SUMMARY
Firefly luciferase-catalyzed light emission from D-luciferin is widely used as a reporter of gene expression and enzymatic activity both in vitro and in vivo. Despite the power of bioluminescence for imaging and drug discovery, light emission from firefly luciferase is fundamentally limited by the physical properties of the D-luciferin substrate. We and others have synthesized aminoluciferin analogs that exhibit light emission at longer wavelengths than D-luciferin and have increased affinity for luciferase. However, although these substrates can emit an intense initial burst of light that approaches that of D-luciferin, this is followed by much lower levels of sustained light output. We have previously postulated that this behavior is due to product inhibition. Here we describe the creation of mutant luciferases that yield improved sustained light emission with aminoluciferins in both lysed and live mammalian cells, allowing the use of aminoluciferins for cell-based bioluminescence experiments.
INTRODUCTION
Light emission from firefly luciferase is fundamentally limited by its access to D-luciferin and the inherent photophysical properties of the D-luciferin substrate (Figure 1A) (Reddy et al., 2010). Replacement of the 6’-hydroxyl group of D-luciferin with a 6’-amino group results in red-shifted light emission (White et al., 1966) and higher affinity for luciferase, but lower maximal light emission and lower cell-permeability (Shinde et al., 2006). Although D-luciferin is the superior substrate for maximal light emission under most conditions, the unique chemistry of 6’-aminoluciferin has expanded the scope of luciferase applications. For example, the liberation of 6’-aminoluciferin from “dark” pro-luciferin protease substrates has been exploited to allow the coupled bioluminescent detection of protease activity, both in vitro (Monsees et al., 1994; Moravec et al., 2009) and in vivo (Shah et al., 2005; Dragulescu-Andrasi et al., 2009; Hickson et al., 2010; Scabini et al., 2011).
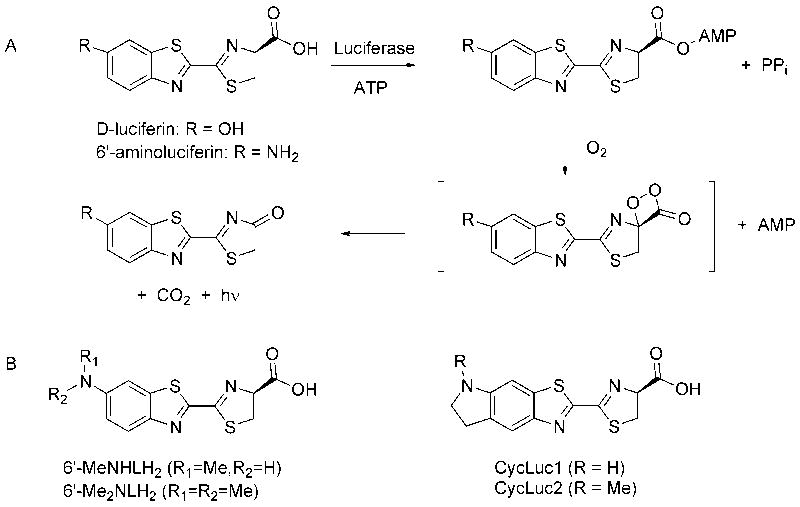
Figure 1. Luciferase mechanism and substrates.
(A) Firefly luciferase catalyzes the formation of an activated AMP ester of its native substrate, D-luciferin. Subsequent oxidation within the luciferase binding pocket generates an excited state oxyluciferin molecule that is responsible for light emission. (B) Synthetic alkylated aminoluciferin substrates used in this study.
Recently, we and others have reported that 6’-alkylaminoluciferins can also be substrates for luciferase (Woodroofe et al., 2008; Reddy et al., 2010; Takakura et al., 2010). These substrates generally have even higher affinity for luciferase than 6’-aminoluciferin, and emit light at even longer wavelengths. Many modifications are tolerated, including long-chain 6’-alkylaminoluciferins, 5’,6’-cyclic alkylaminoluciferins, and even dialkylaminoluciferins (Figure 1B). Synthetic modulation of the properties of these molecules thus presents an opportunity to develop new bioluminescent probes and to optimize luciferase light output for different applications. However, with wild-type Photinus pyralis firefly luciferase, most of these substrates give a rapid burst of light followed by weak sustained emission (Reddy et al., 2010).
The detergent-stable proprietary mutant luciferase Ultra-Glo (Promega) is capable of high sustained light emission with aminoluciferin substrates, particularly in combination with the P450-Glo buffer (Woodroofe et al., 2008; Reddy et al., 2010). The use of aminoluciferins with this luciferase and buffer therefore has potential for novel in vitro screening applications, such as the coupled detection of enzymatic activity (Fan and Wood, 2007). However, Ultra-Glo is a proprietary luciferase reagent that is not available as a genetic construct that can be expressed in cells. Furthermore, the detergent stability of Ultra-Glo and the use of the P450-Glo buffer are important for the light emission behavior. Cellular and in vivo applications such as the detection of gene expression (de Wet et al., 1987) and bioluminescent imaging (Prescher and Contag, 2010) necessitate a genetically-encodable luciferase that is capable of efficient utilization of aminoluciferins under physiological buffer conditions.
We postulated that aminoluciferin substrates possess all of the key photophysical and structural features necessary to give rise to bioluminescence, but are primarily limited as luciferase substrates due to product inhibition (Reddy et al., 2010). Here we report the construction of mutant luciferases capable of sustained and selective light emission with aminoluciferins, thereby expanding the available tools for bioluminescence imaging.
RESULTS
Rational mutation of luciferase
Our initial efforts to improve luciferase light emission with alkylaminoluciferin substrates focused on rational mutation of phenylalanine 247 in the luciferin binding pocket. This residue is involved in a π-stacking interaction with D-luciferin (Branchini et al., 2003; Nakatsu et al., 2006). Mutation of this residue to leucine and alanine has been previously reported by Branchini et al., who found that F247L lowers the affinity for D-luciferin but does not impair catalysis, while F247A is severely impaired in both Km and Vmax (Branchini et al., 2003). We therefore anticipated that F247L would maintain catalytic function but allow improved product dissociation, helping to relieve product inhibition and allowing improvement in the sustained light emission from alkylaminoluciferin substrates.
Surprisingly, we found that the F247L mutation improves maximal sustained light emission from 6’-aminoluciferin by 4.9-fold but has only a small positive effect on light emission from CycLuc1 (Figure 2; Figure S1). Instead, we found that the F247A mutation, which has a marked negative effect on light output from both D-luciferin and 6’-aminoluciferin, gave dramatically improved sustained light output from CycLuc1 (Figure 2; Figure S1). The improved light output of this mutant comes at the cost of a substantially increased Km value. While this is not a concern under conditions where saturating concentrations of substrate can be applied (e.g., in vitro), a high Km is expected to limit light output in live cells (Figure S1C), where only low intracellular concentrations of luciferin substrates are achieved (de Wet et al., 1987; Craig et al., 1991; Shinde et al., 2006). We therefore sought alternate sites for mutation that could potentially increase light output yet retain a low Km.
Figure 2. Characterization of phenylalanine 247 mutants.

Dose-response curves for purified luciferases (WT, F247L, F247A, F247S, and F247V) were generated with D-Luciferin, 6’-NH2LH2 and CycLuc1 at concentrations from 0.122-125 μM. The assays were performed in triplicate and are represented as the mean +/- SEM. Note that the emission scale for D-luciferin is 4-fold higher than 6’-NH2LH2 and CycLuc1. Comparison of each mutant in lysed and live CHO cells is detailed in Figure S1.
Screening for Mutant Luciferases with Improved Utilization of Aminoluciferins
Because mutation of phenylalanine 247 was found to affect substrate utilization, but not in an easily predictable manner, we opted to perform saturating mutagenesis at this and other active site residues, and then screen for mutant luciferases with the desired properties. This approach has the advantage of rational site selection, but is not restricted by preconceived notions of suitable replacement residues. Based on the crystal structure of Luciola cruciata luciferase (Nakatsu et al., 2006), we identified a number of amino acids that interact with D-luciferin, ATP, or play a role in creating the overall local structure of the binding pocket (Figure 3A; Table S1). In addition to phenylalanine 247, we selected arginine 218, threonine 251, leucine 286, tyrosine 340, and serine 347 for saturating mutagenesis. Arginine 218 is proximal to the 4’- and 5’-carbons of D-luciferin and forms part of the van der Waals surface that encompasses the luciferin substrate (Nakatsu et al., 2006). Mutations at this site were expected to provide increased “wiggle room” for substrates, particularly those with 5’-modifications (i.e., CycLuc1 and CycLuc2). Leucine 286 is a candidate residue for interaction with alkyl sidechains on aminoluciferin substrates, while tyrosine 340 forms part of the ATP site and is located at the interface between the ATP and luciferin binding pockets. The methyl group of the threonine 251 sidechain makes a van der Waals interaction with the benzothiazole of the luciferin substrate, while serine 347 forms a hydrogen bond to the benzothiazole nitrogen through the intermediacy of a water molecule (Nakatsu et al., 2006). We reasoned that mutation at these sites could potentially lead to improved continuous light emission from aminoluciferins by altering substrate alignment and/or improving product dissociation. Because aminoluciferins have higher affinity for luciferase, the loss of interactions with the benzothiazole could also confer selectivity over D-luciferin, particularly if the affinity and proper orientation of alkylaminoluciferins can be maintained by ancillary interactions that are not available to D-luciferin.
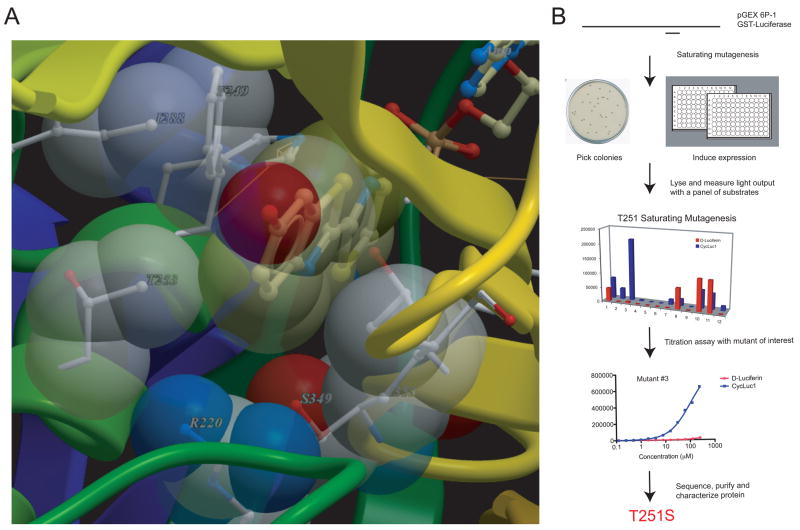
Figure 3. Creation of mutant luciferases.
(A) Mutation sites were selected based on proximity to the luciferin substrate in the crystal structure of Luciola cruciata luciferase (PDB 2D1R). (B) Selected residues were subjected to saturating mutagenesis. Mutant luciferase-expressing bacteria were screened for light emission with CycLuc1 and those that exhibited improved properties were sequenced (Table S1) and the mutant protein purified for further characterization.
At each selected site, we used the degenerate codon NNK to perform saturating site-directed mutagenesis in the wild-type Photinus pyralis luciferase (Figure 3B). At some sites we also introduced alanine point mutations (see Methods). After cloning of the mutant luciferases into bacteria, colonies were picked and grown in 96-well plates (Figure 3B). Luciferase expression was induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) and the bacteria were lysed. Light emission from each lysate was measured at a luciferin concentration of 25 μM. For those lysates that gave improved and/or selective light output with CycLuc1, the plasmid encoding the mutant luciferase was sequenced (Table S1). Mutation at some sites failed to produce any obvious improvement in luciferase performance. For example, mutation of tyrosine 340 in the nucleotide binding pocket primarily yielded inactive luciferases. In hindsight, Y340 makes a hydrogen-bonding interaction with D420 that may be critical for luciferase function (Nakatsu et al., 2006). A secondary assay was also performed on the bacterial extracts to measure light output as a function of CycLuc1 concentration (Figure 3B). Mutants that demonstrated improved light output, low Km, and/or selectivity for CycLuc1 in this assay were expressed as recombinant proteins and purified for further characterization.
Characterization of Mutant Luciferases In Vitro
Screening for mutants of phenylalanine 247 identified F247S and F247V as proteins with greatly improved light output from CycLuc1, but still with relatively high Km values (Figure 2). Mutations at several of the other chosen sites (R218K, T251S, L286M, S347T, and S347A) improved light emission with aminoluciferins, while retaining Km values below that of the native substrate D-luciferin. For these purified mutant luciferases, light output as a function of substrate concentration was measured with D-luciferin and the aminoluciferin substrates shown in Figure 1B, with nonlinear regression fitting to the Michaelis-Menten equation (Figure 4; Table S2). For most of the luciferase mutants, the rank of light output was D-Luc > CycLuc1 > 6’-NH2LH2 > 6’-MeNHLH2 > CycLuc2 > 6’-Me2NLH2. In all cases, the aminoluciferins exhibited much lower Km values than D-luciferin. The R218K mutant was the most generally beneficial mutation, yielding the greatest light output for most aminoluciferin substrates, including a 20-fold improvement in the maximal sustained emission from CycLuc2 while retaining a very low Km of 0.3 μM (Figure 4; Table S2). Similarly, the maximal sustained light emission from CycLuc1 was increased 14-fold to 12.5% of that of D-luciferin with WT luciferase, yet at a Km of 1.8 μM that is still substantially lower than that of D-luciferin. The R218K mutation has been previously described in the context of D-luciferin, and is known to raise the Km but cause minimal disruption of catalytic activity (Branchini et al., 2001).
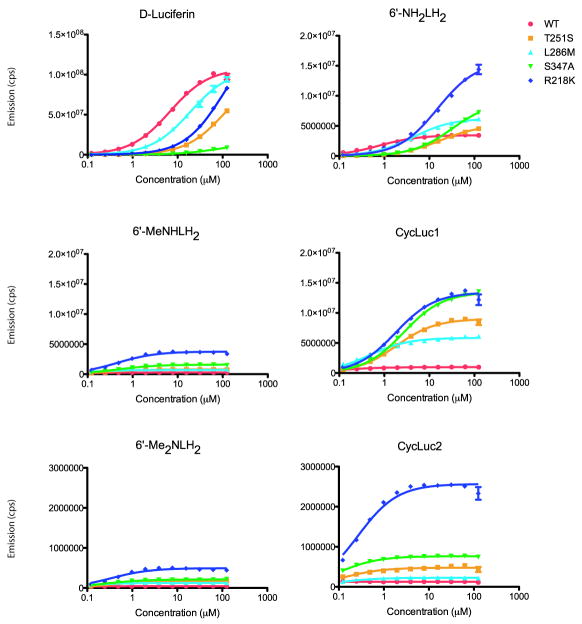
Figure 4. Characterization of luciferase mutants.
Dose-response curves for purified luciferases (WT, T251S, L286M, S347A, and R218K) are shown with each luciferin substrate. The assays were performed in triplicate and are represented as the mean +/- SEM. The Y-axes have been calibrated to allow comparison of mutant emission for each substrate. For comparison between substrates, note the difference in scale. Each curve was fit to the Michaelis-Menten equation by nonlinear regression (GraphPad 5.0) to determine apparent Km and Vmax values (Table S2). Comparison of the light emission of S347T and S347A and the combination of S347A with other mutations is detailed in Figure S2.
The most selective luciferase mutants were S347A and S347T. Because the activity of the purified S347T luciferase was found to decline rapidly when stored at 4°C, S347A was used in preference to S347T for further characterization (Figure S2A). The S347A luciferase yielded a 14-fold increase in light emission for CycLuc1, equivalent to that of the R218K mutant, while simultaneously decreasing the maximal emission from D-luciferin by 7.5-fold and raising the Km for D-luciferin by >10-fold (Figure 4; Table S2). To determine whether this selectivity could be further increased, we created several combinations of the S347A mutation with T251S, L286M, or F247 mutants (Figure S2B). All of these mutants showed increased discrimination against D-luciferin, giving even lower light output than with S347A alone. On the other hand, the S347A/L286M double mutant further increased the maximal sustained light emission from CycLuc1 by two-fold relative to S347A alone, albeit with a ~10-fold increase in the Km to 22 μM.
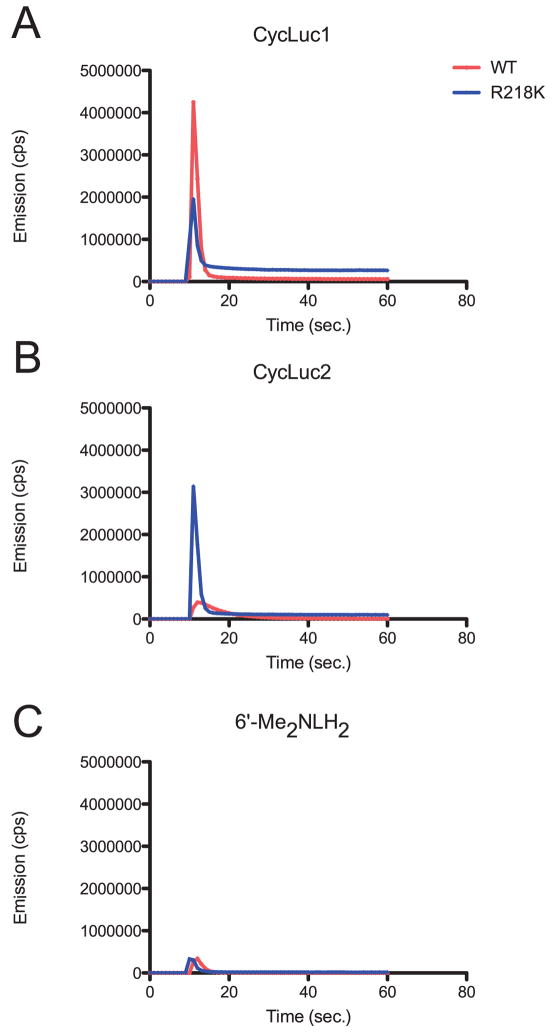
Rapid injection experiments were performed to reveal the burst kinetic behavior of the mutant luciferases (Figure 5; Figure S3). As we have observed for the wild-type protein, most alkylaminoluciferin substrates gave a rapid initial burst of light, followed by a substantial decrease in the rate of light output (Reddy et al., 2010). Most of the mutant luciferases identified here shared this general behavior, although the decrease in the rate of light output was less severe than that of the wild-type protein, resulting in a higher level of sustained light emission (Figure 5A; Figure S3). The most striking finding was that the initial rate of light output for the dialkylaminoluciferin CycLuc2 with the mutant R218K is considerably increased relative to the wild-type protein (Figure 5B; Figure S3). In contrast, the burst kinetic profile for the corresponding acyclic dialkylaminoluciferin 6’-Me2N-LH2 was largely unchanged (Figure 5C; Figure S3).
Figure 5. Burst kinetic profiles with WT and R218K luciferases.
Purified luciferases (10 nM) were rapidly injected at the 10-second time point into 10 μM of CycLuc1 (A), CycLuc2 (B), or 6’-Me2NLH2 (C) and the light emission was recorded. Burst emission behavior for all substrates and characterized mutants is detailed in Figure S3, with emission wavelengths reported in Table S3.
The bioluminescence emission wavelength of aminoluciferins is red-shifted relative to D-luciferin with all of the tested luciferases (Table S3). Because the PMT in the Turner Veritas plate reader is less sensitive to the red-shifted light emission of the aminoluciferin substrates than the green light emission of D-luciferin, this likely leads to an underestimation of the true light output for aminoluciferins. We did not attempt to correct for this difference, which also varies among mutants. For example, the R218K luciferase caused a slight red-shift in the bioluminescence of all luciferins: D-luciferin yields maximal emission at 567 nm, CycLuc1 at 609 nm, and CycLuc2 at 621 nm (Table S3). In contrast, the L286M mutant results in a 5-13 nm blue-shift in the emission of all aminoluciferins, but a diametrically-opposed 13 nm red-shift in D-luciferin light emission (Table S3). Interestingly, the S347A mutant gives discrete emission peaks for all of the aminoluciferins, but anomalous bimodal emission for D-luciferin (Branchini et al., 2003).
Characterization of Mutants in Mammalian Cells
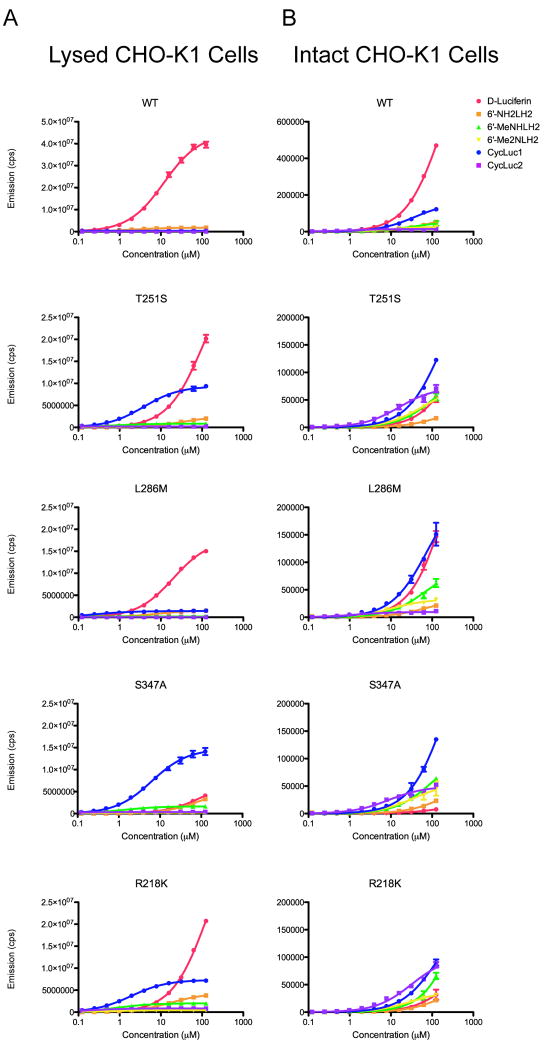
The mutant luciferases that performed best in vitro were cloned into pcDNA3.1 and transfected into CHO-K1 cells. Light emission from mutant luciferases in CHO cell lysates generally mirrored the results with purified proteins (Figures 3-4; Figures S1, S4). The lone exception was L286M, which gave lower light output than expected, possibly indicating that this protein has a lower expression level and/or stability in the cellular context (Figures 3-4; Figure S4). Unsurprisingly, D-luciferin was the best substrate for wild-type luciferase (Figure 6A). However, for the R218K and T251S mutant luciferases, CycLuc1 light output was superior to D-luciferin at substrate concentrations below ~30 μM. In the case of the S347A mutant, CycLuc1 exceeded the light output of D-luciferin over the entire concentration range (Figure 6A). All other substrates had considerably lower levels of light emission in cell lysates, with the exception of 6’-aminoluciferin with the mutant F247L (Figure S1B).
Figure 6. Dose-response profiles from luciferases in expressed in CHO-K1 cell lysates and in live cells.
CHO-K1 cells were transiently transfected with pcDNA3.1 vectors expressing WT, T251S, L286M, S347A, or R218K firefly luciferase. Dose-response curves for each luciferase with D-Luciferin, 6’-NH2LH2, 6’-MeNHLH2, 6’- Me2NLH2, CycLuc1 and CycLuc2 were generated at concentrations from 0.122-125 μM using lysates from the transfected cells (A) or using the intact live cells (B). The assays were performed in triplicate and are represented as the mean +/- SEM. Note that the WT emission scale is larger than that of the mutants by 2-fold and 3-fold for cell lysates and live cells, respectively. Substrate-by-substrate comparison of the light emission from each mutant in cell lysates is detailed in Figure S4.
The light output from luciferase in live cells is much lower than that of lysed cells, as has been previously observed for D-luciferin (de Wet et al., 1987; Craig et al., 1991; Shinde et al., 2006) and 6’-aminoluciferin (Shinde et al., 2006). Moreover, there is less difference in the relative levels of light emission between different substrates. Even with the wild-type luciferase, CycLuc1 emitted 26% of the light of D-luciferin at 125 μM and its light output exceeded that of 6’-aminoluciferin (Figure 6B). CycLuc1 was the superior substrate for T251S, R218K, and particularly S347A luciferase (18-fold higher light output than D-luciferin at 125 μM). Remarkably, live-cell light emission from CycLuc2 exceeded that of both CycLuc1 and D-luciferin with T251S, S347A, and particularly R218K luciferase over a broad concentration range, despite the relatively poor light output from CycLuc2 in cell lysates (Figure 6).
DISCUSSION
We have found that mutation of firefly luciferase can dramatically improve aminoluciferin substrate utilization and selectivity, and allow the use of these substrates to monitor luciferase expression in cells and cell lysates. For example, the F247L mutant improves light output from 6’-aminoluciferin by almost five-fold and can be recommended for improved imaging of this substrate (Figure 2; Figure S1). This has particular significance for bioluminescence assays of protease activity that rely on detection of this substrate (Monsees et al., 1994; Shah et al., 2005; Fan et al., 2007; Dragulescu-Andrasi et al., 2009; Hickson et al., 2010; Scabini et al., 2011). Light output from all aminoluciferins was greatly increased by the R218K mutant (e.g., 14-fold for CycLuc1, 20-fold for CycLuc2), suggesting this mutant as a starting point for measuring light emission from these and other novel alkylated aminoluciferins. Moreover, mutation of serine 347 to threonine or alanine gives similarly improved light emission from CycLuc1 but also exhibits strong selectivity for CycLuc1 over D-luciferin both in vitro and in live cells (Figures 4, 6, S2, S4). Combination of mutations such as the ones described here is expected to allow further enhancement of aminoluciferin light output and selectivity. For instance, we have found that the S347A/L286M double mutant gives twice the light output of S347A with CycLuc1, and further discriminates against D-luciferin (Figure S2B).
The rate at which light is generated by luciferase is dependent on several factors, including the rate of adenylation and oxidation to afford the oxyluciferin excited state, the quantum yield of light emission, and the rate of product release. To gain insight into the underlying mechanisms by which light emission from aminoluciferins is improved with these luciferase mutants, we monitored the rate of light output over the first 50 seconds following substrate addition (Figure 5; Figure S3). For CycLuc1 and 6’-MeNH-LH2, all mutants result in a rapid initial burst that is similar to that observed for the wild-type protein (Reddy et al., 2010), suggesting that there are no substantive improvements in the ability to form the respective luciferyl-AMP, its subsequent oxidation to afford the excited-state oxyluciferin, or the quantum yield of light emission. Rather, higher light output was observed after the initial burst (Figure 5A; Figure S3), implicating a reduction in product inhibition as the primarily factor responsible for the observed improvement in sustained light emission. Even with D-luciferin, the molecular basis for product inhibition is still unresolved, potentially including contributions from both dehydroluciferyl-AMP (L-AMP) and oxyluciferin (Fraga, 2008). Lowered product inhibition could result from a simple reduction in affinity for aminoluciferin substrates and their corresponding products. Alternatively, it is possible that some mutants function in part by reducing the formation of the corresponding L-AMP analog. A better molecular understanding of the nature of the product inhibition with these substrates may help guide future optimization efforts.
Notably, both the burst and sustained light emission from CycLuc1 exceeds that of 6’-MeNH-LH2 with all of the luciferases we have characterized, and is consistent with a role for cyclization in optimizing aminoluciferin light output (Reddy et al., 2010). Like CycLuc1, light emission from CycLuc2 is superior to its acyclic counterpart 6’-Me2N-LH2 for every luciferase we have tested. In contrast to CycLuc1, we find that the burst emission with CycLuc2 is substantially more rapid and intense with the mutant luciferase R218K than with wild-type (Figure 5B; Figure S3), indicating an improvement in one or more of the enzymatic steps required to form this excited-state oxyluciferin (e.g., adenylation and/or oxidation). We postulate that the bulk and rigidity of CycLuc2 enforces a sub-optimal alignment for the production of the excited-state oxyluciferin in the wild-type luciferase. Enlarging the luciferin binding pocket with the R218K mutation improves the alignment of CycLuc2 within the active site, allowing more rapid production of an efficient light-emitting oxyluciferin excited state (Figure 5B). By comparison, the more flexible substrate 6’-Me2N-LH2 gives a rapid but weak burst for all mutants (Figure 5C; Figure S3). We hypothesize that the free bond rotation about the aryl amine bond of 6’-Me2N-LH2 can facilitate the alignment necessary for the rapid formation of the oxyluciferin, but also limits the efficiency of light output from this excited-state molecule (Figure 5C).
For light emission from live cells, the cell-permeability and Km of the substrate are important for access to the intracellular luciferase and the efficiency of light output under sub-saturating concentrations. CycLuc2 has a lower Km than CycLuc1 (Table S2) and is predicted to be more cell-permeable than CycLuc1 because of the replacement of a polar amine proton with a methyl group (cLogP of 2.5 versus 2.0). These differences are therefore likely to explain the better relative performance of CycLuc2 in live versus lysed cells. Moreover, this suggests that optimization of cell permeability and light output at low substrate concentrations – rather than just maximal light output – may be particularly important for imaging in live cells and organisms, because insufficient substrate is delivered into the live cell to achieve maximal light emission.
The bioluminescence emission wavelength of D-luciferin from different beetle luciferases (firefly, click beetle, railroad worm) and their mutants ranges from ~540-622 nm (Viviani et al., 1999; Branchini et al., 2010). Beyond simple changes in the polarity of the environment of the light emitter (Morton et al., 1969), explanations for this sensitivity include the rigidity of the active site (Nakatsu et al., 2006) and perturbations in the ionic interactions of the phenolate of D-luciferin (Hirano et al., 2009). The emission behavior of aminoluciferins can lend insight into this question because they lack an ionizable phenolate (Figure 1). With the mutant R218K, all luciferins exhibit a modest red-shift in emission wavelength (Table S3). Because oxyluciferins are asymmetric charge-transfer molecules, some shared solvatochromatic sensitivity to the polarity of the luciferin binding site is expected (Naumov et al., 2009). However, D-luciferin exhibits anomalous emission behavior with several mutants: the L286M mutation causes a red-shift in the emission of D-luciferin but a blue-shift in the emission of all aminoluciferins, and D-luciferin uniquely gives bimodal emission with S347A (Table S3). These differences are consistent with a role for the ionization state and ionic interactions of the phenol in determining the bioluminescence emission wavelength when D-luciferin is the substrate.
The efficient chemical generation of light by firefly luciferase has been widely used as a sensitive reporter system for gene expression (de Wet et al., 1987; Prescher et al., 2010). However, the application of bioluminescence detection as a general optical reporter of cellular status has lagged behind that of fluorescence. Chemical modification of the luciferin substrate can red-shift the emission wavelength of bioluminescence beyond that of D-luciferin (Reddy et al., 2010). Moreover, the combination of synthetic luciferins and mutant luciferases described here not only extends the wavelength of bioluminescence emission and broadens the scope of substrates that can be used for bioluminescent assays both in vitro and in mammalian cells, but also suggests that orthogonal luciferases could be developed to allow multiplexing of bioluminescent signals. Further chemical modification of luciferin substrates and corresponding mutation of firefly and other beetle luciferases is therefore anticipated to be a fruitful avenue for expanding the power and scope of bioluminescence assays and imaging.
Significance
Aminoluciferin, in which the 6’-hydroxyl group of D-luciferin is replaced by a 6’-amino group, is a firefly luciferase substrate that is widely used for bioluminescent protease assays. Recently we and others have discovered that a wide variety of modifications of the 6’-amino group retain bioluminescent light emission, including alkylation of the 6’-amino group and 5’,6’-fused ring structures. This family of synthetic aminoluciferin substrates exhibits desirable properties such as light emission at longer wavelengths than D-luciferin and increased affinity for luciferase, but is limited by low sustained light output after an initial flash of light. Here we have described the creation of mutant luciferases that yield improved sustained light emission with aminoluciferins, both in vitro and in lysed and live mammalian cells. Light output from 6’-aminoluciferin is greatly increased with the luciferase mutant F247L, which has implications for the use of this substrate in bioluminescent protease assays. The luciferase mutant R218K gives broadly improved light output with a wide variety of aminoluciferins, including monoalkylated, dialkylated, and cyclic aminoluciferin substrates. Significant discrimination between luciferin substrates was observed for many mutants, suggesting that orthogonal luciferases could be developed to allow multiplexing of bioluminescent signals. For example, the cyclic alkylaminoluciferin CycLuc1 is a better substrate for S347A luciferase than D-luciferin. Aminoluciferins possess unique physical and photophysical properties that have yet to be fully exploited in bioluminescent assays and imaging, and these mutant luciferases broaden the scope and utility of their application.
EXPERIMENTAL PROCEDURES
Materials
Chemicals for synthesis were from Aldrich unless otherwise noted. Data were plotted with GraphPad Prism 5.0. NMR spectra were recorded on a Varian Mercury 400 MHz NMR. Small molecule mass spectral data were recorded on a Waters QTOF Premier. Protein structures were displayed with ICM-Browser (MolSoft). Bioluminescence measurements were taken in a Turner Veritas luminometer and were not corrected for differences in the wavelength sensitivity of the PMT. Emission spectra and burst kinetic data measurements were recorded on a Spex FluoroMax-3 fluorimeter. D-Luciferin was from Anaspec and 6’-aminoluciferin was from Marker Gene Technologies Inc. CycLuc1, CycLuc2, 6’-MeNH-LH2, and 6’-Me2NLH2 were synthesized as previously described (Reddy et al., 2010). Protein concentrations were determined using Coomassie Plus (Thermo Scientific). Immobilized glutathione (Thermo Scientific) was used for GST-tagged protein purification. Unless otherwise stated, all protein purification steps were carried out at 4°C.
Plasmid constructs
The Photinus pyralis firefly luciferase gene was PCR-amplified from pGL3 (Promega) and cloned into the BamHI and NotI sites of pGEX6P-1 for bacterial expression. Mutant proteins were created using the Quickchange site-directed mutagenesis kit (Stratagene). Saturating mutagenesis was performed at sites predicted to impact or alter substrate binding, including: F247, T251, L286, S347, R337, R218, Y340, and I351, using the degenerate codon NNK. The point mutants S347A, L286A, A348G, E311A, I351A, and R337A were made independently.
Mutant screening
Screening for mutant luciferases was performed in the E. coli strain JM109, which has high transformation efficiency and plasmid recovery, as well as reasonable levels of inducible protein expression. Other bacterial strains (OMNI, DH5α, BL21(DE3), XL1Blue, XL10Gold), gave less consistent results. Following saturating mutagenesis, 500 μL of the E. coli strain JM109 was transformed and plated on 5 Luria-Bertani (LB) plates that contained 50 μg/mL carbenicillin. Colonies were picked and used to inoculate two 96-well plates where each well contained 150 μL of LB with 50 μg/mL carbenicillin. Inoculated plates were incubated at 37 °C overnight. Cells were induced with 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and incubated at RT overnight. Five μL of the bacterial protein extraction reagent SoluLyse (Genlantis) was then added to 50 μL of induced cells followed by a 10-minute incubation at RT. Two μL of lysate was then added to 60 μL of a solution containing 25 μM substrate in 20 mM Tris pH 7.6, 0.1 mM EDTA, 8 mM MgSO4, 4 mM ATP, and 1 mM TCEP in each well of a white 96-well plate (Costar 3912). The bioluminescence emission was measured in a Turner Veritas luminometer after a delay of 2-5 minutes. Readings were taken every 30 seconds for a total of 10 runs. Mutant lysates that displayed improved and/selective light emission for CycLuc1 at 25 μM were then used to measure light output as a function of CycLuc1 concentration ranging from 0.122-250 μM. These validated mutants were sequenced and subjected to further characterization as described below.
Protein expression
Mutant luciferases were expressed as GST-fusion proteins from the vector pGEX6P-1 in the E. coli strain JM109. Cells were grown at 37 °C until the OD600 reached 0.5-1, induced with 0.1 mM IPTG, and incubated at 20 °C overnight. Cells were pelleted at 5000 rpm in a Sorvall 2C3C Plus centrifuge (H600A rotor) at 4°C for 10 minutes, then flash-frozen in liquid nitrogen and purified immediately or stored at -80°C.
Luciferase purification
E. coli cell pellets from one liter of culture were thawed on ice, resuspended in 25 mL Lysis Buffer (50 mM HEPES pH 7.4, 500 mM NaCl, and 0.5% Tween-20) containing 1 mM PMSF, and disrupted by sonification (Branson Sonifier). Dithiothreitol (DTT) was added at 10 mM and the resulting cell lysate was clarified by centrifugation at 35K rpm in a Beckman 50.2Ti rotor for 30-45 minutes. The supernatant was batch-bound to immobilized glutathione for 1h at 4°C, and the beads were washed with Lysis Buffer containing 10 mM DTT, followed by Wash Buffer (50 mM Tris pH 8.1, 250 mM NaCl and 10 mM DTT), and finally with Storage Buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM TCEP). Twenty units of PreScission Protease (GE Healthcare) were added and incubation continued for 4 hours or overnight at 4°C to cleave the GST-fusion and elute the untagged luciferase protein.
Substrate dose-response assays with purified protein
Luminescence assays were initiated by adding 30 μL 2x substrate in either buffer A (20 mM Tris pH 7.6, 0.1 mM EDTA, 8 mM MgSO4, 4 mM ATP, and 1 mM TCEP) or buffer B (20 mM Tris pH 7.6, 0.1 mM EDTA, 8 mM MgSO4, 4 mM ATP, 6 mg/mL BSA, and 33 mM DTT) to 30 μL of 20 nM purified luciferase in 20 mM Tris pH 7.6, 0.1 mM EDTA, 1 mM TCEP and 0.4 mg/mL BSA in a white 96-well plate (Costar 3912). Measurements were performed 3 minutes post-substrate addition in a Turner Veritas luminometer with final substrate concentrations ranging from 0.122-125 μM.
Cell Culture
Chinese hamster ovary-K1 (CHO-K1) cells were grown in a CO2 incubator at 37°C with 5% CO2 and were cultured in F-12K Nutrient mixture (GIBCO) supplemented with 10% fetal bovine serum and 100 U/mL penicillin/streptomycin.
Transfections
Mutant luciferases F247A, F247L, F247V, F247S, T251S, L286M, S347A, S347T, and R218K were cloned into the BamHI-NotI site of pcDNA 3.1 and transfected into CHO-K1 cells for live and lysed cell experiments. Transient transfections were performed at room temperature using Lipofectamine 2000 on cells plated at 60-75% confluency in 96-well white tissue culture treated plates (Costar 3917) for intact cell assays, or 6-well plates for lysed cell assays. For intact cells, 0.075 μg DNA/well was transfected and for lysed cells, 2.25 μg DNA/well was transfected. Assays were performed in triplicate 24 hours post-transfection.
Intact Cell Assays
Transfected CHO cells were washed with HBSS and overlaid with 60 μL substrate in HBSS. Titration assays were performed 3 minutes post-substrate addition in a Turner Veritas luminometer with final substrate concentrations ranging from 0.122-125 μM.
Lysed Cell Assays
Transfected CHO cells were washed with HBSS and lysed for 10 minutes at RT with 1 mL 1x Passive Lysis Buffer (Promega) per well. Cells from one and a half wells of a 6-well plate were used to test one substrate in triplicate at the 12 concentrations tested. Luminescence assays were initiated by adding 30 μL 2x substrate in 20 mM Tris pH 7.6, 0.1 mM EDTA, 8 mM MgSO4, 4 mM ATP, 6 mg/mL BSA, and 33 mM DTT to 30 μL of lysate in a white 96-well plate (Costar 3912). Titration assays were performed 3 minutes post-substrate addition in a Turner Veritas luminometer with final substrate concentrations ranging from 0.122-125 μM.
Bioluminescence Emission Scans and Burst Kinetics Assays
Each purified protein in 20 mM Tris pH 7.6, 0.1 mM EDTA, 1 mM TCEP and 0.4 mg/mL BSA was added to 2x substrate in 20 mM Tris pH 7.6, 0.1 mM EDTA, 8 mM MgSO4, 4 mM ATP, and 1 mM TCEP in a cuvette. Final protein concentrations were 10 nM and substrate concentrations were 10 μM.
Bioluminescence Emission Scans
Luciferase was rapidly injected into a cuvette containing substrate, and the emission from 400-800 nm was recorded in a SPEX FluoroMax-3 fluorimeter with closed excitation slits 10 seconds post-injection.
Burst Kinetics Assays
Measurements were taken in a Spex FluoroMax-3 fluorimeter every second with a 0.1 second integration time at the maximal emission wavelength for each luciferase/substrate pair. Ten seconds after commencing the assay, protein was rapidly injected into the cuvette containing substrate to observe both the burst and the steady-state light emission over the first minute.
Supplementary Material
HIGHLIGHTS.
Mutation of luciferase can improve light emission from all aminoluciferins.
F247L luciferase improves light emission from 6’-aminoluciferin.
CycLuc1 is a better substrate for S347A luciferase than D-luciferin.
Live cell light emission is highly dependent on cell-permeability and Km.
Acknowledgments
This work was supported by grants from the NIH (CA127196 and EB013270). We thank Walter Thompson for making F247L and F247A point mutants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Branchini BR, Magyar RA, Murtiashaw MH, Portier NC. The role of active site residue arginine 218 in firefly luciferase bioluminescence. Biochemistry. 2001;40:2410–2418. doi: 10.1021/bi002246m. [DOI] [PubMed] [Google Scholar]
- Branchini BR, Southworth TL, Murtiashaw MH, Boije H, Fleet SE. A mutagenesis study of the putative luciferin binding site residues of firefly luciferase. Biochemistry. 2003;42:10429–36. doi: 10.1021/bi030099x. [DOI] [PubMed] [Google Scholar]
- Branchini BR, Ablamsky DM, Davis AL, Southworth TL, Butler B, Fan F, Jathoul AP, Pule MA. Red-emitting luciferases for bioluminescence reporter and imaging applications. Anal Biochem. 2010;396:290–297. doi: 10.1016/j.ab.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Craig FF, Simmonds AC, Watmore D, McCapra F, White MR. Membrane-permeable luciferin esters for assay of firefly luciferase in live intact cells. Biochem J. 1991;276(Pt 3):637–641. doi: 10.1042/bj2760637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragulescu-Andrasi A, Liang G, Rao J. In vivo bioluminescence imaging of furin activity in breast cancer cells using bioluminogenic substrates. Bioconjug Chem. 2009;20:1660–1666. doi: 10.1021/bc9002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F, Wood KV. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol. 2007;5:127–136. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- Fraga H. Firefly luminescence: A historical perspective and recent developments. Photochem Photobiol Sci. 2008;7:146–158. doi: 10.1039/b719181b. [DOI] [PubMed] [Google Scholar]
- Hickson J, Ackler S, Klaubert D, Bouska J, Ellis P, Foster K, Oleksijew A, Rodriguez L, Schlessinger S, Wang B, et al. Noninvasive molecular imaging of apoptosis in vivo using a modified firefly luciferase substrate, Z-DEVD-aminoluciferin. Cell Death Differ. 2010;17:1003–1010. doi: 10.1038/cdd.2009.205. [DOI] [PubMed] [Google Scholar]
- Hirano T, Hasumi Y, Ohtsuka K, Maki S, Niwa H, Yamaji M, Hashizume D. Spectroscopic studies of the light-color modulation mechanism of firefly (beetle) bioluminescence. J Am Chem Soc. 2009;131:2385–2396. doi: 10.1021/ja808836b. [DOI] [PubMed] [Google Scholar]
- Monsees T, Miska W, Geiger R. Synthesis and characterization of a bioluminogenic substrate for alpha-chymotrypsin. Anal Biochem. 1994;221:329–34. doi: 10.1006/abio.1994.1421. [DOI] [PubMed] [Google Scholar]
- Moravec RA, O’Brien MA, Daily WJ, Scurria MA, Bernad L, Riss TL. Cell-based bioluminescent assays for all three proteasome activities in a homogeneous format. Anal Biochem. 2009;387:294–302. doi: 10.1016/j.ab.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Morton RA, Hopkins TA, Seliger HH. Spectroscopic properties of firefly luciferin and related compounds; an approach to product emission. Biochemistry. 1969;8:1598–1607. doi: 10.1021/bi00832a041. [DOI] [PubMed] [Google Scholar]
- Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Structural basis for the spectral difference in luciferase bioluminescence. Nature. 2006;440:372–376. doi: 10.1038/nature04542. [DOI] [PubMed] [Google Scholar]
- Naumov P, Ozawa Y, Ohkubo K, Fukuzumi S. Structure and spectroscopy of oxyluciferin, the light emitter of the firefly bioluminescence. J Am Chem Soc. 2009;131:11590–11605. doi: 10.1021/ja904309q. [DOI] [PubMed] [Google Scholar]
- Prescher JA, Contag CH. Guided by the light: visualizing biomolecular processes in living animals with bioluminescence. Curr Op Chem Biol. 2010;14:80–89. doi: 10.1016/j.cbpa.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Reddy GR, Thompson WC, Miller SC. Robust light emission from cyclic alkylaminoluciferin substrates for firefly luciferase. J Am Chem Soc. 2010;132:13586–13587. doi: 10.1021/ja104525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scabini M, Stellari F, Cappella P, Rizzitano S, Texido G, Pesenti E. In vivo imaging of early stage apoptosis by measuring real-time caspase-3/7 activation. Apoptosis. 2011;16:198–207. doi: 10.1007/s10495-010-0553-1. [DOI] [PubMed] [Google Scholar]
- Shah K, Tung C-H, Breakefield XO, Weissleder R. In vivo imaging of S-TRAIL-mediated tumor regression and apoptosis. Mol Ther. 2005;11:926–931. doi: 10.1016/j.ymthe.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Shinde R, Perkins J, Contag CH. Luciferin derivatives for enhanced in vitro and in vivo bioluminescence assays. Biochemistry. 2006;45:11103–12. doi: 10.1021/bi060475o. [DOI] [PubMed] [Google Scholar]
- Takakura H, Sasakura K, Ueno T, Urano Y, Terai T, Hanaoka K, Tsuboi T, Nagano T. Development of luciferin analogues bearing an amino group and their application as BRET donors. Chem Asian J. 2010;5:2053–2061. doi: 10.1002/asia.201000219. [DOI] [PubMed] [Google Scholar]
- Viviani VR, Bechara EJ, Ohmiya Y. Cloning, sequence analysis, and expression of active Phrixothrix railroad-worms luciferases: relationship between bioluminescence spectra and primary structures. Biochemistry. 1999;38:8271–8279. doi: 10.1021/bi9900830. [DOI] [PubMed] [Google Scholar]
- de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EH, Worther H, Seliger HH, McElroy WD. Amino Analogs of Firefly Luciferin and Biological Activity Thereof. J Am Chem Soc. 1966;88:2015–2019. [Google Scholar]
- Woodroofe CC, Shultz JW, Wood MG, Osterman J, Cali JJ, Daily WJ, Meisenheimer PL, Klaubert DH. N-Alkylated 6’-aminoluciferins are bioluminescent substrates for Ultra-Glo and QuantiLum luciferase: new potential scaffolds for bioluminescent assays. Biochemistry. 2008;47:10383–10393. doi: 10.1021/bi800505u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.