath), 4 ml of 0.01 M HAuCl4, 0.6 ml of 0.01 M silver nitrate (Sigma-Aldrich, St. Louis, Missouri), and 0.64 ml of 0.1 M ascorbic acid (Sigma-Aldrich, St. Louis, Missouri) were added, and the mixture was swirled after the addition of each reactant. After the addition of 50 μl of gold seed, the bottle was capped, inverted five times, and incubated at 29°C overnight. The dimensions of the gold nanorods were measured at 67.1 ± 8.9 nm in length and 32.0 ± 6.1 nm in diameter (n = 114) using TEM (Tecnai G2 Twin, FEI, Hillsboro, Oregon). Assuming a perfectly cylindrical nanorod shape, as well as the complete conversion of HAuCl4 to Au, the concentration of the parent gold nanorod suspension was estimated to be 1.3 × 10−10 M.
Gold nanorods were conjugated according to published procedures13 with anti-EGFR (E2156, clone 225, Sigma-Aldrich, St. Louis, Missouri, 1.56 mg/ml stock solution) or with mouse IgG1 κ isotype control (16-4714-85, eBioscience, 1.54 mg/ml stock solution) as a negative control, as follows: 1 ml of the gold nanorod suspension was centrifuged twice (10,000 rpm for 5 min) and resuspended in 1 ml of 1 mM NaCl. Polystyrene sulfonate (200 μl, 10 mg/ml in 1 mM NaCl, MW 18,000, Polysciences, Inc.) was added to the nanorods to cause the GNR surface potential to become negative, allowing for Ab binding, and expected to decrease the overall cytotoxixity of the nanorods. The suspension was then placed on a shaker for 20 min. The nanorods were centrifuged at 10,000 rpm for 5 min and resuspended in 1 ml of 20 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4. Four μl of 1.56 mg/ml mouse monoclonal anti-EGFR were added, and the suspension was placed on an oscillator for 30 min. Ab concentrations were determined using a NanoDrop Spectrophotometer (ND1000, NanoDrop, Wilmington, Delaware). Optimal Ab concentration was determined by testing for suspension stability (i.e., least aggregation) under various amounts. The nanorods were centrifuged at 10,000 rpm for 5 min and then finally resuspended in 0.5 ml of phosphated buffered saline (PBS) containing 5 mg/ml bovine serum albumin. Absorption spectra of nanorods are taken before and after conjugation to verify a shift in local surface plasmon resonance (LSPR) wavelength due to the bound Ab.14
Anti-IGF-1R nanosphere conjugates consist of 0.5 ml of 100 nm diameter silver colloid (15709-20SC, Ted Pella, Inc., Redding, California) solution diluted with 62.5 μl of 20 mM HEPES buffer and 16.1 μl of 1.40 mg/ml anti-IGF-1R (MS-301-PABX, ABD Serotec, Raleigh, North Carolina) solution diluted with 485 μl of 20 mm HEPES buffer. HER-2 Ab nanosphere conjugates consist of 0.5 ml of 60 nm diameter Au colloid (15709-20, Ted Pella, Inc., Redding, California) solution diluted with 62.5 μl of 20 mm HEPES buffer and 14.4 μl of 1.04 mg/ml HER-2 Ab (MS-301-PABX, Labvision, Fremont, California) solution diluted with 485 μl of 20 mm HEPES buffer. Both types of labels were conjugated following the same protocol. 100 nm K2CO3 was used to adjust the pH of each solution to 7.0 ± 0.2. Solutions were then mixed on an oscillator for 20 min at 190 cycles/min. Following mixing, the solution was tested for Ab-NP conjugation by removing 100 μl and subsequent addition of 5 μl of 10% NaCl. Incomplete Ab coverage would result in a bluish color change due to particle aggregation. To prevent nonspecific binding of proteins to the remaining NP surface, 100 μl of 1% polyethylene glycol (PEG, P2263, Sigma-Aldrich, St. Louis, Missouri) was added to the suspension and then allowed to interact. Fifteen minutes of centrifuging at 6000 rpm was used to remove excess PEG. Supernatant was removed, and the NP pellet was resuspended with 0.5 ml of PBS. These steps were then repeated to ensure complete removal of PEG, with minimal removal of NPs. Absorption spectra of the nanospheres are taken before and after conjugation to verify a shift in the LSPR wavelength due to the bound Ab.14 This protocol is an adaptation of methods developed by the Drezek, Richards–Kortum, and Sokolov groups.15, 16, 17
Cell Lines
MDA-MB-468 human breast adenocarcinoma cells [receptor expression of positive for EGFR, negative for IGF 1R, and negative HER 2 is noted as EGFR (+) / IGF-1R(−) / HER-2(−)] were incubated at 37°C and 5% CO2 using Modified Eagle's Medium (MEM) Alpha, with 10% fetal bovine serum (FBS) 1% penicillin streptomycin.18, 19, 20 HER-2(+) SK-BR-3 human breast adenocarcinoma cells were incubated at 37°C and 5% CO2 using McCoy's 5A Medium Modified, with 10% FBS 1% penicillin streptomycin.19, 21 EGFR (+) / IGF-1R(low) / HER-2(low) expressing A549 human alveolar adenocarcinoma cells were incubated at 37°C and 5% CO2 using F-12 Nutrient Mixture (Ham), 10% FBS, and 1% penicillin streptomycin. A second population of FACS selected high IGF-1R expressing A549 cells. A third population of A549 cells were additionally transfected with pcDNA 3.1 HER-2 and FACS selected for high HER-2 expression. EGFR (−) / IGF-1R(low) / HER-2(+) MDA-MB-453 human breast carcinoma cells were incubated at 37°C using Leibovitz L-15 Medium, with 10% FBS and 1% penicillin streptomycin.19, 20, 21 IGF-1R(low) / HER-2(−) MCF-7 human breast adenocarcinoma cells were incubated at 37°C and 5% CO2 using MEM Alpha, 10% FBS, and 1% penicillin streptomycin.20, 22, 23 IGF-1R(−) / HER-2(+) BT-474 human breast carcinoma cells were incubated at 37°C and 5% CO2 using Dulbecco's Modified Eagle Medium, 10% FBS, and 1% penicillin streptomycin.20, 22, 23 EGFR (−) / IGF-1R(low) / HER-2(−) COLO-320DM human colon adenocarcinoma cells were incubated at 37°C and 5% CO2 using RPMI Medium 1640, 10% FBS, and 1% penicillin streptomycin.19 All cell lines were obtained from the ATCC through the Duke Cell Culture Facility.
ELISA Measurements
Enzyme-linked immunosorbant assay (ELISA) techniques were utilized to quantify receptor expression levels and confirm the levels reported in literature. Each cell line was screened three times for the three receptors (EGFR, HER-2, IGF-1R) as necessary for each experiment, using commercially available ELISA kits. Kits used include: EGFR (full-length) Human ELISA Kit (KHR9061, Invitrogen, Carlsbad, California), Human HER-2 Immunoassay Kit (Total) (KHO0701, Invitrogen, Carlsbad, California), and STAR IGF-1R ELISA Kit (17-481, Millipore, Billerica, Massachusetts). For EGFR and HER-2 measurements, 1:100 dilutions of 1 μg of membrane protein were loaded to ELISA wells. For IGF-1R, 1:20 dilutions of 1 μg of membrane protein were loaded, due to the lower expression levels of this receptor. A summary of the receptor expression levels of the cell lines used as measured via ELISA is presented in Table 1.
Fluorescence-Activated Cell Sorting
Fluorescence-activated cell sorting (FACS) was used on two cell lines to increase the receptor expression levels in cell populations for the experiments described here. A549 cells, which express lower levels of IGF-1R, were sorted for higher expression of the receptor. Furthermore, a population of A549 cells transfected with HER-2 gene was also sorted to increase HER-2 expression. Samples of 5 × 106 cells were prepared for sorting and incubated with either 20 μl of anti-HER-2/neu FITC (340553, BD Biosciences, San Jose, California) or 10 μl mouse anti-human CD221 (anti-IGF-1R) Alexa Fluor 488 (MCA2344A488T, AbD Serotec, Raleigh, North Carolina) per 106 cells. Control cells were incubated with identical concentrations of IgG conjugates [mouse IgG1 FITC (555748, BD Biosciences, San Jose, California) or mouse IgG1 negative control Alexa Fluor 90 488 (MCA928A488, AbD Serotec, Raleigh, North Carolina)] at identical cell suspension concentrations. All steps were performed in minimal light to prevent photobleaching prior to sorting. Cell populations were placed on an oscillator for 22 min at 100 rotations/s. Tubes were then spun down in a centrifuge for 2 min at 1200 rpm. Excess supernatant was removed and cells were washed with 2% FBS/PBS solution. Wash steps were then repeated a second time, and cells were resuspended in the 2% FBS/PBS solution at a concentration of 107 cells/ml to ensure optimal sorting times. All cell populations were then covered in foil and placed in the refrigerator. Sorting was conducted within a 3 h window after labeling. In the case of HER-2 gene transfected A549 cells, the top 5.18% of HER-2 expressers were sorted (Fig. 1). In the case of IGF-1R expressing A549 cells, the top 13.17% of IGF-1R expressers were sorted for IGF-1R molecular specificity experiments (Fig. 1).
Figure 1.
(a) FACS results for A549 cells indicating top 5.18% of HER-2 expressers; (b) FACS results for A549 cells indicating top 13.17% of IGF-1R expressers.
Cell Treatment
After overnight incubation of chambers plated with 80 K cells, media was removed from cell samples designated for experiments. A solution consisting of 0.5 ml PBS/5% FBS and 0.5 ml NP conjugate suspension was then added. Next, cells were incubated for 20 min, allowing for adequate interaction between conjugates and cells. After removal of the solution, cells were washed twice with media. In the case of dual and triple tag labeling, this procedure was then repeated with the additional NP conjugates. Experiments were then immediately conducted using the hyperspectral microscopy system. Results of previous titration experiments were used to select the optimal NP-Ab conjugate concentrations used for the experiments.24
Image Acquisition
A hyperspectral microscopy system (Fig. 2) was used to conduct experiments on cells labeled with NP Ab conjugates.25 The system was built into a Zeiss Axiovert 200 inverted microscope. A super-continuum light source (Fianium), was coupled into an acousto-optic tunable filter (AOTF) to provide a narrowband output with a tunable wavelength. The average spectral bandwidth of the AOTF varied with center wavelength (3.5 nm typical at 515 nm). The filtered light then passes through a custom epi-illumination darkfield light train.26 Briefly, collimated light passes through an axicon, producing a ring of light at the back focal plane of a 40× objective (Zeiss) which illuminates the sample. Transmitted light is lost, while reflected light returns at the same high angle as the original ring of illumination. An aperture blocks this reflected light, only allowing scattered light to be imaged, enabling darkfield imaging. This epi-illumination setup prevents the collection of forward scattered light from the cells, and therefore improves the contrast of NP scattering. Scattered light from the sample is collected and recorded at each selected wavelength by an imaging CCD (Cascade 650, Photometrics). Hyperspectral data cubes were acquired from 450 to 700 nm by sequentially tuning the bandpass of the AOTF, with individual images integrating for 30 ms. Each cell was imaged over a reduced region of interest (170 × 170 pixels) to minimize acquisition time. Two data cubes were acquired for each cell, with 5 and 1 nm increments, taking a total acquisition time of ∼2.4 and ∼12.0 s, respectively. Scattering spectra for each cell are obtained by averaging the pixel values within the region of interest (ROI) at each wavelength. Source-correction is performed by normalizing the signal by source intensity, as measured using a diffuse reflectance standard (WS-1, Ocean Optics, Dunedin, Florida).
Figure 2.
(a) Hyperspectral darkfield microscopy system for analysis of live cells in culture. The system is comprised of a Zeiss inverted microscope, Cascade:650 imaging CCD, Fianium SC450-2 supercontinuum light source, and Crystal Technologies acousto-optic tunable filter. (b) Spectral output of the AOTF at various wavelengths.
Results
Anti-EGFR Gold Nanorods
The first set of experiments was performed to evaluate the molecular specificity of each label. MDA-MB-468 cells, which overexpress EGFR,18, 19, 20 were incubated with the anti-EGFR gold nanorod conjugates. Hyperspectral data cubes were recorded for individual cells. The distribution of the peak wavelengths of spectra averaged over each cell were fit to Gaussian distributions. This led to an average peak scattering wavelength of 664.0 ± 9.3 nm for anti-EGFR gold nanorods (N = 139) (Fig. 3 and Table 2). Controls for this label included: 1. incubating EGFR negative MDA-MB-453 cells with anti-EGFR gold nanorod conjugates;19, 20 2. exposing MDA-MB-468 cells to gold nanorods conjugated with nonspecific IgG1 monoclonal Ab; and 3. imaging MDA-MB-468 cells without any label. Previous studies from this group have investigated binding of unlabeled NPs and shown them to be minimal for the range of incubation times examined here.27 These three control experiments showed insignificant scattering intensity compared to the experimental case (p < 0.001), demonstrating the molecular specificity of the label.
Figure 3.
(a) Representative image and (b) scattering spectrum of a single MDA-MB-468 cell bound with anti-EGFR gold nanorods. (Scale bar = 10 μm.) (c) Distribution of peak scattering peak wavelengths of MDA-MB-468 cells (N = 150) bound with anti-EGFR gold nanorods (67.1 ± 6.1 × 32.0 ± 8.9 nm). Distribution fit has peak wavelength of 664.0 ± 9.3 nm.
Table 2.
Summary of single label experiments. Given uncertainties are the standard deviation of the measurements.
| Cell line | Target receptor | NP type | Number of cells | Peak wavelength (nm) |
|---|---|---|---|---|
| MDA-MB-468 | EGFR | Gold nanorods | N = 139 | 664.0 ± 9.3 |
| A549/IGF-1R | IGF-1R | Silver nanospheres | N = 106 | 520.8 ± 13.5 |
| SK-BR-3 | HER-2 | Gold nanospheres | N = 147 | 587.0 ± 11.9 |
Anti-IGF-1R Silver Nanospheres
The molecular specificity of the anti-IGF-1R silver nanospheres was tested with a similar procedure. A549/IGF-1R human alveolar adenocarcinoma cells which overexpress IGF-1R,20, 28 were incubated with the silver nanosphere tag. Average scattering spectra recorded from the entire cell area were collected and fit to a Gaussian distribution. This showed an average scattering peak wavelength of 520.8 ± 13.5 nm (N = 106) (Fig. 4 and Table 2). Controls for this label included: 1. incubating MDA-MB-468 human breast adenocarcinoma cells, which do not express IGF-1R,20, 29 with the anti-IGF-1R silver nanosphere labels; 2. incubating A549/IGF-1R cells with IgG1 Ab silver nanospheres; and 3. A549/IGF-1R cells without any label. These three control experiments showed insignificant scattering intensity compared to the experimental case (p < 0.001), demonstrating the molecular specificity of the label.
Figure 4.
(a) Representative image and (b) spectrum of single A549/IGF-1R cell bound with anti-IGF-1R immunolabeled silver nanospheres. (Scale bar = 10 μm.) (c) Distribution of peak scattering wavelength of A549/IGF-1R cells bound with anti-IGF-1R 100 nm silver nanospheres. Distribution fit has peak wavelength of 520.8 ± 11.3 nm (N = 102).
HER-2 Ab Gold Nanospheres
SK-BR-3 human breast adenocarcinoma cells19, 21 that overexpress HER-2 were exposed to HER-2 Ab 60 nm gold nanospheres. Peak scattering distributions were fit with Gaussian distributions, indicating an average peak scattering wavelength of 587.0 ± 11.9 nm (N = 147) (Fig. 5 and Table 2). Note that the high degree of HER-2 expression for these cells causes a redshift in both the appearance and scattering spectrum of these labels. Controls for this experiment included: 1. incubating HER-2 nonexpressing MDA-MB-468 human breast adenocarcinoma cells21, 23, 31 with HER-2 Ab nanospheres; 2. incubating SK-BR-3 cells with IgG1 Ab gold nanospheres; and 3. SK-BR-3 cells with no label. These control experiments showed insignificant scattering intensity compared to the experimental case (p < 0.001), demonstrating the molecular specificity of the label.
Figure 5.
(a) Representative image and (b) spectrum of a single SK-BR-3 cell bound with HER-2 Ab labeled 60 nm gold nanospheres. (Scale bar = 10 μm.) Note that plasmonic coupling causes a redshift in the scattered light. (c) Distribution of peak scattering wavelength of SK-BR-3 cells bound with HER-2 Ab labeled 60 nm gold nanospheres. Distribution fit has peak wavelength of 587.0 ± 11.9 nm (N = 122). Adapted from Ref. 30.
EGFR and HER-2 Dual Labels
Following the tests for the molecular specificity of each individual label, tests were performed to evaluate the effectiveness of dual tag systems. To test the immunophenotype indicating capabilities of the EGFR/HER-2 dual label, EGFR(+)/HER-2(−) MDA-MB-468 cells were exposed to both the anti-EGFR nanorods and the HER-2 Ab gold nanospheres consecutively. These scattering spectra showed an average peak originating from the long axis of the nanorods (663.7 ± 6.6 nm, N = 50). The same experiment with EGFR(−)/HER-2(+) MDA-MB-453 cells produced a peak associated with the HER-2 Ab conjugated gold nanospheres (586.9 ± 24.1 nm, N = 52). Although there was a slight redshift in this peak, most likely due to plasmonic coupling, it did not overlap with the scattering spectra from the nanorods. EGFR(−)/HER-2(−) COLO-320DM cells showed no significant scattering peaks.
To demonstrate dual labeling, EGFR(+)/HER-2(+) A549/HER-2 cells were tagged consecutively with anti-EGFR nanorods and anti-HER2 gold nanospheres. The anti-EGFR tag produced a peak scattering wavelength of 657.8 ± 15.8 nm (N = 132) while the anti-HER2 tag produced a peak scattering wavelength of 551.4 ± 8.6 nm (N = 35) (Fig. 6). These two peaks showed significant statistical differences (p < 0.001) from one another during consecutive labeling. The anti-EGFR scattering distributions of individually labeled A549/HER-2 cells were compared with those from the dual-labeled population and showed no significant change (p = 0.724). However, a p-value of 0.045 was determined when comparing the HER-2 Ab peak distributions from both individual and dual label cases, indicating a statistically significant change. No significant changes in these distributions were observed when reversing the order of the two labels. (See Table 3.)
Figure 6.
(a) Representative image and (b) spectrum of a single A549 / HER-2 cell exposed to anti-EGFR nanorods and HER-2 nanospheres consecutively. (Scale bar = 10 μm.) (c) Distribution of peak scattering from A549 / HER-2 cells exposed to anti-EGFR gold nanorods and HER-2 Ab nanospheres consecutively.
Table 3.
Summary of EGFR/HER-2 dual label experiments. P-values in the left column compare dual label experiments to corresponding individual labeling experiments. P-values in the right column compare two neighboring cases. Given uncertainties are the standard deviation of the measurements.
| Cell line (expt/receptor) | Peak wavelength (nm) | Number of cells | p-value | |
|---|---|---|---|---|
| MDA-MB-468 (EGFR) | 663.7 ± 6.6 | N = 50 | ||
| MDA-MB-453 (HER-2) | 586.9 ± 24.1 | N = 52 | ||
| A549/Her-2 (individual EGFR) | 656.1 ± 9.5 | N = 73 | ||
| <0.001 | ||||
| A549/Her-2 (individual HER-2) | 542.7 ± 21.3 | N = 75 | ||
| A549/Her-2 (dual label EGFR peak) | 657.8 ± 15.8 | N = 132 | 0.724 | |
| <0.001 | ||||
| A549/Her-2 (dual label HER-2 peak) | 551.4 ± 8.6 | N = 35 | 0.045 | |
EGFR and IGF-1R Dual Labels
Similar experiments were performed for dual tagging with EGFR and IGF-1R labeled NPs. When exposed to anti-EGFR nanorods and IGF-1R silver nanospheres (in that order), EGFR(+)/IGF-1R(−) MDA-MB-468 cells showed a single scattering peak associated with the long axis of the nanorods (669.9 ± 11.2 nm, N = 48). In contrast, EGFR(−)/IGF-1R(low) MDA-MB-453 cells showed a single scattering peak associated with the anti-IGF-1R conjugated nanospheres (517.4 ± 8.5 nm, N = 49).
To evaluate this dual labeling approach, EGFR(+)/IGF-1R(low) A549 cells were incubated consecutively with anti-EGFR nanorods and anti-IGF-1R silver nanospheres (Fig. 7). The bound anti-EGFR nanorods exhibited a peak scattering wavelength of 658.3 nm ± 17.6 nm (N = 102) while the bound anti-IGF-1R nanospheres exhibited a peak scattering wavelength of 533.1 ± 15.2 nm (N = 81), a statistically significant difference with a p value of <0.001. In contrast, when A549 cells were individually labeled with either anti-EGFR gold nanorods or anti-IGF-1R silver nanospheres, the anti-EGFR label exhibited a peak scattering wavelength of 648.4 ± 14.5 nm (N = 73), while the anti-IGF-1R label exhibited a peak of 520.5± 11.6 nm (N = 75). These two peak distributions are also statistically different with a p value of <0.001, indicating distinct scattering behavior, as expected. When the dual label scattering peaks were compared to the scattering spectra from cells labeled individually with either anti-EGFR or anti-IGF-1R labels, a p value of <0.001 was determined for both labels. This indicates that there was a statistically significant change in the scattering peak distributions of either tag caused by the incubation of cells with the second type of Ab functionalized NPs. (See Table 4.)
Figure 7.
(a) Representative image and (b) spectrum of a single A549 cell exposed to anti-EGFR nanorods and anti-IGF-1R nanospheres consecutively. (Scale bar = 10 μm.)
Table 4.
Summary of EGFR/IGF-1R dual label experiments. P-values in the left column compare dual label experiments to corresponding individual labeling experiments. P-values in the right column compare two neighboring cases. Given uncertainties are the standard deviation of the measurements.
| Cell line (expt/receptor) | Peak wavelength (nm) | Number of cells | p-value | |
|---|---|---|---|---|
| MDA-MB-468 (EGFR) | 669.9 ± 11.2 | N = 48 | ||
| MDA-MB-453 (IGF-1R) | 517.4 ± 8.5 | N = 49 | ||
| A549 (individual EGFR) | 648.4 ± 14.5 | N = 73 | ||
| <0.001 | ||||
| A549 (individual IGF-1R) | 520.5 ± 11.6 | N = 75 | ||
| A549 (dual label EGFR) | 658.3nm ± 17.6 | N = 102 | <0.001 | |
| <0.001 | ||||
| A549 (dual label IGF-1R) | 533.1 ± 15.2 | N = 81 | <0.001 | |
However, when the order in which cells were incubated with the two Ab functionalized NPs was reversed, different scattering peaks were observed (Fig. 8). The bound anti-EGFR nanorods exhibited a peak scattering wavelength of 643.0 ± 5.3 nm (N = 14), while the bound anti-IGF-1R nanospheres exhibited a peak scattering wavelength of 523.1 ± 6.9 nm (N = 46). More interestingly, the peak distributions observed were much narrower compared to the distributions exhibited in the original sequence of incubation. When compared to cells individually labeled with either anti-EGFR or anti-IGF-1R NPs, there was not a statistically distinct difference, with a p value = 0.759 determined for the IGF-1R label and a p value of 0.201 determined for the EGFR label. This shows that for this incubation sequence, the peak scattering distributions of either tag are not affected by incubation of cells with the second tag. A significant reduction in the number of cells containing detectable amounts of anti-EGFR label (N = 14 versus N = 46 for anti-IGF1R), suggests that the reversed order may block the binding of the anti-EGFR label.
Figure 8.
Distribution of peak scattering from A549 cells exposed to anti-EGFR gold nanorods and anti-IGF-1R nanospheres (a) consecutively, and (b) in reverse order. A blueshift of 41.4 nm is obererved in the peak scattering wavelength of the anti-EGFR label after reversing the order. Similarly, a redshift of 2.6 nm is observed for the anti-IGF-1R label after reversing the order.
HER-2 and IGF-1R Dual Labels
Similar experiments were also performed for a dual HER-2/IGF-1R labeling system. IGF-1R(low)/HER-2(−) MCF-7 cells were incubated consecutively with HER-2 Ab gold nanospheres and anti-IGF-1R silver nanospheres. The measured spectra for these cells show scattering peaks associated with the anti-IGF-1R silver spheres (520.4 ± 11.0 nm, N = 49). IGF-1R(−)/HER-2(+) BT-474 cells20, 22, 23 when labeled in the same manner, produce a scattering peak associated with the HER-2 Ab gold nanospheres (566.8 ± 15.8 nm, N = 49). IGF-1R(−)/HER-2(−) MDA-MB-468 cells showed no significant scattering peaks.
To evaluate this dual label system, IGF-1R(+)/HER-2(+) A549/HER-2 cells were labeled with both anti-IGF-1R and HER-2 Ab tags consecutively. However, only one scattering peak (not clearly associated with either label) was observed (Fig. 9). This can be attributed to the close spectral proximity of the scattering peaks for these two labels (approximately 30 nm apart). Due to this effect, the scattering from the individual labels could not be analyzed with this labeling system. Reversing the ordering of these labels likewise did not produce sufficiently distinct peaks to permit analysis.
Figure 9.
(a) Representative image and (b) spectrum of a single A549/HER-2 cell exposed to HER-2 Ab nanorods and anti-IGF-1R nanospheres consecutively. (Scale bar = 10 μm.)
EGFR, HER-2, and IGF-1R Triple Label
With the dual labeling experiments suggesting the potential to further expand this imaging modality to three different labels that target three different receptors, experiments were next performed to evaluate the feasibility of this approach. A549/HER-2 cells were chosen for these experiments, as they have been shown to express all three receptors (Table 1). These cells were first exposed to either anti-EGFR nanorods, HER-2 Ab gold nanospheres, or anti-IGF-1R silver nanospheres in individual labeling experiments to verify receptor expression. These experiments indicated an average scattering peak wavelength of 656.1 ± 9.5 nm (N = 73) for the EGFR label, 521.8±7.9 nm (N = 75) for the IGF-1R label, and 542.7±21.3 nm (N = 75) for the HER-2 label. Comparison of any two of these three distributions produced statistically significant distinctions (p < 0.001), confirming the presence of all three targeted receptors. Previous experiments with negative controls, described above, confirmed the molecular specificity for each of these labels.
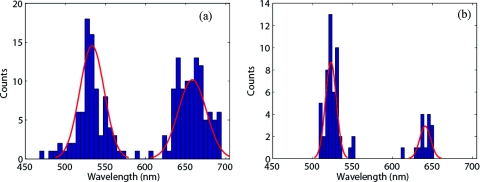
To evaluate the triple labeling system, A549/HER-2 cells were exposed to the anti-EGFR nanorods, anti-IGF-1R silver nanospheres, and HER-2 Ab gold nanospheres consecutively. The anti-EGFR label produced a peak scattering wavelength of 661.3±12.7 nm (N = 127), while the HER-2 Ab label produced a peak scattering wavelength of 555.8 ± 7.2 nm (N = 44). However, there was no discernible peak associated with the anti-IGF-1R label, which was expected to appear in the 500 to 550 nm range (Fig. 10 and Table 5). This is most likely due to the high expression of HER-2 for this cell line, so that scattering by the HER-2 label scattering likely masked scattering from the IGF-1R silver nanospheres. While some individual cells did express enough IGF-1R to enable sufficient labeling to exhibit a distinct scattering peak (Fig. 11), this spectral feature was not consistently observed. The two detectable peak wavelength distributions were determined to be statistically distinct (p < 0.001). The inability to successfully resolve the third spectral peak prevented a detailed analysis of the effect of changes in the sequence of incubation on labeling of cells by for the three label system. (See Table 5.)
Figure 10.
(a) Representative image and (b) spectrum of a single A549 / HER-2 cell exposed to anti-EGFR nanorods, anti-IGF-1R nanospheres, and HER-2 Ab nanospheres consecutively. (Scale bar = 10 μm.) (c) Distribution of peak scattering from A549 / HER-2 cells exposed to anti-EGFR nanorods, anti-IGF-1R nanospheres, and HER-2 Ab nanospheres consecutively.
Table 5.
Summary of EGFR/IGF-1R/HER-2 triple label experiments. P-values given for individual label experiments compares differences to the other individual label experiments. P-values given for each triple label experiment compares differences to corresponding individual labeling experiments. Given uncertainties are the standard deviations of the measurements (n/a – not applicable).
| Cell line (expt/receptor) | Peak wavelength (nm) | Number of cells | p-value |
|---|---|---|---|
| A549/HER-2 (individual EGFR) | 656.1 ± 9.5 | N = 73 | <0.001 |
| A549/HER-2 (individual IGF-1R) | 521.8 ± 7.9 | N = 75 | <0.001 |
| A549/HER-2 (individual HER-2) | 542.7 ± 21.3 | N = 75 | <0.001 |
| A549/HER-2 (triple label EGFR) | 661.3 ± 12.7 | N = 127 | <0.001 |
| A549/HER-2 (triple label IGF-1R) | Not observed | n/a | |
| A549/HER-2 (triple label HER-2) | 555.8 ± 7.2 | N = 44 | <0.001 |
Figure 11.
(a) Representative image and (b) spectrum of a single A549 / HER-2 cell exposed to anti-EGFR nanorods, anti-IGF-1R nanospheres, and HER-2 Ab nanospheres consecutively. (Scale bar = 10 μm.)
Discussion
This study has demonstrated the simultaneous imaging and analysis of three different cellular receptors using plasmonic NP labels. The major advantages of molecular imaging using plasmonic NPs include a lack of photobleaching and minimal cytotoxicity issues compared to organic fluorophores and quantum dots. While previous studies have used gold nanorods of various aspect ratios to produce labels within separate spectral windows,32 the use of different shapes and compositions of plasmonic NPs allows for the further extension of the spectral range across the visible region. The use of these three labels allows for immunophenotype characterization of three receptors overexpressed in many cancer cell lines. We have previously demonstrated the quantitative measurement of receptor expression levels using gold nanospheres.33 Similar quantitative measurements of multiple receptor levels should be possible with the methods presented here. More than three receptors could be included in this labeling system by choosing NP tags of varying sizes and shapes to further expand the range of distinct spectral peaks available.
Cells expressing the receptors of interest were incubated with both dual and triple plasmonic NP labels. In these experiments, the scattering peak distributions for each receptor were statistically distinct (p < 0.001) from the distributions for other receptors in both EGFR/HER-2 and EGFR/IGF-1R experiments. This indicates a clean separation between the scattering windows of each tag. Measurements of these two spectrally separated peaks allows for determination of a relative immunophenotype based on the two tagged receptors. Furthermore, this allows one to simply assess the color of the NPs within the cell as a visual estimate of receptor expression. This capability could be exploited to allow for differentiation of cancer cell type based on receptor expression as well as potentially delineating malignant tumors within a surgical setting. Visual observation could be useful for providing a quick, qualitative estimate of the relative expression levels of tagged receptors without the need for the detailed spectral measurements and data analysis required to quantitatively assess cancer cell immunophenotoype.
During the EGFR / IGF-1R / HER-2 triple label experiments and the EGFR / IGF-1R (in one labeling order) dual label experiments, significant differences were seen between the scattering peak distributions of the multiple label experiments when compared to the individual label experiments. (To a lesser degree of statistical significance, a change was also seen in the EGFR/HER-2 dual label experiments for both labeling orders) This suggests that a slight change in the dielectric environment was caused by the introduction of the second or third label. This can be explained by a change in solute concentration which can influence dielectric environment or cross-talk between the activated receptors. Plasmonic coupling of particles bound to proximal receptors may also cause this difference. Further experiments with techniques such as polarization mapping34 may be necessary to separate changes due to dielectric medium alterations from changes due to plasmonic coupling. This technique isolates the redshifted scattering peak due to plasmonic coupling of a single NP pair by exploiting the fact that it is always associated with the polarization aligned with the long-axis of a NP pair. In general, peak shifts due to changes in the dielectric medium would not exhibit polarization dependence in the scattering spectra. In the EGFR/HER-2 and EGFR/IGF-1R experiments, peak scattering distributions were not statistically different when compared to the individual label trials. This indicates a minimal change in the dielectric medium or minimal plasmonic coupling, suggesting that this effect is receptor specific and with further analysis, may provide insight into an underlying biological mechanism.
Data from the HER-2/IGF-1R dual label and triple label experiments also showed overlap of the scattering peaks scattering between the anti-IGF-1R and HER-2 Ab labels. This resulted in a lack of a distinct scattering peak associated with the anti-IGF-1R label for cells exposed to all three labels. The average peak scattering wavelengths of these two tags during individual labeling were only separated by 30 nm. A potential solution of this problem would be to use silver nanospheres with a smaller diameter to further blueshift their scattering peak. The relatively higher levels of scattering from the HER-2 Ab label may have also masked the lower level of IGF-1R expression in BT-474 cells. In future experiments, it will be necessary to choose labels with narrower spectral widths or more widely spaced scattering peaks to avoid this overlap. One option would be to use gold nanorods of varying lengths but constant widths, extending the range of scattering peaks associated with the long axis into the NIR range. Other conformations, such as gold nanoshells, may also be used to further separate these spectral windows.35
Reversing the sequence of incubation of the two types of NPs in the dual EGFR/IGF-1R label experiment produced an interesting change in the measured scattering distributions, as it caused a significant decrease in the intensity of scattering associated with the anti-EGFR tag. The experiments for both labeling orders were performed consecutively from the same cell populations and NP batches, signifying that the change was solely caused by the reversed sequence. The acquired images show a distinct difference in the relative number of bound anti-EGFR label when using the reverse order. However, in most cases there were only about 20 silver nanospheres seen bound to a given cell. This would not be nearly enough for the larger silver spheres to physically block the binding of the smaller gold spheres. A study by Desbois–Mouthon et al. points to a possible cause of the observed spectra.36 They show that the activation of IGF-1R leads to the release of amphiregulin, an EGFR-ligand, which binds to EGFR. It may be possible that a similar process occurs in these alveolar adenocarcinoma cells, where amphiregulin has been released and competes for binding sites with the anti-EGFR conjugated nanospheres. Further experiments, beyond the scope of this work, are clearly necessary to fully understand this intriguing behavior.
Through these experiments, multiple advantages of plasmonic NPs as labels in multiplexed molecular imaging are evident compared to fluorescent molecules. The first major advantage is the wide spectral range of scattering peaks available for each tag. By extending these peaks into the NIR range, a system of immunolabeled NPs could be developed that distinctly label a larger number of receptors than is possible with fluorescent molecules, which are generally limited to the visible range. Furthermore, plasmonic NPs, unlike fluorescent tags, are also sensitive to the refractive index surrounding dielectric medium. This unique feature of NP labels implies that changes in the cell such as solute concentration or receptor cross talk may be analyzed by monitoring shifts in the peak scattering wavelength. These shifts can also provide information on the signal pathways which are activated when multiple types of receptors are bound to their ligands. For example, it may be possible to monitor receptor dimerization by examining the plasmon coupling between adjacent NP.30 Thus, plasmonic NPs possess unique properties that may provide a significant advantage in multiplexed molecular imaging compared to fluorophores by widening the available spectral range to allow more distinct labels and by accessing additional information regarding cell signaling pathways and receptor mechanisms.
Conclusions
This study evaluated the ability to use three different functionalized plasmonic NPs to simultaneously image multiple cellular receptors. The spectral separation between the scattering peaks of these three labels allows for the characterization of cell immunophenotype. NP labels are also capable of measuring changes in the local environment of the receptors. In some cases, statistically significant differences were detected between the peak wavelength of scattering distributions of individual labels and the same labels used in dual and triple label experiments. This behavior may be due to plasmonic coupling between particles bound to proximal receptors, or changes in the dielectric medium of the cell, reflecting receptor interactions. Both processes would lead to a significant shift in the NP's peak scattering wavelength. With either interpretation, it is possible that this information may be used to better understand dynamics of cell receptor pathways. The ability to concretely separate the effects of these two processes would enable development of more advanced tools for intracellular biosensing applications.
Acknowledgments
We would like to acknowledge the NSF (CBET-0651622) for supporting this research. KS acknowledges support of NIH training grant (T32-EB001040).
References
- Wang H. and Vo-Dinh T., “Multiplex detection of breast cancer biomarkers using plasmonic molecular sentinel nanoprobes,” Nanotechnology 20(6), 065101 (2009). 10.1088/0957-4484/20/6/065101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derfus A. M., Chan W., and Bhatia S. N., “Probing the cytotoxicity of semiconductor quantum dots,” Nano Lett. 4(1), 11–18 (2004). 10.1021/nl0347334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubertret B., Skourides P., Norris D. J., Noireaux V., Brivanlou A. H., and Libchaber A., “In vivo imaging of quantum dots encapsulated in phospholipid micelles.”Science 298(5599), 1759–1762 (2002). 10.1126/science.1077194 [DOI] [PubMed] [Google Scholar]
- Kelly K., Coronado E., Zhao L., and Schatz G., “The optical properties of metal NPs: The influence of size, shape, and dielectric environment,” J. Phys. Chem. B 107(3), 668–677 (2003). 10.1021/jp026731y [DOI] [Google Scholar]
- Marmor M., Skaria K., and Yarden Y., “Signal transduction and oncogenesis by ErbB/HER receptors,” Int. J. Radiat. Oncol., Biol., Phys. 58(3), 903–913 (2004). 10.1016/j.ijrobp.2003.06.002 [DOI] [PubMed] [Google Scholar]
- Nicholson R., Gee J., and Harper M., “EGFR and cancer prognosis,” Eur. J. Cancer 37(4), 9–15 (2001). 10.1016/S0959-8049(01)00231-3 [DOI] [PubMed] [Google Scholar]
- Diaco R., Practical Considerations for the Design of Quantitative PCR Assays in PCR Strategies, Innis M. A., Gelfand D. H., Sninsky J. J., Eds., pp. 84–110, Academic Press, San Diego, CA: (1995). [Google Scholar]
- Ellis M., Garmoudi F., and Cullen K., “Insulin Receptors (IGF1 and IGF2),” Encyclopedia of Cancer, Vol. 2, pp. 927–939 (1997).
- Riedemann J. and Macaulay V. M., “IGF1R signalling and its inhibition,” Endocrine-Related Cancer 13(1), S33–S43 (2006). 10.1677/erc.1.01280 [DOI] [PubMed] [Google Scholar]
- Skirnisdóttir I., Sorbe B., and Seidal T., “The growth factor receptors HER-2/neu and EGFR, their relationship, and their effects on the prognosis in early stage (FIGO I-II) epithelial ovarian carcinoma,” Int. J. Gynecological Cancer 11(2), 119–129 (2008). 10.1046/j.1525-1438.2001.00086.x [DOI] [PubMed] [Google Scholar]
- Chen K., Liu Y., Ameer G., and Backman V., “Optimal design of structured nanospheres for ultrasharp light-scattering resonances as molecular imaging multilabels,” J. Biomed. Opt. 10(2), 024005 (2005). 10.1117/1.1899684 [DOI] [PubMed] [Google Scholar]
- Nikoobakht B. and El-Sayed M., “Preparation and growth mechanism of gold nanorods (NRs) using seed-mediated growth method,” Chem. Mater. 15(10), 1957–1962 (2003). 10.1021/cm020732l [DOI] [Google Scholar]
- Huang X., El-Sayed I., Qian W., and El-Sayed M., “Cancer cells assemble and align gold nanorods conjugated to antibodies to produce highly enhanced, sharp, and polarized surface Raman spectra: a potential cancer diagnostic marker,” Nano Lett. 7(6), 1591–1597 (2007). 10.1021/nl070472c [DOI] [PubMed] [Google Scholar]
- Curry A., Nusz G., Chilkoti A., and Wax A., “Analysis of total uncertainty in spectral peak measurements for plasmonic NP-based biosensors,” Appl. Opt. 46(10), 1931–1939 (2007). 10.1364/AO.46.001931 [DOI] [PubMed] [Google Scholar]
- Sokolov K., Follen M., Aaron J., Pavlova I., Malpica A., Lotan R., and Richards-Kortum R., “Real-time vital optical imaging of precancer using anti-epidermal growth factor receptor antibodies conjugated to gold nanoparticles,” Cancer Res. 63(9), 1999–2004 (2003). [PubMed] [Google Scholar]
- Aaron J., Nitin N., Travis K., Kumar S., Collier T., Park S. Y., Jose-Yacaman M., Coghlan L., Follen M., Richards-Kortum R., and Sokolov K., “Plasmon resonance coupling of metal NPs for molecular imaging of carcinogenesis in vivo,” J. Biomed. Opt. 12(3), 034007 (2007). 10.1117/1.2737351 [DOI] [PubMed] [Google Scholar]
- Loo C., Lowery A., Halas N., West J., and Drezek R., “Immunotargeted nanoshells for integrated cancer imaging and therapy,” Nano Lett. 5(4), 709–712 (2005). 10.1021/nl050127s [DOI] [PubMed] [Google Scholar]
- Filmus J., Pollak M. N., Cailleau R., and Buick R. N., “MDA-468, a human breast cancer cell line with a high number of epidermal growth factor (EGF) receptors, has an amplified EGF receptor gene and is growth inhibited by EGF,” Biochem. Biophys. Res. Commun. 128(2), 898–905 (1985). 10.1016/0006-291X(85)90131-7 [DOI] [PubMed] [Google Scholar]
- Lewis G. D., Figari I., Fendly B., Wong W. L., Carter P., Gorman C., and Shepard H. M., “Differential responses of human tumor cell lines to anti-p185 HER2 monoclonal antibodies,” Cancer Immunol. Immunother 37(4), 255–263 (1993). 10.1007/BF01518520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedemann J., Takiguchi M., Sohail M., and Macaulay V. M., “The EGF receptor interacts with the type 1 IGF receptor and regulates its stability,” Biochem. Biophys. Res. Commun. 355(3), 707–714 (2007). 10.1016/j.bbrc.2007.02.012 [DOI] [PubMed] [Google Scholar]
- Cuello M., Ettenberg S., Clark A., Keane M., Posner R., Nau M., Dennis P., and Lipkowitz S., “Down-regulation of the erbB-2 receptor by trastuzumab (herceptin) enhances tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in breast and ovarian cancer cell lines that overexpress erbB-2,” Cancer Res. 61(12), 4892–4900 (2001). [PubMed] [Google Scholar]
- Chakraborty A., Welsh A., and DiGiovanna M., “Co-targeting the insulin-like growth factor I receptor enhances growth-inhibitory and pro-apoptotic effects of anti-estrogens in human breast cancer cell lines,” Breast Cancer Res. Treat. 120(2), 327–335 (2010). 10.1007/s10549-009-0382-5 [DOI] [PubMed] [Google Scholar]
- Kramer-Marek G., Kiesewetter D. O., and Capala J., “Changes in HER2 expression in breast cancer xenografts after therapy can be quantified using PET and 18F-labeled affibody molecules,” J. Nucl. Med. 50(7) 1131–1139 (2009). 10.2967/jnumed.108.057695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow M. J., et al. , “Plasmonic flow cytometry by immunolabeled nanorods,” Cytometry, Part A 79A(1), 57–65 (2011). 10.1002/cyto.a.20994 [DOI] [PubMed] [Google Scholar]
- Crow M. J. and Wax A., “Development of a novel hyperspectral darkfield microscopy system for characterization of NP sensors,” Biomedical Optics, OSA Technical Digest, BSuE6 (2008). [Google Scholar]
- Curry A., Hwang W., and Wax A., “Epi-illumination through the microscope objective applied to darkfield imaging and microspectroscopy of NP interaction with cells in culture,” Opt. Express 14(14), 6535–6542 (2006). 10.1364/OE.14.006535 [DOI] [PubMed] [Google Scholar]
- Curry A., Crow M., and Wax A., “Molecular imaging of epidermal growth factor receptor in live cells with refractive index sensitivity using dark-field microspectroscopy and immunotargeted NPs,” J. Biomed. Opt. 13(1), 014022 (2008). 10.1117/1.2837450 [DOI] [PubMed] [Google Scholar]
- Gong Y., Yao E., Shen R., Goel A., Arcila M., Teruya-Feldstein J., Zakowski M. F., Frankel S., Peifer M., Thomas R. K., Ladanyi M., and Pao W., “High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507),” PLoS ONE 4(10), e7273 (2009). 10.1371/journal.pone.0007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Ly Y., Hollenberg M., and DeFea K., “A beta-arrestin-dependent scaffold is associated with prolonged MAPK activation in pseudopodia during protease-activated receptor-2-induced chemotaxis,” J. Biol. Chem. 278(36), 34418–34426 (2003). 10.1074/jbc.M300573200 [DOI] [PubMed] [Google Scholar]
- Crow M., Seekell K., Ostrander J. H., and Wax A., “Monitoring of receptor dimerization using plasmonic coupling of gold NPs.” ACS Nano (2011) (under review). [DOI] [PubMed]
- Hancock M., Langton B., Chan T., Toy P., Monahan J., Mischak R., and Shawver L., “A monoclonal antibody against the c-erbB-2 protein enhances the cytotoxicity of cis-diamminedichloroplatinum against human breast and ovarian tumor cell lines.” Cancer Res. 51(17), 4575–4580 (1991). [PubMed] [Google Scholar]
- Yu C., Nakshatri H., and Irudayaraj J., “Identity profiling of cell surface markers by multiplex gold nanorod probes.” Nano Lett. 7(8), 2300–2306 (2007). 10.1021/nl070894m [DOI] [PubMed] [Google Scholar]
- Crow M., Grant G., Provenzale J., and Wax A., “Molecular imaging and quantitative measurement of epidermal growth factor receptor expression in live cancer cells using immunolabeled gold NPs,” AJR, Am. J. Roentgenol. 192(4), 1021–1028 (2008). 10.2214/AJR.07.3535 [DOI] [PubMed] [Google Scholar]
- Crow M., Seekell K., and Wax A., “Polarization mapping of NP plasmonic coupling,” Opt. Lett. 36(5), 757–759 (2011). 10.1364/OL.36.000757 [DOI] [PubMed] [Google Scholar]
- Shi W., Sahoo Y., Swihart M., and Prasad P., “Gold nanoshells on polystyrene cores for control of surface plasmon resonance,” Langmuir 21(4), 1610–1617 (2005). 10.1021/la047628y [DOI] [PubMed] [Google Scholar]
- Desbois-Mouthon C., Cacheux W., Blivet-Van Eggelpoël M. J., Barbu V., Fartoux L., Poupon R., Housset C., and Rosmorduc O., “Impact of IGF-1R/EGFR cross-talks on hepatoma cell sensitivity to gefitinib,” Int. J. Cancer 119(11), 2557–2566 (2006). 10.1002/ijc.22221 [DOI] [PubMed] [Google Scholar]