Abstract
Prostate cancer is the most commonly diagnosed non-skin malignancy in the United States and presents with a wide range of aggressiveness from extremely slow-growing to highly aggressive. There is a clinical need to determine the metastatic potential of the primary tumor to design the most appropriate treatment plan ranging from watchful waiting to more aggressive, invasive surgical treatments. In this study we have developed a nanoparticle based imaging agent that targets SPARC (Secreted Protein Acidic Rich in Cysteine), a molecular marker of prostate cancer metastatic potential. Previous studies by this group used phage display to identify a peptide with high binding affinity and specificity for SPARC. In this study, the SPARC-targeted peptide sequence was used to design a biomaterial with improved pharmacokinetic properties by attaching it to a biocompatible nanoparticle that is also coupled to a fluorophore for in vivo imaging. Prostate cancer cell lines with varying degrees of SPARC expression were used to show the ability of the targeted nanoparticle to bind specifically to SPARC in vitro and in vivo including the clinically relevant bone and lung metastases. We show that in vivo imaging information correlates with the metastatic potential of the prostate tumor. This prognostic information could enable doctors to stratify patients and design personalized treatment strategies.
Introduction
Approximately one out of every six men will be diagnosed with prostate cancer in the United States. Treatment options for localized disease vary considerably, and range from “watchful waiting” (no active treatment) to radical prostatectomy. However, it is difficult to ascertain which treatment option is appropriate because the ability of the treating physician to accurately predict the tumor’s metastatic potential is limited. Prostatic biopsies are crucial both to diagnosing prostate cancer and in deriving prognostic information. However, prostate biopsies are invasive, time consuming, and are limited by a considerable degree of sampling error [28]. In part due to the problems with prostate biopsies, it is not uncommon for the treatment of prostate cancer to be incorrectly paired with the underlying disease aggressiveness, commonly resulting in patients with slow-growing tumors enduring unnecessary surgery and, more rarely, patients with more aggressive tumors relegated to “watchful waiting” [3]. The development of a biomaterial for imaging that provides accurate insight into the metastatic potential of prostate cancer would be an important step in ensuring that urologists recommend the most appropriate treatment for their patients.
Secreted Protein Acidic and Rich in Cysteine (SPARC), also known as osteonectin or BM-40, is a ~34kDa matricellular glycoprotein that modulates cell-matrix interactions and is expressed in a variety of tissues during development and during tissue repair and remodeling [21]. It is involved in a diverse range of mechanisms and has been shown to disrupt cell spreading and adhesion [30], disassemble focal adhesions [23,30], mediate leukocyte diapedesis [17], induce cell rounding and changes in cellular morphology [9,13,20], regulate cell proliferation [11,12], and it has demonstrated activities in angiogenesis [31], extracellular matrix remodeling [14,36], and tumorigenesis [29]. Furthermore, SPARC has been shown to be involved in the endothelial to mesenchymal transition (EMT), a hallmark of tumor invasiveness [33] and prostate cancer progression [35], by repressing E-cadherin [27].
Recently, analysis of human tumor samples has indicated the presence of high levels of SPARC expression correlating with clinical prognosis and outcome. Indeed, in breast, lung, pancreas, and prostate cancer, high levels of SPARC indicates a worse prognosis [6]. Gene profiling has found SPARC to be part of the invasion-specific cluster in both pancreatic and breast carcinoma [10], and SPARC expression is associated with invasiveness and metastasis in gastric cancer [37]. These findings of SPARC expression correlating with a worse prognosis is not surprising since SPARC has been shown to mediate many of the steps in the metastatic pathway, such as cell rounding, disruption of focal adhesions, and degradation of the extracellular matrix. In fact, its promigratory, anti-adhesive effect on cells has led to SPARC being referred to as the “proinvasive” protein [5]. Studies showing differential expression of SPARC in prostate cancer revealed that SPARC is expressed most highly in bone-metastasizing prostate cancer cell lines [34]. Furthermore, metastatic lesions in bone demonstrated intense SPARC staining [34]. In certain prostate cancer cell lines SPARC has been shown to increase activity of the matrix metalloproteinases [16], a class of molecules strongly associated with tumor invasion and metastasis [24].
Given the role of SPARC in invasion and metastasis we reasoned that it would represent an informative target for imaging agents seeking to stratify prostate cancer patients. In prior work, we identified a peptide sequence with ~100 nm affinity. However, direct labeling resulted in rapid clearance, and non-ideal pharmacokinetics reducing its utility for in vivo imaging or drug delivery applications. In this manuscript, we therefore designed a longer circulating, higher affinity targeted nanoparticle and tested this biomaterial as an imaging agent. We envision that this approach could ultimately be used for a molecular based approach to prostate cancer prognostication and treatment selection as well as specific delivery of cytotoxic agents to SPARC expressing cancer cells.
Materials and Methods
Cell lines
The DU145 cell line was obtained from the American Type Tissue Culture Collection, and LNCaP-C42b cell lines were a generous gift from Dr. Michael Weber, University of Virginia Cancer Center. C42b cells were grown in T-Media supplemented with 5% fetal bovine serum, and DU145s were grown in MEM supplemented with 10% fetal bovine serum. All cultures were maintained in a CO2 humidified incubator at 37°C.
Histology and immunohistochemistry
Prostate disease spectrum (prostatic cancer progression) array was purchased from US Biomax, Inc. (Rockville, MD) and probed with anti-SPARC antibody (NCL-O-NECTIN from Novocastra, Newcastle upon Tyne, UK) at a dilution of 1:20, with standard citrate retrieval. Statistics were performed to compare metastatic to non-metastatic tumor samples using an unpaired t-test. P values of less than 0.05 were considered statistically significant.
Materials characterization
AminoSPARK 680 nanoparticles were purchased from Perkin Elmer (Waltham, MA). Information obtained from a phage display screen previously shown to bind to SPARC with high affinity [18] was used to create a peptide sequence, SPPTGINGGSK(FITC)C (SPARC(FITC)) or SPPTGINGGSK(Biotin)C (SPARC (BIOTIN)), which was conjugated to the nanoparticle. Iron concentration was determined spectrophotometrically. Iron oxide core size was determined by transmission electron microscopy (TEM) on a JEOL 200CX instrument at 200 kV. Dilute nanoparticle solution was spotted onto a copper carbon film TEM grid (Electron Microscopy Sciences, Hatfield PA). Size distribution was calculated using ImageJ software and manually. Surface morphology and the three-dimensional shape of the nanoparticles was imaged by tapping mode atomic force microscopy (AFM) on a Dimension 3000 Scanning Probe Microscope (Veeco Instruments, Plainview, NY) and analyzed using Veeco’s Nanoscope software. Nanoparticles were reacted with N-succinimidyl S-acetylthioacetate (SATA, Molecular Biosciences, Boulder CO), deprotected with hydroxylamine, and then dropcast on Au(111) on mica. Hydrodynamic size of the nanoparticles was determined by dynamic light scattering (DLS) using a Zetasizer (Malvern Instruments, Marlboro, MA).
Specificity of SPARC-NP
C42b (SPARC positive) and DU145 cells (SPARC negative) were incubated with increasing log concentrations of SPARC(BIOTIN)-NP for one hour at 4°C, washed three times with DPBS, then streptavidin-HRP (0.25 ug/mL) was added for 1 hour at room temperature. Wells were washed 5× with 1% BSA/0.1% Tween-20 in PBS then the colorimetric substrate, tetramethyl benzadine (TMB) was added. Stop solution of 0.1M H2SO4 was added (100 uL) then absorbance values were read on a plate reader (FluoSTAR Optima, BMG Labtech, Durham, NC) at 455nm. EC50s were determined by fitting a sigmoidal dose response curve in GraphPad Prism (GraphPad Software, LaJolla, CA).
In vivo optical imaging experiments
Subcutaneous tumors
Male nude mice (7–9 weeks of age) were purchased from the MGH breeding colony (COX-7) or NCI and maintained in approved pathogen free institutional housing facilities. All experiments were performed according to institutional guidelines. In all experiments, probe was administered intravenously at 20 mg Fe/kg body weight.
For the blood ½ life studies, blood was collected by retro-orbital bleeding at various time points post intravenous administration, and was imaged using a home-built fluorescence reflectance imaging system. Regions of interest were drawn around the samples to calculate mean fluorescent intensities. All samples were compared to normal blood obtained from non-injected mice (N=5 for all groups).
For in vivo quantitation of SPARC, optical imaging of nude mice (n=10) bearing subcutaneously implanted tumors (diameter 5–6 mm) derived from the immortalized human prostate cancer cell lines, DU145 (little or no SPARC) or C42b (high SPARC levels) was performed. Mice were co-injected via tail vein with SPARC-NP-680 and NP-750, which is the control non-targeted nanoparticle. Fluorescence mediated tomography (FMT) imaging was carried out on a commercial imaging system (FMT2500, Perkin Elmer, Waltham MA). Data analysis was completed within the FMT software as previously described [34] by selecting regions of interest encompassing the tumors in the 3-dimensional data sets. Data is expressed as mean fluorescence intensity within the selected volume of interest (VOI).
Bone tumors
To image bone tumors derived from DU145 or C42b cells, an incision was made in anesthetized mice along the left knee cap. With slight pressure, a syringe that was placed at the center of the end of the tibia syringe was rotated to drill a hole through the bone. Once the needle descended 2–3mm the cancer cells were suspended in PBS at a concentration of 1×106/50µl, mixed well and loaded into a 1cc syringe with 26g needle, void of air bubbles. After depositing of tumor cells, the needle was removed and the hole was sealed with bone wax, and the wound repaired with suture. For this study SCID/bg mice (Jax Laboratories) were individually injected with either C42b or Du145 cells, for a total number of 5 mice per cohort and imaged via CT and FMT then sacrificed by the end of 9 weeks. All SCID/bg mice were injected by the University of Virginia Molecular Assessment and Preclinical Studies core.
Lung metastases
To image metastatic disease, mice (n=5) were injected intravenously with 500,000 C42B cells. Three to five days after cell injection, animals were administered SPARC-NP-680 and imaged by FMT for signal detection and x-ray Computed Tomography (CT) for anatomical reference. Mice were anesthetized (Isoflurane 1.5%, O2 2L/min) during imaging with a gas delivery system integrated into a multimodal imaging cassette that holds the mouse during imaging and facilitates coregistration of FMT to CT data through fiducial landmarks on its frame. CT imaging preceded FMT, applying an x-ray source at 80 kVp and 500 uA with an exposure time of 420 ms to acquire 360 projections. The effective 3D CT resolution was 110 um isotropic. During CT acquisition, Isovue-370 was infused at 20 uL/min through a tail vein catheter to highlight pulmonary vasculature. The CT reconstruction protocol performed bilinear interpolation, used a Shepp-Logan filter, and scaled pixels to Hounsfield units. The mouse was then transferred in the multimodal cassette holder to FMT for imaging where acquisition time was typically 5 to 8 minutes. Data were post-processed using a normalized Born forward equation to calculate three dimensional fluorochrome concentration distribution. We then imported data into OsiriX (The OsiriX foundation) to co-register FMT and CT images. Fiducials on the imaging cartridge were visualized and tagged in FMT and CT images with point markers to define their XYZ coordinates. Using these coordinates, data were re-sampled, rotated, and translated to match the image matrices and finally displayed in one hybrid image.
Results
SPARC Expression correlates with prostate cancer grade
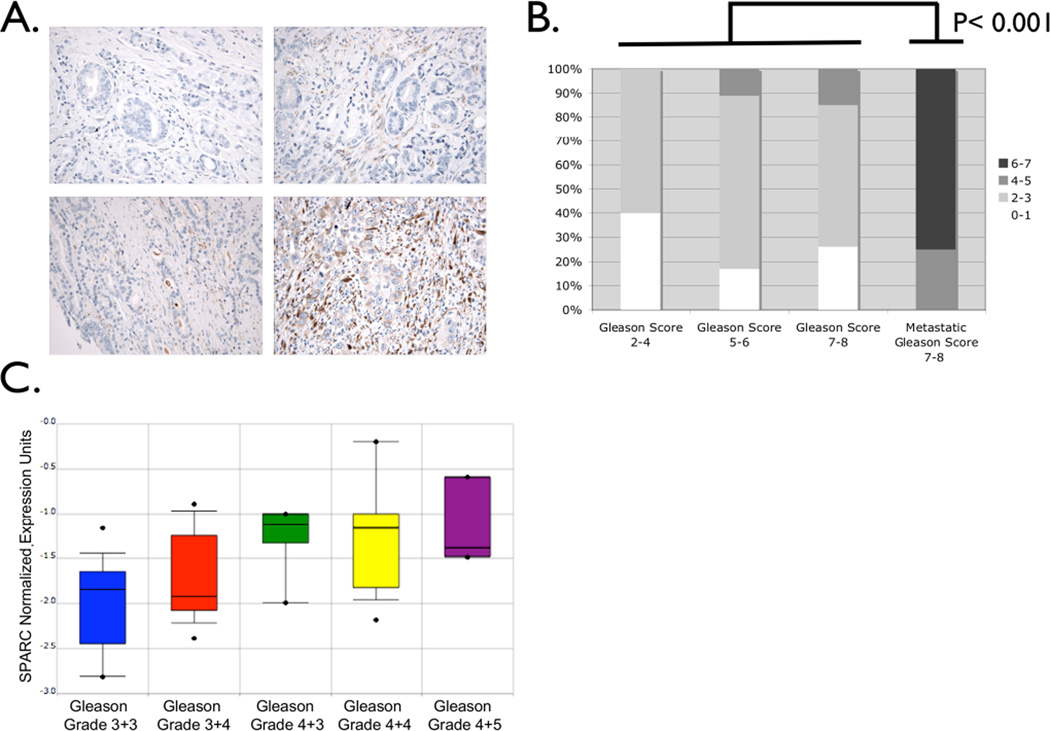
SPARC expression in 53 human prostate cancer samples was measured by imunnohistochemical analysis of tissue microarrays consisting of cores of various cancer grades. SPARC expression was most prominent in the epithelial and stromal compartments of the tumors analyzed (Figure 1A). As prostate cancer becomes advanced as scored by Gleason grade, SPARC expression also increased. As anticipated, metastatic cancer samples expressed the highest levels of SPARC (Figure 1B). Data from Lapointe et al. [22] analyzed using Oncomine [26] showed increasing SPARC expression with prostate cancer progression, confirming our immunohistochemical results (Figure 1C).
Figure 1. SPARC expression is correlated with prostate cancer progression and metastasis.
A. Immunohistochemical analysis of SPARC expression in a prostate cancer tissue microarray of a range of Gleason patterns. Representative staining of cores of Gleason Score 2–4 (upper left), Gleason Score 5–6 (upper right), Gleason Score 7–8 (lower left), and Gleason Score 7–8 (lower right). B. A summary of the expression quantitation in tumoral, stromal, and epithelial compartments scored in intensity from 0–3 and shown here as a single summed score from 0–7. C. Analysis of gene expression data originally obtained by Lapointe et al. illustrates that increasing SPARC expression correlates with increasing Gleason Grade.
Characterization of the SPARC targeted imaging agent
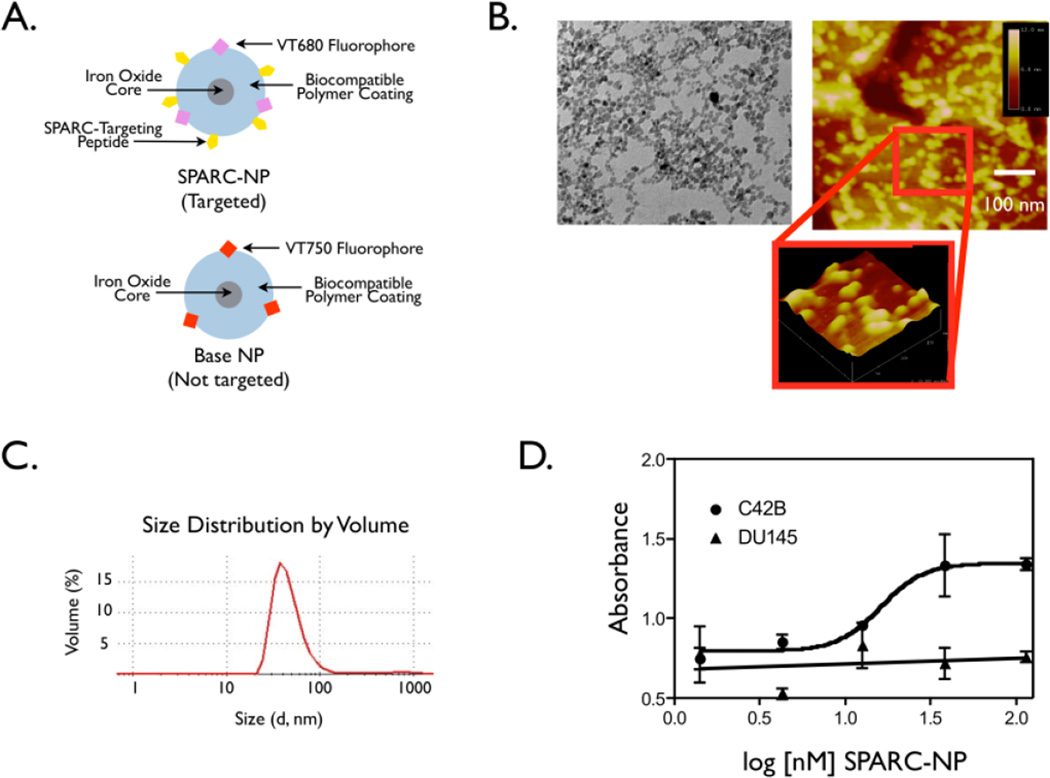
A schematic of the nanoparticles used in this study is shown in Figure 2A. It is a SPARC targeted nanoparticle referred to as SPARC-NP, which is conjugated to a SPARC-targeting peptide [18], as well as a fluorophore at a wavelength of 680 nm for in vivo imaging. One of the main determinants of how a nanoparticle behaves in vivo is its size and shape [7]. TEM of the amino SPARK nanoparticles was used to examine the core size (Figure 2B, left) and tapping mode AFM was used to examine the size and shape of the polymer coating (Figure 2B right). TEM images of the nanoparticles reveal a mean core diameter of 8.75 nm with a standard deviation of 2.49 nm, and the AFM height images show spherical nanoparticles of about 20–30 nm in size. Light scattering, which assesses the size of the polymer-coated nanoparticle in solution, was determined to be 54 nm in diameter (Figure 2C). The difference in diameters between the AFM measurement and the light scattering measurement could be due to a combination of swelling of the polymer coating in solution during the light scattering measurement, and compression of the polymer coating during the tapping mode AFM measurement.
Figure 2. Characterization of SPARC targeted biomaterial.
A. Schematic of the targeted and base nanoparticles. B. TEM micrographs of aminoSPARK nanoparticles. C. AFM height image of aminoSPARK nanoparticles on gold on mica. D. Specificity of the nanoparticles.
Increasing log concentrations of SPARC (Biotin) NP were incubated with either C42b SPARC positive cells or DU145 SPARC low or negative prostate cancer cells. After incubation with streptavidin-HRP and addition of TMB, absorbance was read on a plate reader.
To determine whether conjugation of SPARC binding peptide to the nanoparticles changed specificity, we decided to utilize cell lines with various levels of SPARC expression [34]. C42bs are in vivo-derived sublineages of LNCaPs that mark cancer progression milestones; specifically bone metastasis. C42b cells have the highest level of SPARC expression, however, DU145s exhibit little to no SPARC expression and are thus an important negative control in this study [34]. In vitro cell uptake assays were used to determine specificity of the imaging agent for SPARC expression. SPARC (BIOTIN)-NP had an avidity of 15.6 nM when incubated with the SPARC positive C42b cells. Importantly, SPARC (BIOTIN)-NP demonstrated negligible binding (Figure 2D) to SPARC negative, control cells, DU145 (Figure 2D).
In vivo studies
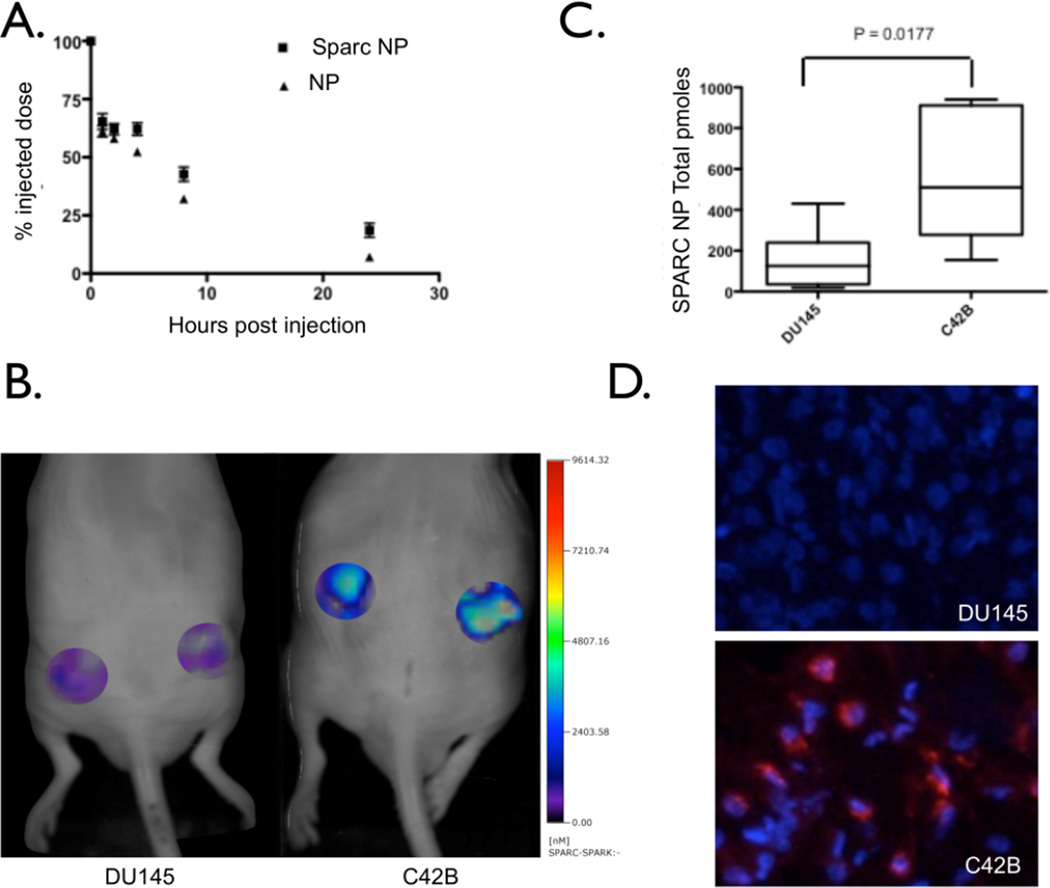
As part of the in vivo characterization of the nanoparticles, we determined the blood half-life of SPARC-NP-680. Conjugation of the peptide to the base nanoparticle did not significantly alter the vascular clearance as blood-half life studies show similar clearance of the SPARC-NP and base NP from the blood pool (7.22 hrs Figure 3A). To determine whether we could distinguish the levels of SPARC expression in prostate tumors, C42b and DU145 tumor-bearing mice underwent fluorescent molecular tomographic (FMT) imaging (Figure 3B). Mice were co-injected with SPARC-NP-680 and NP-750. Mean SPARC-NP-680 within tumors of varying SPARC expression are plotted in Figure 3C, and show that increased SPARC expression correlates with increased probe concentration. In contrast, control, non-targeted NP-750 minimally accumulated in DU145 or C42b tumors (Figure S1). Indeed, there was a 45.9 fold increase in uptake in C42b derived tumors of targeted SPARC-NP-680 to non-targeted NP-750 agents. There was no statistical difference between NP-750 uptake in the DU145 or C42b derived tumors (ratio of 1.1 C42b to DU145 NP-750). After imaging, tumors were harvested and sectioned for correlative histology. Tumors derived from C42b cells demonstrate targeted probe accumulation (Figure 3D). In contrast, tumors derived from the SPARC negative cell line, DU145, had significantly less SPARC targeted probe uptake (Figure 3 B,3C), and upon histological inspection, tumor cells had low or absent SPARC-NP-680 accumulation (Figure 3D).
Figure 3. In vivo characterization and imaging.
A. Blood ½ life studies with both SPARC NP and base NP. B. FMT imaging of DU145 (left) or C42b (right) subcutaneous prostate tumors. C. Quantitation of SPARC NP accumulation and binding in the DU145 or C42b tumors. D. Correlative fluorescence microscopy of either DU145 or C42b tumors. Blue – DAPI nuclear stain, Red – SPARC NP.
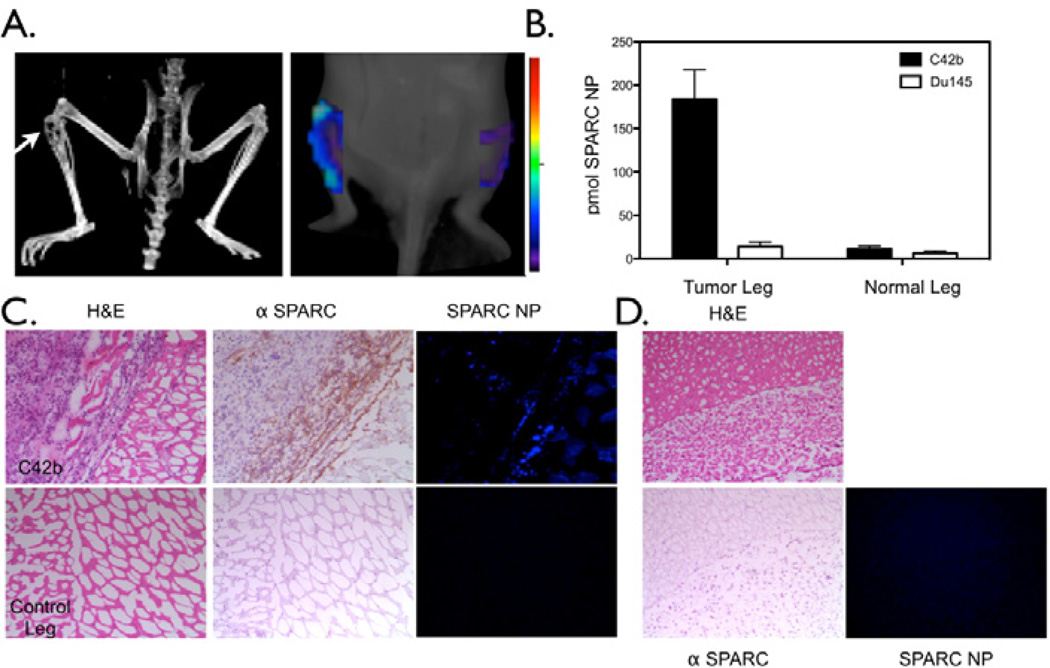
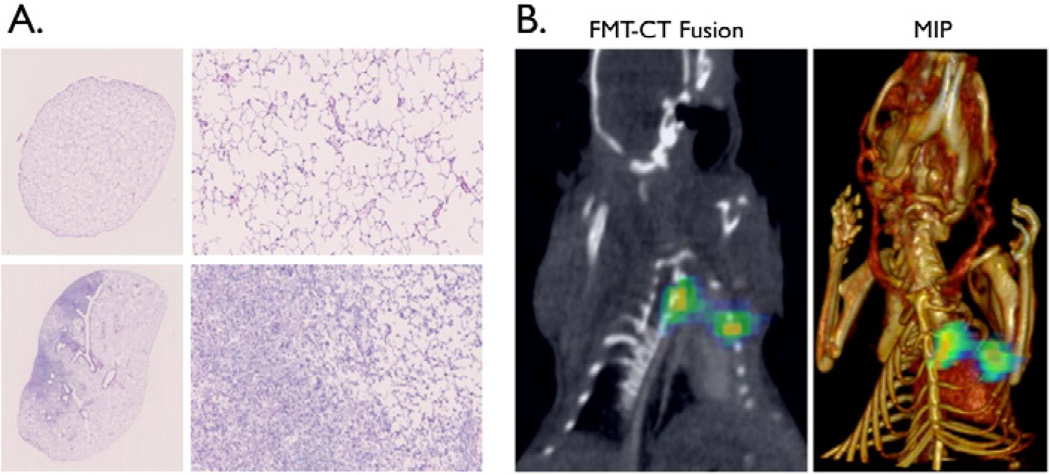
As prostate cancer has an increased tendency to metastasize to the bone and indeed, the bone is almost twice as likely when compared to other distant organs to have prostate cancer metastases [32], we wanted to determine whether the targeted SPARC specific nanoparticle based imaging agent could non-invasively detect bone tumors derived from prostate cancer cells. C42b or DU145 cells were implanted into the tibia of mice and imaged via tail vein injection of SPARC NP. Animals were imaged by FMT for SPARC expression and CT for anatomical imaging (Figure 4A). SPARC NP was able to image SPARC expressing prostate cancer in the bone with a 16.1 fold higher binding to C42b SPARC positive derived bone tumors vs. normal leg (Figure 4B). Further, there was a 10.6 higher binding of SPARC NP to the SPARC positive bone derived tumors when compared with SPARC low (DU145) bone tumors (Figure 4B). Similarly to the subcutaneous tumor findings, non-targeted control NP-750 had minimal accumulation in both the DU145 or the C42b derived tumors (Figure S2). Correlative histology confirmed the presence of SPARC expression in the C42b derived tumors (Figure 4C top) whereas there was little to no expression of SPARC in the control leg tibia (Figure 4C bottom) or the DU145 derived tumors (Figure 4D). SPARC NP uptake as determined by 680 fluorescence correlated with regions of high SPARC expression (Figure 4C). The probe confirmed the imaging findings and specificity for SPARC as there was little or no accumulation detected in control legs or DU145 derived tumors (Figure 4C, 4D). In another application, we demonstrated the imaging of prostate cancer lung metastases with SPARC NP. We employed a mouse model of intravenous injection of DU145 or C42b cells. Mice injected intravenously with DU145 cells did not develop lung tumors even after 6 months. However, mice injected with C42B cells intravenously showed lung tumor growth, mimicking metastatic disease (Figure 5A left). C42b injected mice also show binding of SPARC-NP-680 to the lung tumor, and fused FMT datasets with CT confirm probe signal within the tumor (Figure 5B). Histology confirmed the presence of tumors in the lungs (Figure 5A).
Figure 4. Detection of Bone tumors with SPARC NP.
Animals were injected intratibially with either C42b or DU145 cells. A. CT (left) and FMT (right) imaging of SPARC NP injected animals. Arrow points to the C42b derived tumor. B. Quantitation of SPARC NP accumulation in either C42b or DU145 derived tumors in the bone vs. the control non-tumored leg of the same mouse. C. Correlative histology from legs with C42b tumors or control non-tumored legs harvested and sectioned after the completion of the imaging experiments. Left, H&E, middle anti-SPARC antibody, and Right fluorescence from SPARC NP. D. Correlative histology from legs with DU145 intratibial tumors.
Figure 5. SPARC-NP is able to detect prostate cancer lung metastases.
FMT-CT fusion, illustrating in vivo detection of tumor in the lung. A. The left panel shows fluorescence data from FMT imaging of SPARC-targeted agent in mouse with metastatic tumor in the lung. Right panel is a maximum intensity projection image of the CT and FMT data sets. B. H&E staining of excised lungs corroborates in vivo imaging results. Top panel illustrates normal lung architecture. Bottom panel shows deposition of tumor cells (left) next to normal lung tissue (right). Scalebars are 1mm
Discussion
While prostate cancer affects millions of men worldwide, current methods of prognostication are inaccurate. Biopsies often do not accurately reveal the tumor’s underlying aggressiveness, and many men unnecessarily undergo the most invasive form of treatment, radical prostatectomy [2]. In order to solve this clinical dilemma, we developed a nanoparticle targeting the protein, SPARC and validated its in vivo behavior by imaging. Critically, this targeted biomaterial is able to both identify areas of prostate tumor and provide important biologic information; namely, the degree of SPARC expression, which we and others have demonstrated to correlate with metastatic potential [6,35,37]. Indeed, we have been able to use our SPARC targeted nanoparticle to image not only “primary” tumors but also prostate cancer derived bone tumors and lung metastases. Thus, a SPARC based imaging agent may allow clinicians to not only stratify patients into appropriate treatment regimens but may also allow detection of metastases enabling accurate staging of the tumor burden as well.
The immunohistochemical results from the tissue microarrays show that SPARC expression in prostate cancer is present not only in the tumoral compartment but also in the stromal and epithelial compartments surrounding the tumor. This result was expected, as SPARC is a secreted matricellular protein and a known regulator of tumor-stromal interactions. Indeed, this finding has been corroborated in several other tissue microarray studies as well [4,15,19,25,38]. Increased SPARC expression in tissue microarrays has been correlated with increased tumor metastasis [1], and malignant prostate cancer has been shown to be induced by the stroma itself [8].
The overall goal of this imaging nanoparticle is to reduce the number of unnecessary radical prostatectomies and, conversely, to ensure that highly invasive tumors are treated aggressively. Importantly, SPARC expression imparts prognostic information in other solid tumors as well and may become the target of future molecular therapeutics. Thus, the potential clinical applications of this novel imaging agent extend beyond prostate cancer to a variety of tumor types and therapeutic indications. Furthermore, we anticipate that this biomaterial based approach could be used to target therapeutic and theranostic nanoparticles to SPARC avid cancers, opening the possibilities of more selective therapies.
Supplementary Material
Acknowledgments
We would like to thank Fred Reynolds for his assistance in nanoparticle conjugation. Funding for this work was provided by NIH NCI RO1CA13707 and NIH NIBIB RO1EB010023.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Alonso SR, Tracey L, Ortiz P, Perez-Gomez B, Palacios J, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67:3450–3460. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- 2.Bangma CH, Roemeling S, Schroder FH. Overdiagnosis and overtreatment of early detected prostate cancer. World J Urol. 2007;25:3–9. doi: 10.1007/s00345-007-0145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bill-Axelson A, Holmberg L, Filen F, Ruutu M, Garmo H, et al. Radical prostatectomy versus watchful waiting in localized prostate cancer: the Scandinavian prostate cancer group-4 randomized trial. J Natl Cancer Inst. 2008;100:1144–1154. doi: 10.1093/jnci/djn255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloomston M, Ellison EC, Muscarella P, Al-Saif O, Martin EW, et al. Stromal osteonectin overexpression is associated with poor outcome in patients with ampullary cancer. Ann Surg Oncol. 2007;14:211–217. doi: 10.1245/s10434-006-9128-3. [DOI] [PubMed] [Google Scholar]
- 5.Brekken RA, Sage EH. SPARC, a matricellular protein: at the crossroads of cell-matrix communication. Matrix Biol. 2001;19:816–827. doi: 10.1016/s0945-053x(00)00133-5. [DOI] [PubMed] [Google Scholar]
- 6.Clark CJ, Sage EH. A prototypic matricellular protein in the tumor microenvironment--where there’s SPARC, there’s fire. J Cell Biochem. 2008;104:721–732. doi: 10.1002/jcb.21688. [DOI] [PubMed] [Google Scholar]
- 7.Corot C, Robert P, Idee JM, Port M. Recent advances in iron oxide nanocrystal technology for medical imaging. Adv Drug Deliv Rev. 2006;58:1471–1504. doi: 10.1016/j.addr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Cunha GR, Hayward SW, Wang YZ. Role of stroma in carcinogenesis of the prostate. Differentiation. 2002;70:473–485. doi: 10.1046/j.1432-0436.2002.700902.x. [DOI] [PubMed] [Google Scholar]
- 9.Everitt EA, Sage EH. Expression of SPARC is correlated with altered morphologies in transfected F9 embryonal carcinoma cells. Exp Cell Res. 1992;199:134–146. doi: 10.1016/0014-4827(92)90471-j. [DOI] [PubMed] [Google Scholar]
- 10.Framson PE, Sage EH. SPARC and tumor growth: where the seed meets the soil? J Cell Biochem. 2004;92:679–690. doi: 10.1002/jcb.20091. [DOI] [PubMed] [Google Scholar]
- 11.Funk SE, Sage EH. The Ca2(+)-binding glycoprotein SPARC modulates cell cycle progression in bovine aortic endothelial cells. Proc Natl Acad Sci U S A. 1991;88:2648–2652. doi: 10.1073/pnas.88.7.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funk SE, Sage EH. Differential effects of SPARC and cationic SPARC peptides on DNA synthesis by endothelial cells and fibroblasts. J Cell Physiol. 1993;154:53–63. doi: 10.1002/jcp.1041540108. [DOI] [PubMed] [Google Scholar]
- 13.Goldblum SE, Ding X, Funk SE, Sage EH. SPARC (secreted protein acidic and rich in cysteine) regulates endothelial cell shape and barrier function. Proc Natl Acad Sci U S A. 1994;91:3448–3452. doi: 10.1073/pnas.91.8.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasselaar P, Loskutoff DJ, Sawdey M, Sage EH. SPARC induces the expression of type 1 plasminogen activator inhibitor in cultured bovine aortic endothelial cells. J Biol Chem. 1991;266:13178–13184. [PubMed] [Google Scholar]
- 15.Iacobuzio-Donahue CA, Argani P, Hempen PM, Jones J, Kern SE. The desmoplastic response to infiltrating breast carcinoma: gene expression at the site of primary invasion and implications for comparisons between tumor types. Cancer Res. 2002;62:5351–5357. [PubMed] [Google Scholar]
- 16.Jacob K, Webber M, Benayahu D, Kleinman HK. Osteonectin promotes prostate cancer cell migration and invasion: a possible mechanism for metastasis to bone. Cancer Res. 1999;59:4453–4457. [PubMed] [Google Scholar]
- 17.Kelly KA, Allport JR, Yu AM, Sinh S, Sage EH, et al. SPARC is a VCAM-1 counter-ligand that mediates leukocyte transmigration. J Leukoc Biol. 2007;81:748–756. doi: 10.1189/jlb.1105664. [DOI] [PubMed] [Google Scholar]
- 18.Kelly KA, Waterman P, Weissleder R. In vivo imaging of molecularly targeted phage. Neoplasia. 2006;8:1011–1018. doi: 10.1593/neo.06610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koukourakis MI, Giatromanolaki A, Brekken RA, Sivridis E, Gatter KC, et al. Enhanced expression of SPARC/osteonectin in the tumor-associated stroma of non-small cell lung cancer is correlated with markers of hypoxia/acidity and with poor prognosis of patients. Cancer Res. 2003;63:5376–5380. [PubMed] [Google Scholar]
- 20.Lane TF, Sage EH. Functional mapping of SPARC: peptides from two distinct Ca+(+)-binding sites modulate cell shape. J Cell Biol. 1990;111:3065–3076. doi: 10.1083/jcb.111.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane TF, Sage EH. The biology of SPARC, a protein that modulates cell-matrix interactions. FASEB J. 1994;8:163–173. [PubMed] [Google Scholar]
- 22.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy-Ullrich JE, Lane TF, Pallero MA, Sage EH. SPARC mediates focal adhesion disassembly in endothelial cells through a follistatin-like region and the Ca(2+)-binding EF-hand. J Cell Biochem. 1995;57:341–350. doi: 10.1002/jcb.240570218. [DOI] [PubMed] [Google Scholar]
- 24.Nelson AR, Fingleton B, Rothenberg ML, Matrisian LM. Matrix metalloproteinases: biologic activity and clinical implications. J Clin Oncol. 2000;18:1135–1149. doi: 10.1200/JCO.2000.18.5.1135. [DOI] [PubMed] [Google Scholar]
- 25.Paley PJ, Goff BA, Gown AM, Greer BE, Sage EH. Alterations in SPARC and VEGF immunoreactivity in epithelial ovarian cancer. Gynecol Oncol. 2000;78:336–341. doi: 10.1006/gyno.2000.5894. [DOI] [PubMed] [Google Scholar]
- 26.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robert G, Gaggioli C, Bailet O, Chavey C, Abbe P, et al. SPARC represses E-cadherin and induces mesenchymal transition during melanoma development. Cancer Res. 2006;66:7516–7523. doi: 10.1158/0008-5472.CAN-05-3189. [DOI] [PubMed] [Google Scholar]
- 28.Ruijter E, van Leenders G, Miller G, Debruyne F, van de Kaa C. Errors in histological grading by prostatic needle biopsy specimens: frequency and predisposing factors. J Pathol. 2000;192:229–233. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH703>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 29.Sage EH. Terms of attachment: SPARC and tumorigenesis. Nat Med. 1997;3:144–146. doi: 10.1038/nm0297-144. [DOI] [PubMed] [Google Scholar]
- 30.Sage EH, Bornstein P. Extracellular proteins that modulate cell-matrix interactions. SPARC, tenascin, and thrombospondin. J Biol Chem. 1991;266:14831–14834. [PubMed] [Google Scholar]
- 31.Sage EH, Vernon RB. Regulation of angiogenesis by extracellular matrix: the growth and the glue. J Hypertens Suppl. 1994;12:S145–S152. [PubMed] [Google Scholar]
- 32.Saitoh H, Hida M, Shimbo T, Nakamura K, Yamagata J, et al. Metastatic patterns of prostatic cancer. Correlation between sites and number of organs involved. Cancer. 1984;54:3078–3084. doi: 10.1002/1097-0142(19841215)54:12<3078::aid-cncr2820541245>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 33.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 34.Thomas R, True LD, Bassuk JA, Lange PH, Vessella RL. Differential expression of osteonectin/SPARC during human prostate cancer progression. Clin Cancer Res. 2000;6:1140–1149. [PubMed] [Google Scholar]
- 35.Tran NL, Nagle RB, Cress AE, Heimark RL. N-Cadherin expression in human prostate carcinoma cell lines. An epithelial-mesenchymal transformation mediating adhesion with stromal cells. Am J Pathol. 1999;155:787–798. doi: 10.1016/S0002-9440(10)65177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tremble PM, Lane TF, Sage EH, Werb Z. SPARC, a secreted protein associated with morphogenesis and tissue remodeling, induces expression of metalloproteinases in fibroblasts through a novel extracellular matrix-dependent pathway. J Cell Biol. 1993;121:1433–1444. doi: 10.1083/jcb.121.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang CS, Lin KH, Chen SL, Chan YF, Hsueh S. Overexpression of SPARC gene in human gastric carcinoma and its clinic-pathologic significance. Br J Cancer. 2004;91:1924–1930. doi: 10.1038/sj.bjc.6602213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue LY, Hu N, Song YM, Zou SM, Shou JZ, et al. Tissue microarray analysis reveals a tight correlation between protein expression pattern and progression of esophageal squamous cell carcinoma. BMC Cancer. 2006;6:296. doi: 10.1186/1471-2407-6-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.