Abstract
The pH dependence of native PHM and its M314H variant have been studied in detail. For WT PHM the intensity of the Cu-S interaction visible in the Cu(I) EXAFS data is inversely proportional to catalytic activity over the pH range 3 - 8. A previous model based on more limited data was interpreted in terms of two protein conformations involving an inactive met-on form and an active flexible met-off form which derived its catalytic activity from the ability to couple into vibrational modes critical for proton tunneling. The new studies comparing the WT and M314H variant have led to an evolution of this model where the met-on form has been found to be derived from coordination of an additional Met residue, rather than a more rigid conformer of M314 as previously proposed. The catalytic activity of the mutant decreased by 96% due to effects on both kcat and KM but it displayed the same activity/pH profile with a maximum around pH 6. At pH 8, the reduced Cu(I) form gave spectra which could be simulated by replacing the CuM Cu-S(Met) interaction with a Cu-N/O but the data did not unambiguously assign the ligand to the imidazole side chain of H314. At pH 3.5 the EXAFS still showed the presence of a strong Cu-S interaction, establishing that the met-on form observed at low pH in WT cannot be due to a strengthening of the CuM-methionine interaction, but must arise from a different Cu-S interaction. Therefore, lowering the pH causes a conformational change at one of the Cu centers which brings a new S-donor residue into a favorable orientation for coordination to copper and generating an inactive form. Cys coordination is unlikely since all Cys residues in PHM are engaged in disulfide crosslinks. Sequence comparison with the PHM homologues TBM and DBM suggest that M109 (adjacent to the H-site ligands H107 and H108) is the most likely candidate. A model is presented in which H108 protonates with a pKA of 4.6 to generate the inactive low-pH form with CuH coordinated by M109, H107 and H172.
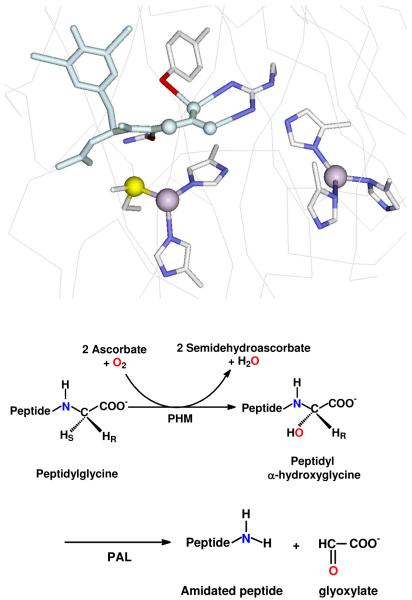
Peptidylglycine monooxygenase (PHM)1 catalyses the first step in the amidation of neuropeptides hormones, converting the glycine-extended propeptide to its α-hydroxyglcine intermediate (1). The catalytic core (residues 42 – 356) termed PHMcc is homologous to that of other copper monooxygenases involved in neurotransmitter biosynthesis such as dopamine β-monooxygenase (DBM) (2) and tyramine β-monooxygenase (TBM) (3). Spectroscopic studies have established structural similarities between their active sites (4-8), but PHMcc is the only member of the group for which crystal structures are available (9-12). Unlike the coupled binuclear centers common to hemocyanins, tyrosinases, catechol oxidases (13-16) and oxygen activating dicopper model complexes (17), the copper centers in PHM are mononuclear, and are separated by 11 A of solvent-filled channel. CuM is coordinated by two histidines and a methionine while CuH is coordinated to three histidine residues (Figure 1).
Figure 1.
Top: view of the active site of PHM showing the CuH (right) coordinated to three histidines and the CuM center (left) centers coordinated to two histidines and a methionine. A substrate molecule (di-iodo-YG) is bound in the site close to the M center. (Taken from pdb file 1OPM). Bottom: reactions catalyzed by the PHM and PAL domains of peptidylglycine α-amidating lyase (PAM).
The detailed mechanism of substrate hydroxylation has been the subject of much debate. It is generally accepted that the enzyme cycles through a reductive phase in which the two copper centers are reduced to Cu(I), and an oxidative phase in which O2 is activated and subsequently hydroxylates the substrate. A structure of the reduced enzyme co-crystalized with a slow substrate has revealed the presence of a dioxygen molecule bound at the CuM center, the bond length of which is consistent with a Cu(II)-superoxo species. A superoxo intermediate is supported by other biochemical (18, 19) and theoretical (15, 20) studies that implicate Cu(II)-O•−2 as the reactive oxygen species. It has been further suggested that the large spatial separation of the Cu centers in PHM prevents formation of the peroxide, and thus allows the potent electrophilic reactivity of the mononuclear Cu(II)-superoxo species to be fully expressed in the form of H-atom abstraction from the substrate (15, 18, 20) to form a mononuclear hydroperoxo species at CuM. Subsequent steps in the reaction pathway are less clear and alternate mechanisms have been proposed that involve long-range electron transfer from CuH either before (18) or after (20) the transfer of an OH group to the substrate radical to form product (17).
Early site-directed mutagenic studies on PHMcc M314X substitutions reported undetectable catalytic activity in the Ile, His, Asp and Cys variants (21, 22), while more recent mutagenic analysis of the homologous Met ligand in TBM (M471) showed similarly low or undetectable activity for His, Asp, and Cys variants(4). These studies established an essential role for M314 in PHM catalysis. Extensive characterization of the Cu-S bond by EXAFS (5-7, 23, 24), led to the finding that the interaction was unusually weak at pHs at or above the pH optimum (5.8), but became much more intense at lower pH. This led to the suggestion that the dynamics of the Cu-S(Met) interaction were controlling catalytic activity via a “Met-on” (inactive) to “Met-off ”(active) structural transition. (24).
In the present paper we explore further the relationship between catalytic activity and the Cu(I)-methionine interactions as a function of pH. Using a complex buffer system containing formate, MES, HEPES and CHES, we have determined the complete pH-activity profile between pH 3 and 8, and correlated this with changes in the strength or shell occupancy of the Cu-S interaction in the EXAFS. These results suggest that the increase in Cu-S interaction is due to coordination of an additional S ligand at low pH mediated by conformational change at one of the Cu centers, which brings a new S-donor residue into a favorable orientation for coordination to copper, thereby generating the inactive form. Cys coordination is unlikely since all Cys residues in PHM are engaged in disulfide crosslinks. Sequence comparison with the PHM homologues TBM and DBM suggest that M109 (adjacent to the H-site ligands H107 and H108) is the most likely candidate. A model is presented in which H108 protonates with a pKA of 4.6 to generate the inactive low-pH form with CuH coordinated by M109, H107 and H172.
Materials and Methods
All buffers and formic acid had a minimum purity of 99% and were acquired from Sigma-Aldrich. Dansyl-YVG was obtained from American Peptide Co. and the bovine liver catalase from Roche.
Expression and Purification of the PHMcc WT and M314H Mutant
The WT and M314H mutant catalytic cores were constructed as reported previously (21) and were isolated and purified from a Chinese Hamster Ovary (CHO) cell line as described (25, 26). The proteins were subjected to two cycles of 8 hour dialysis in 10 mM Tris-acetate buffer (WT at pH 8.4, M314H at pH 8.2) and purified further on a BioCAD 700E Perfusion Chromatography Workstation (Applied Biosystems) by anion exchange on a 10×100 mm column packed with Poros 20 m HQ anion exchange resin. The column was equilibrated with 5 column volumes of 10 mM Tris-acetate buffer at the appropriate pH, loaded with the partially purified enzyme, washed with 2 column volumes of buffer and eluted with a 1-300 mM NaCl gradient over 10 column volumes. The fractions containing PHM were collected, pooled and concentrated by ultrafiltration.
Determination of Copper and Protein Concentration
The purified enzymes were reconstituted with cupric sulfate by slow addition of 2.5 molar equiv followed by exhaustive dialysis against 20 mM ultra-pure sodium phosphate buffer (pH 8.0) containing 25 M Cu(II) ion. The reconstituted enzyme was concentrated by ultrafiltration to approximately 4 mM in Cu(II) and used immediately. This protocol also served to reduce exogenous chloride to levels that were insignificant relative to the protein copper concentration. Protein concentrations were determined using the OD280 on a Cary 50 spectrophotometer at ambient temperature using an extinction coefficient for a 0.1% solution at 280 nm of 0.980. Copper concentrations were determined using a Perkin-Elmer Optima 2000 DV inductively coupled plasma optical emission spectrometer.
Combination Buffer
The following buffers were mixed at 50 mM each and brought to the appropriate pH (3-10) using sodium hydroxide: N-cyclohexyl-2-aminoethanesulfonic acid (CHES), 4-(2-hydroxyethyl)-1-piperazine-ethanesulfonic acid (HEPES), 2-(N-morpholino)-ethanesulfonic acid (MES), and formic acid.
HPLC Separation and Detection of Substrate and Product
Reverse-phase HPLC was performed with a Varian Pro Star solvent delivery module equipped with a Varian Pro Star model 410 autosampler (250-μl syringe, 100-μl sample loop), on a 250 × 4.6-mm Varian Microsorb-MV 100-5 C18 column. Substrate (dansyl-YVG; American Peptide Co.) and product (produced by the PHM-catalyzed reaction) were monitored by their dansyl fluorescence using a Waters 474 scanning fluorescence detector (λEx = 365 nm, λEm = 558 nm). Solvent A was 0.1% trifluoroacetic acid in water, and solvent B was 0.1% trifluoroacetic acid in acetonitrile). Product was separated from substrate via isocratic loading and elution at 25% B in A.
Rates of reactions were determined by monitoring the rate of substrate disappearance as a function of reaction time. A water-jacketed glass reaction vessel at 37 °C was used to perform the reaction. Two mL of combination buffer at varying pHs was introduced to the vessel followed by other reaction components at the following final concentrations: Cu(II) as copper sulfate (5 μM), catalase (26,000 units), ascorbate (10 mM) and dansyl-YVG (variable concentrations between 5 and 400 μM). The reagents were incubated for at least 2 minutes to ensure thorough mixing. The reaction was initiated by addition of enzyme (0.05 - 0.80 μM). In a typical experiment, depending on the pH, 300 μL aliquots were removed every 30 s and placed in a 1.5 mL microcentrifuge tube containing 20 μL of 20% TFA to quench the reaction. Reactions at very low or very high pHs required longer sampling times due to slower reaction rates. Aliquots were then analyzed by HPLC to separate substrate and product and the concentration of substrate remaining at each time point was determined by reference to a standard curve. Simulation of the rate data was performed by non-linear regression, fitting the data to a model which assumes up to four enzyme species connected by deprotonation equilibria. Full details of the derivation and simulation of this model has been described in a previous publication (24).
O2-electrode Activity Assays
Measurement of the rate of oxygen consumption was performed using a Rank Brother’s Oxygen Electrode at 37°C. Each reaction was performed in a total volume of 2 mL in the presence of catalase (amount determined empirically so as to suppress the rate of non-enzymatic oxygen consumption), ascorbate (10 μM), and Cu2+ as copper sulfate (5 μM); using 5, 10, 15, or 20 μL of an 10 μM PHM solution. Once reagents were added, the electrode was capped, and the reaction was initiated by addition of substrate to a maximum concentration of 400 μM using a Hamilton Syringe.
Sample Preparation for XAS
All solutions were first made anaerobic, and all manipulations were carried out in the anaerobic chamber. Samples of the reduced WT enzyme were prepared in a single step by dilution of a 20 μL volume of 2 mM oxidized PHM [4 mM in Cu(II), 20 mM sodium phosphate (pH 8.0)], with 80 μL of the appropriate chloride-free combination buffer containing 5 mM ascorbate and 25% ethylene glycol. In one series of experiments to determine the effect of ascorbate on the EXAFS spectra, 1 mM ascorbate was used in place of 5mM. The sample was transferred to the XAS cuvette, and flash-frozen by rapid immersion in liquid nitrogen. Samples of the M314H variant were prepared in an identical manner.
XAS Data Collection and Analysis
Cu K edge (8980 eV) extended X-ray absorption fine structure (EXAFS) and X-ray absorption near edge structure (XANES) data were collected at the Stanford Synchrotron Radiation Lightsource at 3 GeV with currents between 100 and 80 mA on beam lines 7.3 and 9-3. The beam lines were configured with a liquid nitrogen cooled Si[220] monochromator and a Rh-coated mirror upstream of the monochromator with a 13 keV energy cutoff to reject harmonics. On beam line 9.3 a second Rh mirror downstream of the monochromator was used to focus the beam. Data were collected in fluorescence mode using a high-count-rate Canberra 30-element Ge array detector with maximum count rates below 120 kHz. A Z-1 metal (nickel) oxide filter and Soller slit assembly were placed in front of the detector to reduce the elastic scatter peak. Six to nine scans of a sample containing only sample buffer were collected, averaged, and subtracted from the averaged data for the protein samples to remove Z-1 Kβ fluorescence and produce a flat pre-edge baseline. The samples (80 μL) were measured as aqueous glasses in 25 % ethylene glycol at 15 K. Energy calibration was achieved by reference to the first inflection point of a copper foil (8980.3 eV) placed between the second and third ionization chamber. Data reduction and background subtraction were performed with the program modules of EXAFSPAK (27). Data from each detector channel were inspected for glitches or drop-outs before inclusion in the final average. Spectral simulation was carried out with EXCURVE 9.2 (28-31), as previously described (23). This allowed for inclusion of multiple scattering pathways between the metal center and the atoms of imidazole rings of the histidine residues. The experimental threshold energy, E0, was chosen as 8985 eV. The structural parameters refined during the fitting process included coordination numbers (N), bond distances (R) and Debye-Waller factors (2σ2), which result from both thermal motion and static disorder of the absorber-scatter pairs. The nonstructural parameter,ΔE0 equivalent to a small correction to the threshold energy, was also allowed to vary but was restricted to a common value for every component in a given fit.
Results
pH dependence of the WT protein
In previous work we showed that in a mixed buffer system comprising 50 mM each of acetate, HEPES, and CHES, the maximum PHM activity occurred at pH 7.0. The activity profile could be simulated by a model which assumed interconversion of an inactive protonated form and an active deprotonated form with a pKA of 5.8. The interconversion exactly tracked a structural transition observable by EXAFS, in which a Cu-S interaction assigned to Cu(I)-S(Met) decreased in intensity over a pH range indicative of an identical pKA. This Cu-S interaction was tentatively assigned to the catalytically essential M314 ligand to CuM, where the decrease in intensity was interpreted by a change from a rigid (inactive) species with a small Debye-Waller factor, to a highly flexible (active) species, and it was suggested that the highly flexible or fluxional Cu-S(Met) interaction was important in modulating the dynamics of H-atom tunneling during the H-atom abstraction step of the mechanism. However, it was noted that the EXAFS data could also be interpreted by the coordination of a second Cu-S(Met) species at low pH, although no clues as to the identity of this second inactive species could be inferred. Interpretation of the data was further complicated by the fact that acetate buffer both inhibited the enzyme, and shifted the pH maximum to 7.0 from its usual position of 5.8. When the experiment was performed in MES-HEPES-CHES buffer mix, the pH maximum shifted back to its native value, but the buffering range of MES only permitted data to be collected to pH 5.0. This was sufficient to show that the pKA for the Cu-S transition was also downshifted, but insufficient to show that it fully tracked the catalytic activity under native conditions. Therefore we developed a new buffer system in which formate was substituted for acetate, which activated rather than inhibited the enzyme, and retained the native pH optimum of 5.8. Using this system we measured the EXAFS spectra of the ascorbate-reduced enzyme and compared the pH dependence of the Cu-S “on” to “off” transition with the pH-activity profile.
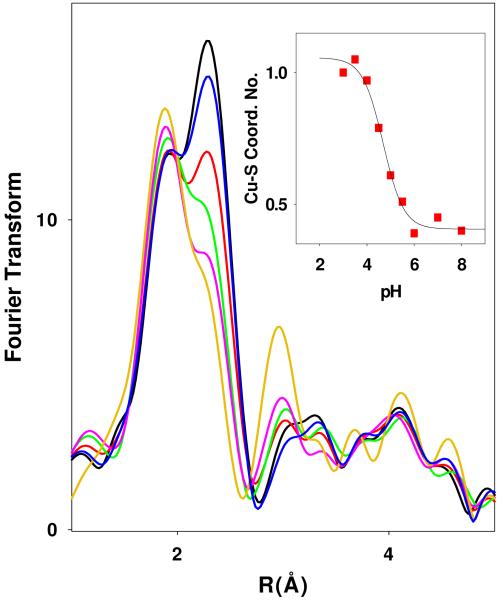
Figure 2 shows the pH dependence of the Fourier transforms from pH 3 to 8 where it can be seen that the intense feature due to Cu-S scattering around 2.25 Å at low pH decreases to a shoulder between pH 5 and 7, and thereafter remains constant. These EXAFS data were fitted using (a) a previously validated structural model involving a mixed Cu-His and Cu-S coordination environment, and (b) a simpler model involving a single shell of Cu-His and Cu-S scattering. At pH 7, the best fit to the data using a split shell model required 0.5 Cu-S at 2.24 Å, 1 Cu-N(His) at 1.85 Å and 1 Cu-N(His) at 1.96 Å which reflected a chemical model in which the two active site Cu centers were refined as separate shells (least squares fitting parameter F= 0.361). This analysis has been discussed in detail in previous communications, and is interpreted in terms of a 3-coordinate (His)2S(Met) M-site with RCu-His=1.96 Å, and a 2-coordinate (His)2 H-site with RCu-His = 1.85 Å. However, although the split shell model generally gives better fits as measured by the value of the fitting parameter F, it is probably not justified by the resolution of the data (k=2-13), where the minimum significant splitting of shells ΔR = π/2Δk = 0.14 Å. Using a single shell of Cu-His scatterers, a comparable fit (F = 0.373) was obtained with 2.5 Cu-His at 1.95 Å, and 0.5 Cu-S at 2.24 Å. It may be argued that this fit is more representative of the reduced form of the native PHM since it accounts for all five of the His ligands, but the data does not easily distinguish between them. Therefore the simpler single Cu-His shell fit was used throughout the analysis.
Figure 2.
Fourier transforms of x-ray absorption data for WT PHMcc between pH 3 and pH 8. Spectra correspond to the pHs (from the bottom) 3.5, 4.0, 4.5, 5.0, 5.5, and 7.0. The inset shows the change in the shell occupancy of the Cu-S interaction (simulated using parameters listed in Table S1 of the supporting information) plotted as a function of pH, and simulated using a pKA=4.69.
Simulation of EXAFS and FT data at pH 3.5 show an increase in the shell occupancy of the Cu-S component from 0.5 to 1.0, and a slight lengthening of the Cu-S bond length from 2.24 to 2.27 Å. The Cu-N(His) shell increases from 1.92 to 1.95 Å. Simulations for all pHs are compared with experimental data in Figures S1 and S2 with metrical details for each pH listed in Table S1 of the Supporting Information. The inset to Figure 2 shows the pH dependence of the Cu-S intensity, where the increased intensity has been simulated by keeping the Debye-Waller term constant at 0.010 Å2 and refining the coordination number of the Cu-S shell. The latter increases from 0.45±0.1 at pH 8 to 1.0±0.1 at pH 3, following a titration curve with pKA = 4.69 ± 0.1.
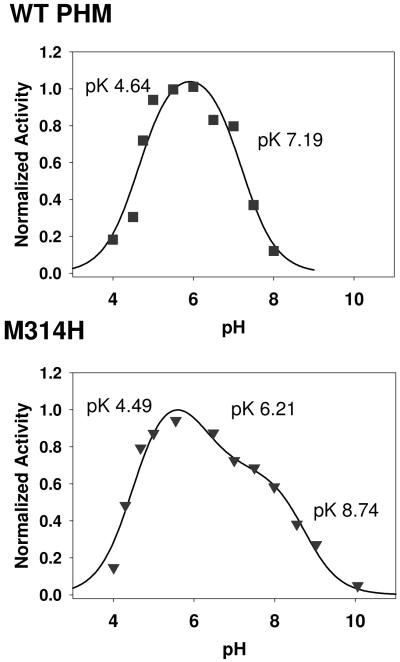
Figure 3(a) shows the pH-activity profile (under saturating conditions of dansyl-YVG, ascorbate and atmospheric oxygen) in a formate-MES-HEPES-CHES mixed-buffer system which provides buffering capacity between pH 2.5 to 10. The profile can be simulated by assuming a single active species which is formed by deprotonation of an inactive precursor in a step with a pKA of 4.64 ± 0.14 and is converted to an inactive state by a further deprotonation event with pKA of 7.20 ± 0.15. The pH of the activity maximum (5.8) is identical to that obtained in 100 mM MES with no additional components, the buffer commonly used for measuring PHM activity. The pKA of the activation phase is identical within experimental error to that obtained from EXAFS for Cu-S structural change.
Figure 3.
Normalized activity versus pH profiles for (a) the WT and (b) M314H variant of PHM. Red squares are experimental data, black lines are simulated data. Activity data were measured using an O2 electrode under saturating conditions of dansyl-YVG, ascorbate and atmospheric oxygen. Activity data were normalized for ease of comparison. The data were analyzed as described previously in reference (24), in terms of an inactive species S1 present at low pH which is transformed into the active state S2 (or active states S2 and S3 in the M314H variant) by deprotonantion steps with pKA values as shown.
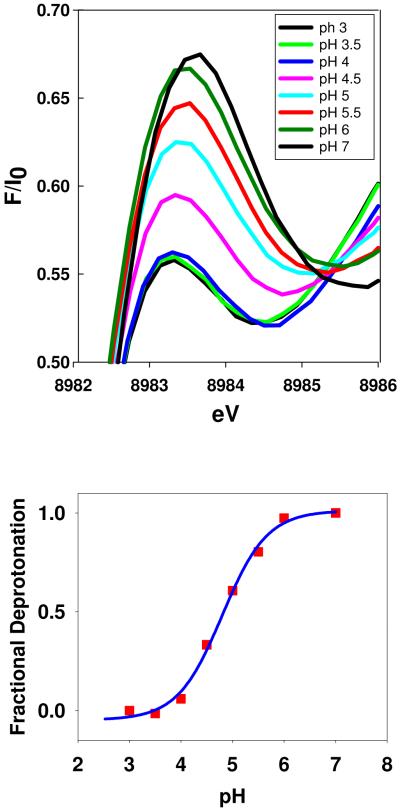
The absorption edge of the WT PHM shows a feature at 8983 eV assigned to a 1s → 4p transition, the intensity of which can often be used as an indicator of coordination number. Two-coordinate complexes exhibit intense features associated with allowed 1s → 4px,y transitions, whereas the intensity is attenuated in 3-coordinate systems due to mixing of s character into the excited state. Cu-S coordination also has the effect of dropping the intensity due to increased covalency and consequent mixing of metal and ligand orbitals, such that the excited state no longer has pure p-character. Inspection of Fig. 4(a) shows that the intensity of the 8983 eV peak decreases with decreasing pH suggesting that the formation of the low-pH species is accompanied by an increase in the average coordination number of the two copper centers. Fig 4(b) shows a fit to the 8983 eV intensity data assuming that the changes are driven by a single deprotonation event. The calculated pKA is 4.8 ± 0.1 which again matches the pKAs of both the Cu-S structural transition and the enzyme activation. We conclude that the structural transition from high to low pH is accompanied by an increase in average coordination number at one or both Cu centers, or alternatively/additionally involves an increase in covalency. The changes observed on the absorption edge are thus fully consistent with the conclusions from EXAFS analysis that a new species is formed at low pH with an increased Cu-S interaction and/or increased Cu-S coordination number. The fact that this new species tracks the activation of the enzyme both under native conditions (this work) and in acetate when the activity profile is shifted by 1.2 units to higher pH (24), provides strong evidence that it is responsible for catalytic activation of the enzyme.
Figure 4.
Top: pH dependence of the 1s → 4p transition on the absorption edge of WT PHMcc. Bottom: the fractional change in 8983 eV intensity (red squares) fitted to a pH titration curve with pKA = 4.8.
The S-donor residue responsible for the Cu-S “on” to “off” transition is not immediately obvious but two possibilities exist. First, as suggested previously, the catalytically essential M-site ligand M314 could exist in two distinct conformations: an inactive strongly coordinated and rigidly bound “on” state which would result in an intense Cu-S interaction with a small DW factor, and an active flexible or fluxional “off” state with correspondingly weaker bonding and higher DW. Second, at low pH a structural transition initiated by protonation of protein side chain could cause a second S donor ligand to coordinate at one of the copper centers. To probe these options, we constructed a PHM variant in which M314 was mutated to histidine. If the Cu-S transition originates from “on” to “off” states of M314, the prediction is that it should be eliminated in the M314H variant. Further, the pH dependence of M314H should be altered at low pH.
Properties of the M314H variant
The activity of the M314H variant was measured either by following oxygen consumption using the O2 electrode, or by dansyl-YVG substrate depletion using HPLC. Table S2 compares K’M for dansyl-YVG, k’cat, and k’cat/K’M for WT and the M314H variant where K’ and k’cat represent apparent kinetic constants measure under saturating ascorbate and atmospheric oxygen concentrations. Figure S3 compares the dansyl-YVG-dependence of the steady-state rate for WT and M314H respectively. Replacing M314 by His resulted in a decrease in k’cat and an increase in K’M equivalent to a 96 percent decrease in the apparent k’cat/K’M. However, activity was not eliminated in the variant indicating that the Met ligand was important but not essential for catalysis. Of further interest, although the catalytic rate was much reduced, the reaction remained 100 percent coupled as judged by stoichiometric conversion of oxygen and substrate into hydroxylated product. Fig. 3(b) shows the normalized pH-activity profiles for the M314H variant (also measured under saturating concentrations of dansyl-YVG, ascorbate and atmospheric oxygen) fitted to a model which assumed a number of states interconverting by proton dissociation steps. The profile differs from the WT in that at least four states appear to be present. At low pH the variant is in an inactive state S1 with converts to active state S2 with a pKA of 4.49 ± 0.13, closely similar to that of the WT protein. Thereafter, a further deprotonation (pKA 6.21 ± 0.5) generates state S3 which also exhibits measurable activity, and is further converted into an inactive form S4 with pKA = 8.74 ± 0.20. It should be noted that since the total activity of S3 is much lower than WT, it could be present yet undetectable in the WT profile. Although differences exist in the high pH range, we can conclude that the formation of the active S2 state appears to follow a similar pH profile as the WT.
Since Met 314 is coordinated in an axial position and is undetectable by EXAFS in the oxidized form of the WT protein, the prediction is that substitution of Met by His should have no affect on the EXAFS spectrum of oxidized M314H. This prediction was borne out by the finding that the EXAFS spectrum of oxidized M314H could be simulated using an identical parameter set as for the WT protein. These parameters are given for data collected at pH 7 in Table 1. The spectrum (Fig S4) is well simulated by four low Z scatterers at 1.96. In the WT protein we showed previously that the fit was consistent with an average of 2.5 His ligands per Cu, with additional 1.5 non-His (solvent) ligands completing the expected 4-coordination in the equatorial plane (23). For M314H there is the additional possibility that His314 might replace a water/hydroxide ligand in the equatorial plane, and M314H oxidized fit B involving three His ligands at each copper (Table 1) shows that such a fit is only slightly worse as judged by the value of the goodness of fit parameter F. The fit could be improved (~20%) by a model in which the additional His ligand was bound at a longer (axial) distance suggesting that His 314 coordinates similar to its Met homologue. We also used XAS to investigate the coordinate structure of the Cu center the reduced M314H variant at pH 7 and pH 3.5. We explored fits that employed either all low-Z scatterers (Cu-N(His) + additional O/N shells), or that also included heavier (Cu-S/Cl) scatterers. Figure 5(a) shows the EXAFS data for pH 7.0, which is best simulated with low-Z scatterers and no S/Cl component. The data fit best to 2.5 Cu-His at 1.91 Å, and is somewhat improved (14%) by inclusion of an addition 0.5 low Z scatterer at 2.5 Å which would be consistent with the coordination of a third His ligand at CuM. However, the correlation between DW factors and coordination numbers generally prohibits estimation of shell occupancy to better than ~25 percent, so that it is difficult to detect the presence of one additional His ligand at CuM in either the oxidized or reduced forms.
Table 1.
Fits obtained to the EXAFS of the M314H variant of peptidylglycine monooxygenase
| Fa | Nob | R (Å)c |
DW (Å2) |
Nob | R (Å)c |
DW (Å2) |
Nob | R (Å)c |
DW (Å2) |
−E0 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Oxidized Proteins | |||||||||||
|
| |||||||||||
| Cu-N(His) d | Cu-O/N e | Cu-S | |||||||||
| pH 7 fit A | 0.403 | 2.5 | 1.96 | 0.015 | 1.5 | 1.96 e | 0.015 | 5.20 | |||
| fit B | 0.478 | 3 | 1.97 | 0.014 | 1 | 1.96e | 0.014 | 4.56 | |||
| fit C | 0.332 | 2.5 | 1.96 | 0.015 | 1.5 | 1.96e | 0.015 | ||||
| 0.5 | 2.33 | 0.006 | 4.98 | ||||||||
| Reduced Proteins | |||||||||||
| Cu-N(His1) d | Cu-O/N | Cu-S | |||||||||
| pH 7 fit A | 0.580 | 2.5 | 1.91 | 0.018 | 0.5 | 2.49 | 0.016 | 0.79 | |||
| pH 7 fit B | 0.664 | 2.5 | 1.90 | 0.018 | 0.57 | ||||||
| pH 3.5 | 0.543 | 2.0 | 1.90 | 0.017 | 0.5 | 2.25 | 0.007 | 0.92 | |||
F is a least-squares fitting parameter defined as
Coordination numbers are generally considered accurate to ± 25%
In any one fit, the statistical error in bond-lengths is ±0.005 Å. However, when errors due to imperfect background subtraction, phase-shift calculations, and noise in the data are compounded, the actual error is probably closer to ±0.02 Å.
Fits modeled histidine coordination by an imidazole ring, which included single and multiple scattering contributions from the second shell (C2/C5) and third shell (C3/N4) atoms respectively. The Cu-N-Cx angles were as follows: Cu-N-C2 126°, Cu-N-C3 −126°, Cu-N-N4 163°, Cu-N-C5 −163°.
First-shell distances of the Cu-N(His) and Cu-N/O (non-His) shells were constrained to be equal in fits to the oxidized proteins.
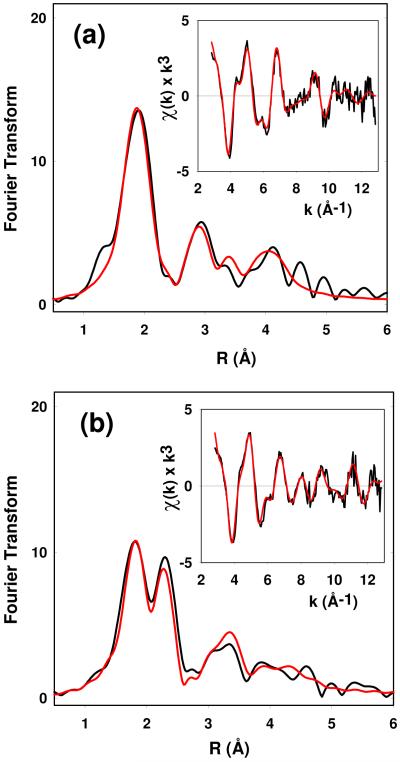
Figure 5.
Experimental and simulated Fourier transforms and EXAFS (insets) for the reduced forms of the M314H variants (a) at pH 7 and (b) at pH 3.5. Parameters used in the fits are listed in Table 1.
The EXAFS of M314H at pH 3.5 is shown in Fig 5(b) with parameters listed in Table 1. The data shows a similar pattern to that observed for the WT protein, viz strong scattering amplitude at ~2.2 – 2.3 Å which can be simulated by 0.5 Cu-S/Cl at 2.25 Å with a DW factor of 0.007 Å2. This result establishes that this Cu-S interaction does not arises from a “met-on” configuration of the active site M314 ligand, and must arise from coordination of a different S-donor ligand at low pH.
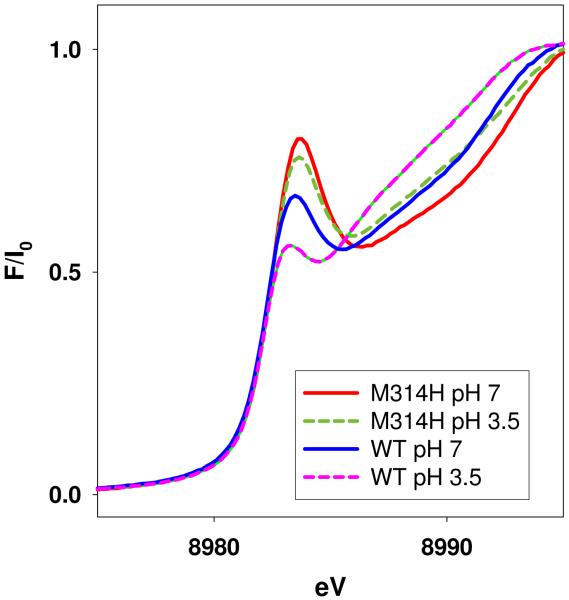
Absorption edges of the reduced M314H samples show 8983eV peaks which are more intense than those of the WT at both high and low pHs (Figure 6). This supports the conclusion that His314 is probably dissociated from CuM.
Figure 6.
Comparison of the 1s → 4p absorption edge transition for WT and M314H PHMcc at pH 7 and 3.5.
The low-pH Cu-S/Cl scatterering could arise from coordination of S from cysteine or methionine, or could be due to coordination of exogenous Cl. The latter possibility is unlikely since (a) the protein was isolated under conditions where exogenous chloride was minimized, and (b) it is chemically unreasonable since HCl is a strong acid, and it is hard to envisage why Cl binding should be strongly influenced by deprotonation of a protein side-chain. Cu-S(cys) coordination is also unlikely since all ten Cys residues in PHMcc form disulfide cross-links. Notwithstanding, we considered the possibility that in the presence of 5 mM ascorbate a disulfide crosslink might be reduced to a bis thiol. C315 is an obvious candidate since it anchors the β-sheet which carries the M314 residue as well as the peptide substrate binding N316 and Y318 (9-12). To probe Cu-S(Cys) coordination, we compared the low pH forms of a 1 mM sample of WT PHMcc reduced with 5 and 1 mM equivalents of ascorbate and obtained identical EXAFS spectra with a fully coordinated Cu-S species (Fig. S5). Since all the reducing equivalents in the 1 mM ascorbate sample were required to reduce the two copper centers to Cu(I), we conclude that in this sample all disulfide crosslinks must be intact. Therefore the low-pH S-donor must be a methionine residue.
Discussion
We have shown that catalytic inactivation of PHM in the pH range 8-3 is accompanied by a structural change between two states of the protein, an active state with a single, weakly bound Cu-S present at higher pH and an inactive state involving a significant increase in the coordination number or strength of the Cu-S interaction at lower pHs. We first reported this effect in an earlier paper where the activity was measured in acetate buffer which shifted the activity maximum from its native value of pH 5.8 to 7, but we were unable to fully determine the titration profile under native conditions due to inaccessabilty of pHs below 5 in MES buffer. In the present paper we have extended the pH range downwards using a formate buffer system, and have been able to show that the Cu-S “off-to-on” structural transition still accurately tracks the inactivation of the enzyme in the pH 8 -3 range. This extends our earlier study into the pH range relevant to the physiological state of the enzyme, and confirms that the “off” state is the catalytically active form. The Cu-S interaction present in the ‘off” state has been assigned to the Cu-S(Met314) (7, 23) where M314 is a ligand to the catalytic M-site and is characterized by a larger than usual Debye Waller factor relative to other Cu(I)-S interactions in Cu-Metx or Cu-HisMet environments (32-36), suggesting a highly dynamic or fluxional environment. Earlier, we suggested that such dynamics might couple into vibrational modes of the catalytic site which were critical for H-tunneling in H-atom abstraction from the substrate, and further that the “on” state might represent more rigid dynamics for Cu-Met314 resulting in loss of coupling into these critical tunneling modes(24). However, two lines of evidence presented in the earlier study cast doubt on whether the ‘on” state was the result of a local conformational effect at the M-site, or arose from a more global conformational effect, perhaps involving changes at the H-site. Specifically, photoreduction in the frozen state at low pH resulted in loss of solvent coordination but failed to induce the “on” state, while the KD for peptide substrate binding close to the M-site was independent of pH. In the present study we tested these predictions in a variant in which the catalytic M314 residue was mutated to histidine and demonstrated that the “off-to-on” transition was still observed in the absence of Met 314. This has led to an evolution of the model in which we conclude that the origin of the strong Cu-S interaction at low pH cannot be a more tightly bound form of the M314 residue, but must arise from an additional Cu-S which binds at one of the two copper centers in the low pH inactive form, but dissociates at higher pH to form the catalytically active species.
In other work (3) we have shown that a similar Cu-S on to off transition occurs in the homologous enzyme TBM, suggesting a functional role for the transition in setting the pH optimum of these enzymes around 5.5. This may be physiologically important since PHM, DBM and TBM are each localized to secretory granules whose internal pH coincides with the pH optimum of the enzymes (37). Sequence comparison of PHM, DBM, TBM and MOX reveal four conserved cysteines and two conserved methionines. All four of the conserved Cys residues occur as disulfide crosslinks in PHM, and in order to coordinate to Cu(I) would have to be reduced to thiols. It is possible that ascorbate at ~5 mM (the concentration used to reduce the copper centers) might also reduce one or more of these disulfide crosslinks, releasing a free thiol. For example reduction of the crosslink between Cys315 and Cys293 would place a reduced thiol adjacent to the M-site Met ligand M314. With this in mind we compared EXAFS data of the low-pH ascorbate-reduced WT enzyme produced with 5-fold versus equimolar concentrations of ascorbate. The mechanism of reduction by ascorbate has been shown in DBM to proceed by 1-electron redox to form the monodehydroascobate radical followed by disproportionation to form dehydroascorbate and regenerated ascorbate products (38). Thus one equivalent of ascorbate is sufficient to reduce both copper centers but is insufficient to reduce additional disulfides. The experimental finding that the Cu-S intensity was identical in five-fold and one-fold ascorbate reduction experiments therefore allows us to rule out cysteine as the origin of the sulfur donor.
PHM, TBM and DBM have two conserved Met residues, M314 and M109 (PHM numbering). M314 is the catalytic Met residue and we have shown in this study that its mutation to histidine does not affect the intensity of the Cu-S feature at low pH. This suggests that M109 may be the S-donor. In MOX the residue is Ile, and interestingly, although the function is unknown, the protein is retained in the ER and does not appear to enter the secretory pathway. Consequently it would have no need to adjust its pH optimum to 5 via a His to Met transition (39). M109 is close to the H site (Figure 7) but extends away from the copper center on the same β-strand that holds the coordinated H108 and H107 ligands. Protonation of either H107 or H108 might induce rearrangement of this strand so as to coordinate the M109 thioether. However, of the additional 14 Met residues (excluding M314 and M109) identified in the PHMcc structure two (M116 and M82) are in the vicinity of the H-site and one (M208) is in the vicinity of the M-site. Therefore, while the M109 assignment is supported by available data, it must be considered as tentative at this time.
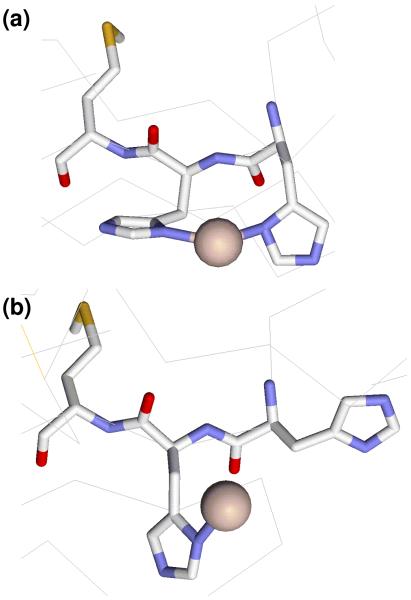
Figure 7.
Structural depiction of two different conformations of the H center in PHMcc showing differences in the orientation of H107, H108 and M109 residues. The H172 ligand has been omitted for clarity. (a) Conformation found in WT PHMcc (pdb 1OPM) with each His ligand coordinated by imidazole-Nδ; (b) conformation observed in the M314I variant (pdb 1YI9) with H107 dissociated.
There is precedent for different conformational states at the H center. A crystal structure of the M314I variant showed an altered conformation, with H107 dissociated from the copper (11). A comparison of this structure with that of the WT oxidized protein is shown in Fig. 7. The WT structure indicates rather poor hybridization of the H107 Nδ donor atom, suggesting a strained conformation induced by the coordination of the contiguous H107 and H108 imidazole side chains which is relieved in the M314I structure by H107 dissociation. Protonation of H108 at low pH would be an alternative way to relieve this strain generating the more stable M109-X-H107 coordination.
The pKa for the catalytic transition is 4.6, within the range determined for protonation of histidine residues coordinated to Cu(I). Protonation and dissociation of a Cu(I)-bound imidazole residue occurs in reduced forms of cupredoxins (40, 41), and Cu/Zn superoxide dismutases (42-44). The simultaneous addition of a proton along with an electron keeps the overall charge at the active site constant, and in the case of cupredoxins has been ascribed to proton-coupled electron transfer. In cupredoxins, the C-terminal histidine ligand is contained within a short loop of structure located between the Cys and Met ligands, and variation of both the sequence and length of this loop have been shown to have profound effects on the value of the pKa for histidine protonation. For example replacement of the native loop of azurin (C112TFPGH117SALM, pKaH117 <2) with shorter loops from amicyanin or plastocyanin produces chimeras with pKAs for protonation of the corresponding histidine of 5.5 and 4.3 respectively. For plastocyanin, the pKAs of the native protein and the chimera in which the native loop (C84SPH87QGAGM92) is replaced with the azurin loop-sequence are 4.7 and 4.9 respectively (45). Taken together, the studies on cupredoxins have demonstrated sensitivity of Cu(I) coordinated histidines to loop length, identity of non-coordinated residues within the loop, and to second-shell effects such as solvation and H-bonding. Similar effects are likely to modulate the acidity of the CuH-coordinating histidine residues in PHM, and provide a reasonable rationale for the observed pKA value.
The apparent kinetic constants for the M314H variant indicate that the catalytic M314 has a complex role. The increase in K’M implies that reversible binding events that precede the first irreversible catalytic step are altered in the mutant while the decrease in k’cat implies additional effects on the irreversible catalytic step. EXAFS studies confirm the loss of the catalytic Cu-S interaction at CuM but provide no evidence either way for coordination of the substituted His residue. The additional deprotonation step observed at high pH may indicate minor mechanistic differences, perhaps due to structural perturbations in the mutant. However, it is clear from this work that catalysis does occur in the His mutant, and product formation remains tightly coupled to oxygen consumption, suggesting that the mechanism remains essentially the same, albeit with reduced catalytic efficiency. Therefore M314 serves to increase catalytic efficiency rather than providing a unique mechanistic role. Further studies on additional methionine variants aimed at establishing the coordination of M109 (and/or other Met residues) and the mechanistic role of M314 are ongoing in our laboratory.
Supplementary Material
Acknowledgement
We thank Drs Betty Eipper and Richard Mains for providing us with cell lines expressing the M314H variant.
Footnotes
Supporting Information available. Supporting information (five figures and two tables) is available free of charge via the Internet at http://pubs.acs.org.
Abbreviations. DBM, dopamine β-monooxygenase; DW, Debye-Waller factor; dansyl-YVG, dansyl-tyrosylvalylglycine; EXAFS, extended x-ray absorption fine structure; FT, Fourier transform; PHM, peptidylglycine monooxygenase; TBM, tyramine β-monooxygenase; XANES, x-ray absorption near edge structure; XAS, x-ray absorption spectroscopy.
References
- 1.Prigge ST, Mains RE, Eipper BA, Amzel LM. New insights into copper monooxygenases and peptide amidation: structure, mechanism and function. Cell. Mol. Life Sci. 2000;57:1236–1259. doi: 10.1007/PL00000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klinman JP. The copper-enzyme family of dopamine β -monooxygenase and peptidylglycine α -hydroxylating monooxygenase: Resolving the chemical pathway for substrate hydroxylation. J. Biol. Chem. 2006;281:3013–3016. doi: 10.1074/jbc.R500011200. [DOI] [PubMed] [Google Scholar]
- 3.Hess CR, Klinman JP, Blackburn NJ. The copper centers of tyramine beta-monooxygenase and its catalytic-site methionine variants: an X-ray absorption study. J. Biol. Inorg. Chem. 2010;15:1195–1207. doi: 10.1007/s00775-010-0677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hess CR, Wu Z, Ng A, Gray EE, McGuirl MA, Klinman JP. Hydroxylase activity of Met471Cys tyramine β -monooxygenase. J. Am. Chem. Soc. 2008;130:11939–11944. doi: 10.1021/ja800408h. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn NJ, Hasnain SS, Pettingill TM, Strange RW. Copper K-EXAFS studies of oxidized and reduced dopamine-β -hydroxylase: confirmation of a sulfur ligand to Cu(I) in the reduced enzyme. J. Biol. Chem. 1991;266:23120–23127. [PubMed] [Google Scholar]
- 6.Pettingill TM, Strange RW, Blackburn NJ. Carbonmonoxy dopamine-β -hydroxylase: structural characterization by FTIR, fluorescence and XAS spectroscopy. J. Biol. Chem. 1991;266:16996–17003. [PubMed] [Google Scholar]
- 7.Boswell JS, Reedy BJ, Kulathila R, Merkler DJ, Blackburn NJ. Structural investigations on the coordination environment of the active-site copper centers of recombinant bifunctional peptidylglycine α -amidating enzyme. Biochemistry. 1996;35:12241–12250. doi: 10.1021/bi960742y. [DOI] [PubMed] [Google Scholar]
- 8.Chen P, Bell J, Eipper BA, Solomon EI. Oxygen activation by the noncoupled binuclear copper site in peptidylglycine α -hydroxylating monooxygenase. Spectroscopic definition of the resting sites and the putative CuIIM-OOH intermediate. Biochemistry. 2004;43:5735–5747. doi: 10.1021/bi0362830. [DOI] [PubMed] [Google Scholar]
- 9.Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Amidation of bioactive peptides: the structure of peptidylglycine α -hydroxylating monooxygenase. Science. 1997;278:1300–1305. doi: 10.1126/science.278.5341.1300. [DOI] [PubMed] [Google Scholar]
- 10.Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Substrate-mediated electron transfer in peptidylglycine α -hydroxylating monooxygenase. Nature Struct. Biol. 1999;6:976–983. doi: 10.1038/13351. [DOI] [PubMed] [Google Scholar]
- 11.Siebert X, Eipper BA, Mains RE, Prigge ST, Blackburn NJ, Amzel LM. The catalytic copper of peptidylglycine alpha-hydroxylating monooxygenase also plays a critical structural role. Biophys. J. 2005;89:3312–3319. doi: 10.1529/biophysj.105.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chufan EE, Prigge ST, Siebert X, Eipper BA, Mains RE, Amzel LM. Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase. J. Am. Chem. Soc. 2010;132:15565–15572. doi: 10.1021/ja103117r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magnus KA, Hazes B, Ton-That H, Bonaventura C, Bonaventura J, Hol WGJ. Crystallographic analysis of oxygenated and deoxygenated states of arthropod hemocyanin shows unusual differences. Proteins Struct. Funct. Genet. 1994;19:302–309. doi: 10.1002/prot.340190405. [DOI] [PubMed] [Google Scholar]
- 14.Gerdemann C, Eicken C, Krebs B. The crystal structure of catechol oxidase: new insight into the function of type-3 copper proteins. Acc. Chem. Res. 2002;35:183–191. doi: 10.1021/ar990019a. [DOI] [PubMed] [Google Scholar]
- 15.Chen P, Solomon EI. O2 activation by binuclear Cu sites: noncoupled versus exchange coupled reaction mechanisms. Proc. Natl. Acad. Sci. U S A. 2004;101:13105–13110. doi: 10.1073/pnas.0402114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matoba Y, Kumagai T, Yamamoto A, Yoshitsu H, Sugiyama M. Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J. Biol. Chem. 2006;281:8981–8990. doi: 10.1074/jbc.M509785200. [DOI] [PubMed] [Google Scholar]
- 17.Lewis EA, Tolman WB. Reactivity of dioxygen-copper systems. Chem. Rev. 2004;104:1047–1076. doi: 10.1021/cr020633r. [DOI] [PubMed] [Google Scholar]
- 18.Evans JP, Ahn K, Klinman JP. Evidence that dioxygen and substrate activation are tightly coupled in dopamine β -monooxygenase: Implications for oxygen activation. J. Biol. Chem. 2003;278:49691–49698. doi: 10.1074/jbc.M300797200. [DOI] [PubMed] [Google Scholar]
- 19.Bauman AT, Yukl ET, Alkevich K, McCormack AL, Blackburn NJ. The hydrogen peroxide reactivity of peptidylglycine monooxygenase supports a Cu(II)-superoxo catalytic intermediate. J. Biol. Chem. 2006;281:4190–4198. doi: 10.1074/jbc.M511199200. [DOI] [PubMed] [Google Scholar]
- 20.Chen P, Solomon EI. Oxygen activation by the noncoupled binuclear copper site in peptidylglycine α -hydroxylating monooxygenase. Reaction mechanism and role of the noncoupled nature of the active site. J. Am. Chem. Soc. 2004;126:4991–5000. doi: 10.1021/ja031564g. [DOI] [PubMed] [Google Scholar]
- 21.Eipper BA, Quon ASW, Mains RE, Boswell JS, Blackburn NJ. The catalytic core of peptidylglycine α -hydroxylating monooxygenase: investigation by site-directed mutagenesis, Cu x-ray absorption spectroscopy, and electron paramagnetic resonance. Biochemistry. 1995;34:2857–2865. doi: 10.1021/bi00009a016. [DOI] [PubMed] [Google Scholar]
- 22.Kolhekar AS, Keutman HT, Mains RE, Quon ASW, Eipper BA. Peptidylglycine α -hydroxylating monooxygenase: active site residues, disulfide linkages, and a two-domain model of the catalytuc core. Biochemistry. 1997;36:10901–10909. doi: 10.1021/bi9708747. [DOI] [PubMed] [Google Scholar]
- 23.Blackburn NJ, Rhames FC, Ralle M, Jaron S. Major changes in copper coordination accompany reduction of peptidylglycine monooxygenase. J. Biol. Inorg. Chem. 2000;5:341–353. doi: 10.1007/pl00010663. [DOI] [PubMed] [Google Scholar]
- 24.Bauman AT, Jaron S, Yukl ET, Burchfiel JR, Blackburn N. pH dependence of peptidylglycine monooxygenase. Mechanistic implications for Cu-methionine binding dynamics. Biochemistry. 2006;45:11140–11150. doi: 10.1021/bi060905a. [DOI] [PubMed] [Google Scholar]
- 25.Jaron S, Blackburn NJ. Does superoxide channel between the copper centers in peptidylglycine monooxygenase? A new mechanism based on carbon monoxide reactivity. Biochemistry. 1999;38:15086–15096. doi: 10.1021/bi991341w. [DOI] [PubMed] [Google Scholar]
- 26.Bauman AT, Ralle M, Blackburn N. Large scale production of the copper enzyme peptidylglycine monooxygenase using an automated bioreactor. Protein Expression and Purification. 2007;51:34–38. doi: 10.1016/j.pep.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 27.George GN. Stanford Synchrotron Radiation Laboratory; Menlo Park, CA: 1990. [Google Scholar]
- 28.Binsted N, Gurman SJ, Campbell JW. Daresbury Laboratory; Warrington, England: 1998. [Google Scholar]
- 29.Gurman SJ, Binsted N, Ross I. A rapid, exact, curved-wave theory for EXAFS calculations. J. Phys. C. 1984;17:143–151. [Google Scholar]
- 30.Gurman SJ, Binsted N, Ross I. A rapid, exact, curved-wave theory for EXAFS calculations. II. The multiple-scattering contributions. J. Phys. C. 1986;19:1845–1861. [Google Scholar]
- 31.Binsted N, Hasnain SS. State of the art analysis of whole X-ray absorption spectra. J. Synchrotron Rad. 1996;3:185–196. doi: 10.1107/S0909049596005651. [DOI] [PubMed] [Google Scholar]
- 32.Loftin IR, Franke S, Blackburn NJ, McEvoy MM. Unusual Cu(I)/Ag(I) coordination of Escherichia coli CusF as revealed by atomic resolution crystallography and X-ray absorption spectroscopy. Protein Sci. 2007;16:2287–2293. doi: 10.1110/ps.073021307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loftin IR, Blackburn NJ, McEvoy MM. Tryptophan Cu(I)-pi interaction fine-tunes the metal binding properties of the bacterial metallochaperone CusF. J. Biol. Inorg. Chem. 2009;14:905–912. doi: 10.1007/s00775-009-0503-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagai I, Liu W, Rensing C, Blackburn NJ, McEvoy MM. Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J Biol Chem. 2007;282:35695–35702. doi: 10.1074/jbc.M703937200. [DOI] [PubMed] [Google Scholar]
- 35.Sarret G, Favier A, Coves J, Hazemann JL, Mergeay M, Bersch B. CopK from Cupriavidus metallidurans CH34 binds Cu(I) in a tetrathioether site: characterization by X-ray absorption and NMR spectroscopy. J Am Chem Soc. 2010;132:3770–3777. doi: 10.1021/ja9083896. [DOI] [PubMed] [Google Scholar]
- 36.Rubino JT, Riggs-Gelasco P, Franz KJ. Methionine motifs of copper transport proteins provide general and flexible thioether-only binding sites for Cu(I) and Ag(I) J. Biol. Inorg. Chem. 2010;15:1033–1049. doi: 10.1007/s00775-010-0663-9. [DOI] [PubMed] [Google Scholar]
- 37.Wu MM, Grabe M, Adams S, Tsien RY, Moore HP, Machen TE. Mechanisms of pH regulation in the regulated secretory pathway. J. Biol. Chem. 2001;276:33027–33035. doi: 10.1074/jbc.M103917200. [DOI] [PubMed] [Google Scholar]
- 38.Diliberto EJ, Jr., Allen PL. Mechanism of dopamine β-hydroxylation. Semidehydroascorbate as the enzyme oxidation product of ascorbate. J. Biol. Chem. 1981;256:3385–3393. [PubMed] [Google Scholar]
- 39.Xin X, Mains RE, Eipper BA. Monooxygenase X, a member of the copper-dependent monooxygenase family localized to the endoplasmic reticulum. J. Biol. Chem. 2004;279:48159–48167. doi: 10.1074/jbc.M407486200. [DOI] [PubMed] [Google Scholar]
- 40.Guss JM, Harrowell PR, Murata M, Norris VA, Freeman HC. Crystal structure analyses of reduced (CuI) poplar plastocyanin at six pH values. J Mol Biol. 1986;192:361–387. doi: 10.1016/0022-2836(86)90371-2. [DOI] [PubMed] [Google Scholar]
- 41.Li C, Sato K, Monari S, Salard I, Sola M, Banfield MJ, Dennison C. Metal-binding loop length Is a determinant of the pKa of a histidine ligand at a type 1 copper site. Inorg. Chem. 2011;50:482–488. doi: 10.1021/ic101413e. [DOI] [PubMed] [Google Scholar]
- 42.Bertini I, Luchinat C, Monnanni R. Evidence of the breaking of the copper-imidazolate bridge in copper/cobalt-substituted superoxide dismutase upon reduction of the Cu(II) centers. J. Am. Chem. Soc. 1985;107:2178–2179. [Google Scholar]
- 43.Blackburn NJ, Hasnain SS, Binsted N, Diakun GP, Garner CD, Knowles PF. An extended-x-ray-absorption-fine-structure study of bovine erythrocyte superoxide dismutase in aqueous solution. Direct evidence for three-coordinate Cu(I) in the reduced enzyme. Biochem. J. 1984;219:985–990. doi: 10.1042/bj2190985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banci L, Bertini I, Cramaro F, Del Conte R, Viezzoli MS. The solution structure of reduced dimeric copper zinc superoxide dismutase. The structural effects of dimerization. Eur J Biochem. 2002;269:1905–1915. doi: 10.1046/j.1432-1033.2002.02840.x. [DOI] [PubMed] [Google Scholar]
- 45.Li C, Banfield MJ, Dennison C. Engineering copper sites in proteins: loops confer native structures and properties to chimeric cupredoxins. J. Am. Chem. Soc. 2007;129:709–718. doi: 10.1021/ja0661562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.