Abstract
Genomic imprinting in mammals is controlled by DNA methylation imprints that are acquired in the gametes, at essential sequence elements called ‘imprinting control regions’ (ICRs). What signals paternal imprint acquisition in male germ cells remains unknown. To address this question, we explored histone methylation at ICRs in mouse primordial germ cells (PGCs). By 13.5 days post coitum (d.p.c.), H3 lysine-9 and H4 lysine-20 trimethylation are depleted from ICRs in male (and female) PGCs, indicating that these modifications do not signal subsequent imprint acquisition, which initiates at ∼15.5 d.p.c. Furthermore, during male PGC development, H3 lysine-4 trimethylation becomes biallelically enriched at ‘maternal’ ICRs, which are protected against DNA methylation, and whose promoters are active in the male germ cells. Remarkably, high transcriptional read-through is detected at the paternal ICRs H19-DMR and Ig-DMR at the time of imprint establishment, from one of the strands predominantly. Combined, our data evoke a model in which differential histone modification states linked to transcriptional events may signal the specificity of imprint acquisition during spermatogenesis.
Keywords: DNA methylation, epigenetic, genomic imprinting, histone methylation, primordial germ cells
Introduction
Mammalian imprinted genes are organised in clusters and their parental allele-specific expression is regulated by essential, CpG-rich, sequence elements called ‘imprinting control regions’ (ICRs; Bartolomei, 2009; Arnaud, 2010). It is not known why some ICRs acquire DNA methylation imprints in the female germ line, while others become methylated specifically in the male germ line. However, in both the germ lines de-novo DNA methyltransferase DNMT3A is involved in imprint acquisition (Kaneda et al, 2004; Kato et al, 2007). The related DNMT3L protein can form complexes with DNMT3A and plays an essential role in imprint acquisition as well (Bourc'his et al, 2001; Hata et al, 2002; Kato et al, 2007). Interestingly, DNMT3L can bind to histone H3 in vitro, but H3 lysine-4 dimethylation, and in particular H3-K4 trimethylation (H3K4me3), was found to prevent its association with chromatin (Ooi et al, 2007). Recent studies show that DNMT3A itself is also sensitive to the H3 lysine-4 methylation status. Its ‘ATRX-DNMT3-DNMTL’ (ADD) domain binds to the H3 tail most efficiently when lysine-4 is unmethylated (Otani et al, 2009; Zhang et al, 2010; Li et al, 2011). Despite a suspected involvement of histone methylation states in the recruitment of DNA methyltransferase complexes, so far no studies have directly assessed chromatin at imprinted loci in germ cells to test this hypothesis.
Many ICRs acquire their DNA methylation in the female germ line (Arnaud, 2010). Two well-known ‘maternal ICRs’ are the CpG island/promoter of the Snrpn gene (Shemer et al, 1997), which controls the Prader–Willi Syndrome imprinted domain on central chromosome 7, and the KvDMR1, a CpG island/promoter that controls the Kcnq1 domain on distal chromosome 7 (Fitzpatrick et al, 2002). Only four differentially methylated regions (DMRs) are known to acquire their methylation during spermatogenesis, a pre-meiotic process that initiates during fetal stages of development (Arnaud, 2010). The best-characterised ‘paternal ICRs’ are the H19 DMR controlling the Igf2-H19 domain on mouse distal chromosome 7 (Tremblay et al, 1995) and the Ig-DMR, which controls the Dlk1-Dio3 domain on mouse distal chromosome-12 (Lin et al, 2003).
Maternal ICRs comprise gene promoters, whereas paternal ICRs are intergenic non-promoter regions. This intriguing distinction might somehow contribute to the specificity of imprint acquisition. For instance, specific histone modifications such as H3K4me3 are linked to promoter activity, at least in somatic cells (Mikkelsen et al 2007; Zhao et al, 2007), and could prevent acquisition of de-novo DNA methylation (Ooi et al, 2007; Ciccone et al, 2009; Zhang et al, 2010). Here, we explore histone methylation in mouse primordial germ cells (PGCs), and in later-stage germ cells, with particular emphasis on the male germ line. A carrier chromatin immunoprecipitation (cChIP) approach was developed, adapted to small batches of FACS-sorted cells, to assess ICRs at critical stages of spermatogenesis. A main finding is that at maternal ICRs protection against DNA methylation in male germ cells correlates with promoter activity and biallelic enrichment of H3 lysine-4 (tri)methylation. At the paternal ICRs H19-DMR and Ig-DMR, in contrast, chromatin is organised differently and here acquisition of DNA methylation correlates with transcriptional read-through. Our data evoke a putative link between transcription, histone methylation, and imprint acquisition during spermatogenesis.
Results
Dynamics of DNA methylation acquisition in male germ cells
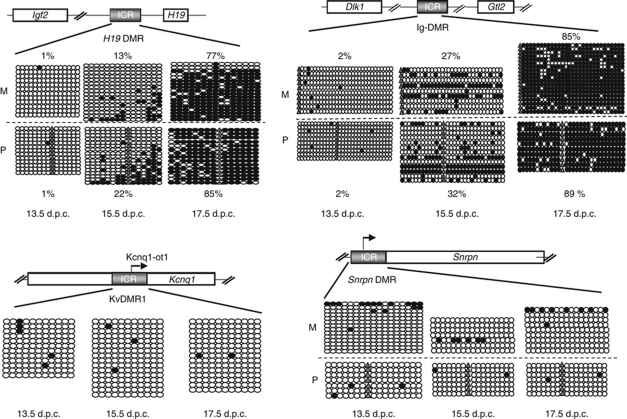
Embryos were derived that were intra-specific hybrid between C57BL/6J and M. m. molossinus strain JF1 (Koide et al, 1998) and transgenic for a GFP gene driven by Oct4 regulatory sequences (Yoshimizu et al, 1999). Gonads were dissected at fetal stages. Batches of FACS-sorted germ cells were obtained for DNA methylation, transcription, and chromatin studies. Single-nucleotide polymorphisms (SNPs) between C57BL/6J and JF1 allowed us to distinguish the parental chromosomes (Henckel et al, 2009). First, we determined by bisulphite sequencing the timing of DNA methylation acquisition in germ cells of male fetuses. At four ICRs analysed, no DNA methylation was detected at 13.5 d.p.c., confirming that complete DNA demethylation had occurred at this stage (Figure 1). At the H19 DMR, initial acquisition of methylation was observed at 15.5 d.p.c., and by 17.5 d.p.c., DNA methylation was almost complete. Less pronouncedly than in some studies (Davis et al, 2000), but comparable to others (Kato et al, 2007; Lee et al, 2010), imprint acquisition was delayed on the maternal compared with the paternal allele. At the Ig-DMR controlling the Dlk1-Dio3 domain (Lin et al, 2003), the timing of imprint acquisition was comparable to the H19 DMR, with partial acquisition at 15.5 d.p.c. and almost full DNA methylation at 17.5 d.p.c. The measured levels of DNA methylation were not much different on the parental chromosomes at 15.5 d.p.c. Also at the DMR of the Gpr1-Zdbf2 imprinted domain on mouse chromosome 1 (Hiura et al, 2010), some CpG dinucleotides had acquired methylation by 15.5 d.p.c, and almost all CpGs were fully methylated at 17.5 d.p.c. (Supplementary Figure S1). The maternal ICRs KvDMR1 and Snrpn DMR, as expected, remained unmethylated in the male germ cells at the stages analysed (Figure 1).
Figure 1.
Dynamics of ICR methylation in male germ cells. Analysed ICRs (grey boxes) and genes (empty boxes) are represented. Representative data from one experiment are shown for each of the regions analysed. Each horizontal row of circles represents the CpG dinucleotides on an individual chromosome. Solid circles depict methylated CpGs, open circles unmethylated CpGs. Parental origin (M, maternal; P, paternal) was determined using SNPs. Grey triangles show CpGs that are absent due to SNPs. For the H19 DMR and Ig-DMR, the measured percentile level of methylation (% methylated CpGs/total CpGs analysed) is indicated for the maternal and the paternal alleles. These levels were reproduced in independently repeated experiments.
cChIP adapted to small numbers of FACS-sorted cells
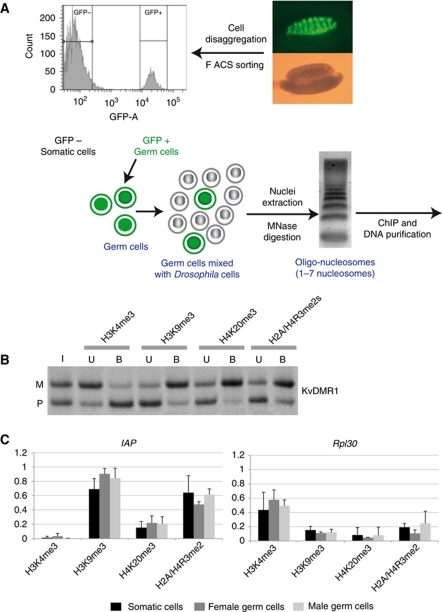
To explore histone methylation in small batches of PGCs, we adapted a cChIP approach (O'Neill et al, 2006). Small batches of FACS-sorted mouse cells were mixed with an excess of Drosophila melanogaster S2 cells, nuclei were purified, followed by partial MNase digestion, purification of chromatin fragments of 1–7 nucleosomes in length, and immunoprecipitation (Figure 2A). First, we checked whether our methodology faithfully revealed the allele specificity of H3 and H4 methylation at ICRs in the negatively sorted (GFP minus) somatic cells. Indeed, using ∼75 000 somatic cells per cChIP, we obtained allelic precipitation at the KvDMR1 (Figure 2B), and at Snrpn DMR, H19 DMR, and the Ig-DMR (Supplementary Figure S2). As in earlier studies on somatic cells (Delaval et al, 2007; Verona et al, 2008; Pannetier et al, 2008; Henckel et al, 2009), H3K9me3 and H4K20me3 were enriched on the DNA-methylated allele, and H3K4me3 on the unmethylated allele. Using smaller batches of FACS-sorted somatic cells, the prepared input chromatin was often biased towards one of the parental alleles, and quantification of the precipitated chromatin was not reproducible between experiments. For our subsequent chromatin studies on 13.5 and 15.5 d.p.c germ cells, therefore, we decided to use 75 000–100 000 cells per cChIP. cChIP could not be performed at 17.5 d.p.c., given the relative difficulty in trypsinising and sorting gonadal cells at this developmentally advanced stage.
Figure 2.
Overview and validation of a carrier ChIP approach. (A) Schematic overview. PGCs were obtained from embryos that were Oct4-GFP transgenic. In the 13.5 d.p.c. male gonad shown, green fluorescence visualises the PGCs. (B) PCR-SSCP analysis after cChIP on somatic control cells with antisera directed against H3K4me3, H3K9me3, H4K20me3, and H2A/H4R3me2s. Results on the KvDMR1 are shown, for input chromatin (I) and antibody-bound (B), and -unbound (U) fractions. In this qualitative assay, the PCR amplifications were close to saturation. (C) Precipitation of H3K4me3, H3K9me3, H4K20me3, and H2A/H4R3me2s in 13.5 d.p.c. female germ cells (grey), male germ cells (white), and somatic control cells (black) at Rpl30 and IAP elements. cChIP was performed, at least, in triplicate. Bound chromatin fractions were quantified by real-time PCR and corrected for background precipitation (percentile precipitation with standard deviation).
Reprogramming of histone methylation at ICRs during PGC development
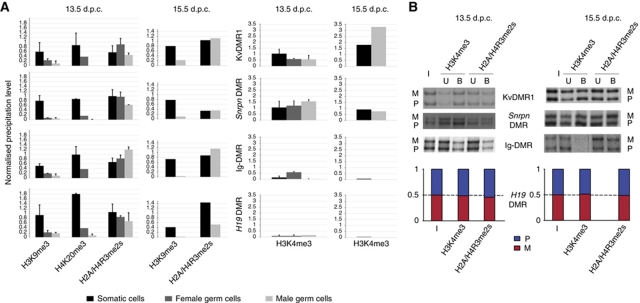
Since in 13.5 d.p.c. germ cells there was no detectable DNA methylation at ICRs, we asked whether associated histone methylations had been lost as well. Locus-specific precipitation levels of repressive histone modifications (H3K9me3, H4K20me3, and H2A/H4R3me2s) were normalised against precipitation at intra-cisternal-A particle (IAP) sequences. These repeat elements exceptionally remain DNA methylated in developing PGCs despite the global DNA demethylation (Lane et al, 2003). Concordantly, H3K9me3, H4K20me3, and H2A/H4R3me2s at IAP elements were similarly high in the 13.5 d.p.c. PGCs as in somatic cells (Figure 2C). At all ICRs analysed, in contrast, the male 13.5 d.p.c. PGCs showed almost complete absence of H3K9me3 and H4K20me3 (Figure 3A). Similarly, at 15.5 d.p.c. we found H3K9me3 to be absent from ICRs in male germ cells. Also in female PGCs, we detected strongly reduced H3K9me3 and H4K20me3 at all four ICRs analysed (Figure 3A). Thus, together with the loss of DNA methylation, the associated H3K9me3 and H4K20me3 had been lost in the developing male and female PGCs as well.
Figure 3.
Reprogramming of histone methylation at ICRs in PGCs. (A) To the left, quantification of H3K9me3, H4K20me3, and H2A/H4R3me2s in male (white) and female (grey) PGCs and somatic control cells (black; a mixture of male and female cells). Relative enrichment is defined as the ratio between the bound (with subtracted background) and the input chromatin, and was normalised to the (bound-background)/input ratio obtained for IAP elements. To the right, quantification of H3K4me3. Relative enrichment was calculated as the ratio between the bound fraction (with subtracted background) and input and was normalised to the (bound-background)/input ratio obtained for Rpl30. Experiments were performed at least in triplicate. Standard deviations are not given for experiments at 15.5 d.p.c., which were performed twice on male PGCs. Female PGCs were not analysed by ChIP at 15.5 d.p.c. (B) PCR-SSCP analysis (KvDMR1, Snrpn DMR, Ig-DMR) or real-time PCR-based allelic discrimination (H19 DMR) after cChIP on male germ cells. At all four ICRs analysed, the histone methylations studied were similarly precipitated from the maternal (M) and the paternal (P) alleles (the ratio between the parental alleles were in all cases smaller than 1.5). In this qualitative assay, the PCR amplifications were close to saturation.
In somatic cells, symmetrical dimethylation on arginine-3 of histones H2A and H4 (H2A/H4R3me2s) is enriched on the DNA-methylated allele of ICRs as well (Verona et al, 2008; Henckel et al, 2009; Supplementary Figure S2). Interestingly, despite the lack of DNA methylation, H2A/H4R3me2s was still present at high levels at all four ICRs analysed in the PGCs (male and female), at 13.5 and 15.5 d.p.c. (Figure 3A). By conventional PCR, followed by electrophoretic discrimination of Single-Strand Conformation Polymorphisms (SSCPs), it was found to be present on both the parental chromosomes at all four ICRs analysed (Figure 3B).
Acquisition of biallelic H3 lysine-4 methylation correlates with protection against DNA methylation
Next, we explored whether H3 lysine-4 methylation could be enriched at maternal ICRs in male germ cells. As an internal control, we chose a housekeeping gene, Rpl30 (Ribosomal protein L30), which is expressed at high levels in both male and female germ cells (Supplementary Figure S3). We focused on H3 lysine-4 trimethylation (H3K4me3) given its direct link with active promoters in somatic cells (Bernstein et al, 2006; Zhao et al, 2007). Indeed, in the PGCs this modification showed high levels of precipitation at Rpl30, as in somatic cells (Figure 2C). In the male PGCs, high levels of H3K4me3 were detected at the maternal ICRs Snrpn DMR and KvDMR1, at both 13.5 and 15.5 d.p.c. (Figure 3A). H3K4me3 was equally present on both the parental chromosomes (Figure 3B).
If H3 lysine-4 methylation prevents acquisition of DNA methylation in vivo, one expects this modification to be absent from paternal ICRs at the time of imprint acquisition in the male germ cells. This is precisely what we found at Ig-DMR and H19 DMR, at 13.5 d.p.c., and even more pronouncedly so at 15.5 d.p.c. (Figure 3A), when paternal DNA methylation establishment initiates (Figure 1).
Transcription is linked to the specificity of imprint acquisition
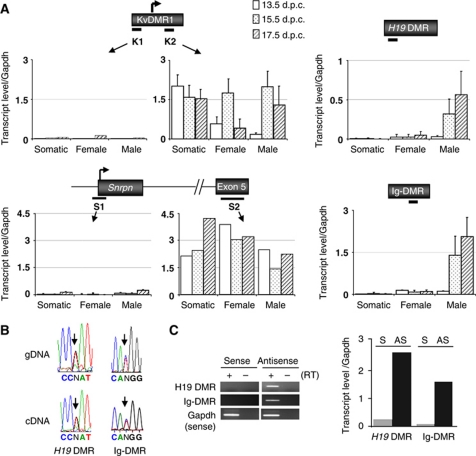
Next, we explored whether the differential histone methylation between ICRs could be linked to transcriptional events. cDNAs were synthesised from total RNAs using random oligonucleotides, followed by quantitative PCR amplification (Supplementary Table S1). First, we analysed the maternal ICRs Snrpn DMR and KvDMR1. Both these CpG island promoters produce mono-allelic, long transcripts in somatic cells, which are readily detected downstream of their transcriptional initiation sites (Bartolomei, 2009). With a primer pair at the 3′ side of the KvDMR1, high levels of transcript were detected in both male and female PGCs, at 13.5, 15.5 and 17.5 d.p.c. (Figure 4A). To assess whether transcription originated from within the KvDMR1, we also amplified the cDNA with a primer pair at the 5′ extremity of the KvDMR1, upstream of the somatic transcription start site. As in somatic cells, no amplification was detected in the germ cells at this region. At the Snrpn DMR, transcripts are readily amplified (from both the parental alleles) between Snrpn exons 1 and 3 (Supplementary Figure S4); similarly, high transcript levels were detected with primers at exon 5 (Figure 4A). A second primer pair, located directly 5′ of its somatic promoter (see Figure 4A), and primers spanning further upstream U1 promoters (Supplementary Figure S4), did not give significant amplification. Although we did not determine the precise transcription start sites, these results suggest that the KvDMR1 and Snrpn DMR act as promoters in PGCs as well. At several other maternal ICRs analysed (ZacI, Grb10, Impact), we also detected promoter activity in the male germ cells at 15.5 and 17.5 d.p.c. (Supplementary Figure S4).
Figure 4.
Biallelic transcription across paternal ICRs at the time of imprint establishment. (A) Quantitative RT–PCR analysis of male and female germ cells and gonad somatic control cells (somatic) at 13.5, 15.5, and 17.5 d.p.c. Black dashes indicate the positions of the amplified regions (see Supplementary Table SI). The H19 DMR and Ig-DMR were each analysed by quantitative PCR at one other region as well, which gave comparable results (Figure 5; Supplementary Figure S6). For KvDMR1 and Snrpn DMR, amplifications were performed upstream (K1 and S1, respectively) and downstream (K2 and S2, respectively) of the known transcription start site (see also Supplementary Figure S4). Measured levels of transcripts for each of the analysed regions are normalised to that of Gapdh. (B) H19 DMR and Ig-DMR germ cell transcripts are biallelic. Sequence traces of RT–PCR products are shown in the lower panel (cDNA), with an arrow indicating SNPs. As control sequences, sequence traces obtained from genomic DNA (gDNA) are shown. (C) Strand-specific RT–PCR analysis. Reverse transcription reactions with 15.5 d.p.c male PGC RNA were performed with primers specific for the sense (S) or antisense (AS) H19-DMR and Ig-DMR transcripts, respectively. As a positive control, reactions were multiplexed with a primer set specific for Gapdh, further used to normalise the data. Non-quantitative (left panel) and quantitative (right panel) analyses are shown.
Next, we explored transcription at the H19 DMR and the Ig-DMR (Figure 4A). In agreement with earlier reports (Drewell et al, 2002; Schoenfelder et al, 2007), only low levels of transcription were detected at the H19 DMR in somatic tissues and embryos (Supplementary Figure S5). Hardly any transcription was detected at the H19 DMR and the Ig-DMR in the FACS-sorted somatic control cells either. In female PGCs, only weak transcription was observed at the two paternal ICRs at the three developmental stages analysed (Figure 4A; Supplementary Figure S5).
A strikingly different picture emerged analysing male germ cells. Whereas at 13.5 d.p.c., virtually no transcription is detected at the Ig-DMR and H19 DMR, the signal increased tremendously by 15.5 d.p.c., at the onset of imprint acquisition, to levels comparable to those of the housekeeping genes Gapdh and Rpl30. The high expression of Ig-DMR and H19 DMR RNAs persisted at 17.5 d.p.c. (Figure 4A; Supplementary Figure S5), when almost full DNA methylation had been acquired (Figure 1). Similarly high levels were observed with primers pairs at the 5′ and the 3′ side of each of the ICRs (Figure 5A; Supplementary Figure S6). Sequencing of amplification products indicated that the germ cell transcripts at the Ig-DMR and H19 DMR originated from both the parental chromosomes (Figure 4B). By analysing cDNA, reverse transcribed using stand-specific oligonucleotides, next we determined from which of the two DNA strands the male germ cell transcripts originated. At both the Ig-DMR and the H19 DMR, transcription was detected from one strand predominantly. At the H19 DMR, transcription originated predominantly from the side where the close-by H19 gene is located. At the Ig-DMR, transcription was found to come from the side where the Gtl2 gene is located (Figure 4C).
Figure 5.
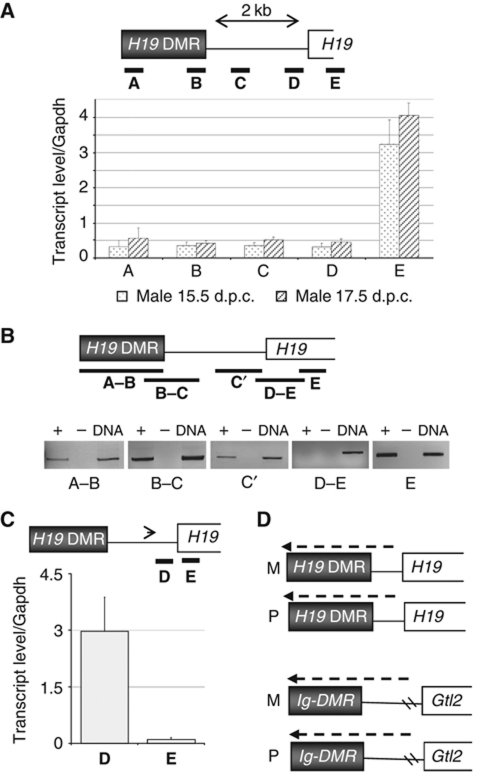
Characterisation of H19 DMR and Ig-DMR non-coding RNAs in male germ cells. (A) Quantitative RT–PCR analysis at 15.5 and 17.5 d.p.c. Five regions (regions A–E, black bars) were analysed and measured transcript levels were normalised to that of Gapdh. (B) RT–PCR in male PGCs at 15.5 d.p.c. (followed by gel electrophoresis). Five regions (A–B, B–C, C′, D–E, E; black bars) were analysed. Region D–E, spanning the H19 promoter, failed to amplify in multiple experiments. +, With reverse transcriptase (RT); −, without RT; DNA, control amplification on genomic DNA. (C) Quantification following strand-specific RT–PCR. The arrowhead indicates the relative position of the strand-specific oligonucleotide used to produce cDNA. Measured levels of transcripts were normalised to that of Gapdh. (D) Schematic summary of biallelic transcription across the H19 DMR and Ig-DMR in male germ cells at the time of imprint establishment. Transcripts are in the opposite direction of the main nearby genes, H19 and Gtl2, respectively.
In further analyses, transcription was also detected between the H19 DMR and the H19 gene. With four primer pairs in the H19 gene-upstream region A–D, comparably high transcript levels were detected, at both 15.5 and 17.5 d.p.c. (Figure 5A). Amplification of several longer RT–PCR fragments (Figure 5B) strongly suggests that these amplification events correspond to a single transcript, which is predominantly nuclear in its localisation (Supplementary Figure S7).
Importantly, no RT–PCR product was amplified in a region (region D–E, Figure 5B) overlapping the promoter of H19 gene. Furthermore, at exon-1 of H19 (region E) transcription was much higher than at the upstream regions A–D (Figure 5A), also suggesting that independent transcription events occur upstream and downstream of the H19 promoter, respectively. Consistently, by strand-specific RT–PCR dedicated to amplify (H19) antisense transcription events, much higher levels of transcription were detected upstream (region D) than downstream (region E) of the H19 promoter (Figure 5C). Combined, these data suggest that transcription across the H19 DMR predominantly initiates upstream of the H19 gene, which itself is highly transcribed in the male PGC cells as well. However, expression of the RNAs crossing the H19 DMR seems not linked to the expression of the H19 gene. In female PGCs, H19 is more highly expressed than in male PGCs, whereas the antisense transcript crossing the H19 DMR is expressed at highest levels in male PGCs. In somatic control cells, H19 is expressed 25-fold higher than in the male PGCs, and yet, the upstream non-coding RNA (ncRNA) is not detected (Supplementary Figure S8).
Because of repeat sequences in this region, it was difficult to estimate the precise start site of the Ig-DMR transcripts in the male PGCs. However, transcription was detected at a region between the Ig-DMR and Gtl2 as well, and cDNA amplification was readily achieved across this ICR (Supplementary Figure S6). Combined, our data indicate high levels of biallelic transcription across the Ig-DMR and H19 DMR ICRs, on one of the two strands predominantly (Figure 5D). We assume that these are ncRNAs, given the absence of significant open reading frames (of >100 bp) in the regions concerned. The concordant levels of transcription across regions A–D for the H19 DMR, and A-C the Ig-DMR (Figure 5A; Supplementary Figure S6) suggest that these ncRNAs are unspliced. Indeed, bio-informatic analysis (Genescan) shows that the H19 and Ig-DMR and their flanking regions do not comprise canonical splice donor and acceptor sites.
Discussion
Our main finding is that maternal and paternal ICRs have distinct states of histone methylation and transcription at the time of imprint acquisition in male germ cells. At maternal ICRs, a strong correlation is found between promoter activity, H3 lysine-4 methylation and in-vivo protection against DNA methylation. Biallelic transcriptional read-through, and absence of H3K4me3, correlate with acquisition of DNA methylation at the paternal ICRs H19 DMR and Ig-DMR. These data evoke the possibility that histone methylation and transcriptional events are linked to the specificity of imprint acquisition in male germ cells.
Given the relatively high numbers of FACS-sorted cells that were still required, we focused our cChIP studies on histone modifications that could potentially influence DNA methylation. We found that maternal ICR promoters were biallelically active in male PGCs and this correlated with biallelic enrichment of H3K4me3, a mark which prevents recruitment of DNMT3A–DNMT3L complexes in vitro (Ooi et al, 2007; Zhang et al, 2010). We did not explore H3K4me2, since we failed to achieve allelic precipitation at ICRs in somatic control cells, indicating that the different sera tested were not good-enough for cChIP (Henckel et al, data not shown). Whether, similarly as in somatic cells (Mikkelsen et al, 2007; Zhao et al, 2007), all active promoters are marked by H3K4me3 in PGCs, is not known. However, high levels of H3K4me3 were detected at the control housekeeping genes and at the transcriptionally active H19 gene as well. Gene promoters are generally active in male germ cells at the time of imprint acquisition (Guo et al, 2004). Indeed, at several other maternal ICRs (ZacI, Grb10, Impact), we detected promoter activity in the male germ cells as well (Supplementary Figure S4). Whether promoter-associated H3-lysine-4 methylation is maintained through post meiotic stages of spermatogenesis as well, is not known. However, at the KvDMR1 and Snrpn DMR, chromatin is enriched in H3K4me2/3 in post meiotic spermatocytes and elongating spermatids as well (Delaval et al, 2007).
In non-mammalian model species, including Neurospora crassa (Tamaru et al, 2003), H3K9me3 facilitates acquisition of DNA methylation. Our quantitative measurements following cChIP showed that by 13.5 d.p.c., this repressive mark was completely lost from ICRs in male (and female) germ cells. Although a recent ChIP study detected H3K9me3 at the H19 DMR in male PGCs at this developmental stage, no quantitative data were included to assess its degree of enrichment (Lee et al, 2010). The complete depletion of H3K9me3 at ICRs observed in our study contrasts with the presence of H3K9me3 at pericentric chromatin, which does not decrease significantly during PGC development, at least till 13.5 d.p.c. (Seki et al, 2005). Also, at IAP elements we detected persistence of H3K9me3. At all ICRs analysed, we also noted depletion of H4K20me3 in the male (and female) PGCs. These repressive histone marks, therefore, do not signal subsequent acquisition of DNA methylation, at least not at the paternal ICRs.
The loss of H3K9me3 and H4K20me3 in PGCs complements our previous observation in mouse embryos that repressive histone modifications at ICRs depend on the presence of DNA methylation (Henckel et al, 2009). Consequently, the loss of H3K9me3 and H4K20me3 in PGCs could be linked to the active, wide-spread removal of DNA methylation which occurs from ∼10 to 12.5 d.p.c. (Hajkova et al, 2002). Conversely, the deposition of these repressive marks onto the methylated alleles of ICRs most likely occurs after fertilisation only, in the developing embryo. In agreement with this scenario, the DNA methylation imprints at paternal ICRs are not associated with H3K9me3 and H4K20me3 in spermatocytes and spermatids (Delaval et al, 2007).
H2A/H4R3me2s was recently shown to be required for DNA methylation acquisition at the silenced Globin genes in primary erythroid progenitors, and this PRMT5-mediated H2A/H4 arginine methylation constituted a direct binding target of DNMT3A (Zhao et al, 2009). Since in early germ cells, BLIMP1 associates with PRMT5 to control global levels of H2A/H4R3me2s (Ancelin et al, 2006), we explored whether there could be enrichment of this mark at imprinted loci. Indeed, high levels of biallelic H2A/H4R3me2s were detected at ICRs in 13.5 and 15.5 d.p.c. germ cells. Although H2A/H4R3me2s has been hypothesised to facilitate DNMT3A recruitment at ICRs in male PGCs (Jelinic et al, 2006), it would not provide the signal indicating which ICRs need to become methylated since it was detected at both maternal and paternal ICRs.
In the male germ cells, we detected high levels of transcription through the H19 DMR and Ig-DMR at 15.5 d.p.c., at the beginning of imprint establishment. Transcription levels were still high at 17.5 d.p.c., when methylation acquisition was almost complete. One hypothesis, therefore, could be that the acquired CpG methylation had led to enhanced transcription through these ICRs. A recent study on neurogenic gene loci provides evidence for such a mechanism in which DNA methylation enhances transcription (Wu et al, 2010). In an earlier study on post meiotic male germ cells (which have full DNA methylation at paternal ICRs), we did not detect transcription across the Ig-DMR and H19-DMR (Delaval et al, 2007). An alternative, non-exclusive, hypothesis is that transcription across the paternal ICRs facilitates imprint acquisition in the male PGCs. Insights into such a mechanism have recently been obtained relative to imprint acquisition in the female germ line. At the Gnas locus on mouse distal chromosome 2, DNA methylation acquisition in growing oocytes requires transcription across the ICR (Chotalia et al, 2009). How, precisely, transcription could contribute to acquisition of de-novo methylation in germ cells is unknown, but insights have emerged from studies on somatic cells. X-linked genes, for instance, have higher levels of DNA methylation on the transcriptionally active than on the inactive X chromosome (Hellman and Chess, 2007). The observed strand-preference of transcription across both H19 DMR and Ig-DMR could be relevant in relation to recent work on rDNA genes, showing RNA-dependent acquisition of DNA methylation is linked to the formation of RNA–DNA triplexes (Schmitz et al, 2010). Together with insights from other model systems, our combined data make us to propose a working model (Figure 6) in which a specific histone modification state and transcriptional read-through, producing long ncRNAs, guide the acquisition of DNA methylation at paternal ICRs.
Figure 6.
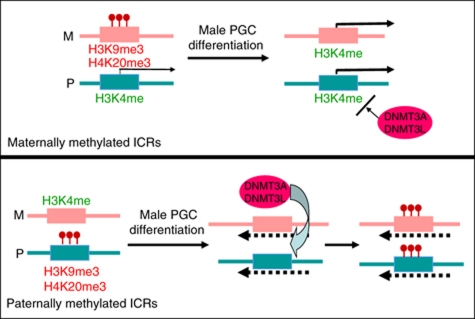
Model for imprint acquisition in male germ cells. At the maternally methylated ICRs (upper part), which comprise promoters, during male PGC differentiation there is loss of repressive histone methylation and acquisition of biallelic H3 K4 methylation. The latter correlates with biallelic promoter activity and prevents recruitment of DNMT3A–DNMT3L complexes to the DNA. At paternally methylated ICRs (lower part), there is also loss of allele-specific repressive histone methylation during male PGC differentiation, as well as a complete depletion of already-low levels of H3K4me3. Possibly involving facilitating histone modifications, such as H2A/H4R3me2s, this histone modification state allows recruitment of DNMT3A–DNMT3L complexes, the signal for which could be provided by transcription through these intergenic ICRs (dashed lines). Our data indicate that the model could be pertinent in particular for the Ig-DMR and the H19 DMR.
Imprinting acquisition in transgenic contexts has been observed for large constructs which comprised the ICR and sequences up to the 3′ part of the H19 gene (Cranston et al, 2001). When ectopically inserted, the H19 DMR itself is not sufficient for DNA methylation acquisition in male germ cells, although methylation can sometimes be acquired during early development (Matsuzaki et al, 2009). This finding agrees with a role of the observed transcriptional read-through initiating upstream of the H19 gene in PGCs.
It seems unlikely that all paternal ICRs use the same imprinting mechanism. The Rasgrf1 locus on mouse chromosome 9 is clearly different from the others, and was not included in this study. It does not have an equivalent in humans and its methylation acquisition requires not only DNMT3A, but also DNMT3B (Kato et al, 2007). Acquisition of the paternal imprint at this retrotransposed locus involves small RNAs and depends on the expression of Mili, Miwi2, and MitoPLD, all components of the PIWI-interacting RNA (piRNA) pathway (Shoji et al, 2009; Watanabe et al, 2011). Methylation at the H19 DMR and Ig-DMR, in contrast, is not regulated by components of the piRNA pathway, and no small RNAs were detected in male germ cells at these ICRs (Shoji et al, 2009; Watanabe et al, 2006, 2011). Thus, whereas imprint acquisition at the Ig-DMR and H19 DMR shows striking similarities to the mechanisms thought to control imprint establishment in oocytes (Chotalia et al, 2009; Ciccone et al, 2009), the process could be different at other paternal ICRs.
Materials and methods
Germ cell collection
Gonads were trypsinised in the presence of 0.2 mg/ml of DNAse-I, filtered through 40 μm cell strainers (BD Falcon™) and cells were sorted using a FACS-Aria™ (BD Biosciences). Both GFP-positive (germ cells) and GFP-negative (somatic control) cells were collected. On average, ∼7000 germ cells were obtained per gonad.
Carrier ChIP
Drosophila melanogaster S2 cells were washed with PBS and mixed with mouse cells of interest (10 million S2 cells per 100 000 mouse cells). Nuclei were extracted as described (Umlauf et al, 2004; Henckel et al, 2009), resuspended in 200–300 μl of ‘Nuclei PURE Storage Buffer’ (SIGMA, S9183), and stored at −80°C. Batches of nuclei were thawed on ice and pooled to get ∼75 000–100 000 mammalian cells of interest per ChIP. Chromatin was digested for 3 min at 37°C with MNase (at 7.5 U/ml, USB/Affymetrix), purified as described (Umlauf et al, 2004; Henckel et al, 2009), and precleared with 50 μl of protein-A sepharose overnight at 4°C before ChIP. Immunoprecipitations were performed in a 500-μl volume overnight at 4°C, with 10 μg of antibodies bound to Dynabeads protein-A magnetic beads (Invitrogen). Details on antibodies are in Supplementary Table S2. Bead-bound chromatin was washed with 1 ml of three successive washing buffers (Henckel et al, 2009), three times for each, and DNA was purified using NucleoSpin Extract II kit (Macherey Nagel). Primers used for qualitative and quantitative PCR analysis are provided in Supplementary Table S1. cChIP data were obtained from at least three (and up to five) assays, performed on independent chromatin preparations, except for the data on male germ cells at 15.5 d.p.c. (Figure 3A), which derived from two independent experiments.
Quantitative analysis of immunoprecipitated DNA by real-time PCR amplification
Input and antibody-bound fractions were quantified by real-time PCR amplification with a SYBR Green mixture (Qiagen) using an MX3000 apparatus (Stratagene). Background precipitation levels were determined by performing mock precipitations with a non-specific IgG antiserum (Sigma C-2288), and were only a fraction of that observed in the precipitations with specific antisera. Bound/Input ratios were calculated and were normalised, according to the antiserum used, against precipitation at IAP elements or Rpl30.
Bisulphite sequencing and RNA expression analysis
DNA extraction and bisulphite sequencing were conducted on batches of ∼15 000 cells, as described (Henckel et al, 2009). For each region analysed, CpG methylation profiles were obtained from at least three independent bisulphite treatments, with analysis of two or more independent PCR products per treatment. For each amplicon, methylation patterns were also assessed by digestion with ‘diagnostic’ restriction endonucleases (the ‘Cobra’ approach) followed by direct sequencing (data not shown). The two different approaches gave concordant results.
For reverse transcription, total RNA was extracted from batches of 30 000–100 000 cells using Trizol Reagent (Invitrogen). After digestion with RNase-free DNase-I, first-strand cDNA was generated by reverse transcription with Superscript-II (Invitrogen), or Quantiteck Reverse transcript Kit (Qiagen), using randomised primers. Duplicate sets of control samples were produced with the reverse transcriptase omitted to detect amplification from contaminating DNA. PCR amplifications on the cDNAs were performed using gene-specific primers (Supplementary Table S1). RT–PCR products were quantified by real-time PCR amplification with a SYBR Green mix (Qiagen) using an MX3000 apparatus (Stratagene). Gapdh transcript levels were used for normalisation (Supplementary Table S1). RNA expression analyses were repeated at least three times (and up to six times).
Supplementary Material
Acknowledgments
We thank Jérôme Cavaillé and Michael Weber for reading the manuscript; Azim Surani (Gurdon Institute, Cambridge, UK) and Wolf Reik (The Babraham Institute, Cambridge, UK) for the Oct4-GFP mice; and Myriam Boyer-Clavel and Cedric Mongellaz from the Montpellier RIO Imaging platform (MRI) for their help with cytometry. This work was grant supported by the Agence National de Recherche (ANR blanc ‘EMPREINTE’), the European Chemical Industry Council (CEFIC) Long Research Initiative (EMSG49-CNRS-08), and La Ligue Contre le Cancer. AH acknowledges a PhD Fellowship received from the Association de Recherche contre le Cancer (ARC).
Author contributions: PA and RF designed the study; AH, PA, and SKK performed the experiments; KC supervised and organised the mouse embryology work; PA, RF and AH analysed the data; RF wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA (2006) Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol 8: 623–630 [DOI] [PubMed] [Google Scholar]
- Arnaud P (2010) Genomic imprinting in germ cells: imprints are under control. Reproduction 140: 411–423 [DOI] [PubMed] [Google Scholar]
- Bartolomei MS (2009) Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev 23: 2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH (2001) Dnmt3L and the establishment of maternal genomic imprints. Science 294: 2536–2539 [DOI] [PubMed] [Google Scholar]
- Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G (2009) Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev 23: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T (2009) KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461: 415–418 [DOI] [PubMed] [Google Scholar]
- Cranston MJ, Spinka TL, Elson DA, Bartolomei MS (2001) Elucidation of the minimal sequence required to imprint H19 transgenes. Genomics 73: 98–107 [DOI] [PubMed] [Google Scholar]
- Davis TL, Yang GL, McCarrey JR, Bartolomei MS (2000) The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet 9: 2885–2894 [DOI] [PubMed] [Google Scholar]
- Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R (2007) Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J 26: 720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewell RA, Arney KL, Arima T, Barton SC, Brenton JD, Surani MA (2002) Novel conserved elements upstream of the H19 gene are transcribed and act as mesodermal enhancers. Development 129: 1205–1213 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ (2002) Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet 32: 426–431 [DOI] [PubMed] [Google Scholar]
- Guo R, Yu Z, Guan J, Ge Y, Ma J, Li S, Wang S, Xue S, Han D (2004) Stage-specific and tissue-specific expression characteristics of differentially expressed genes during mouse spermatogenesis. Mol Reprod Dev 67: 264–272 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA (2002) Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117: 15–23 [DOI] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E (2002) Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129: 1983–1993 [DOI] [PubMed] [Google Scholar]
- Hellman A, Chess A (2007) Gene body-specific methylation on the active X chromosome. Science 315: 1141–1143 [DOI] [PubMed] [Google Scholar]
- Henckel A, Nakabayashi K, Sanz LA, Feil R, Hata K, Arnaud P (2009) Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals. Hum Mol Genet 18: 3375–3383 [DOI] [PubMed] [Google Scholar]
- Hiura H, Sugawara A, Ogawa H, John RM, Miyauchi N, Miyanari Y, Horiike T, Li Y, Yaegashi N, Sasaki H, Kono T, Arima T (2010) A tripartite paternally methylated region within the Gpr1-Zdbf2 imprinted domain on mouse chromosome 1 identified by meDIP-on-chip. Nucleic Acids Res 38: 4929–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinic P, Stehle JC, Shaw P (2006) The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol 4: e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H (2004) Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429: 900–903 [DOI] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H (2007) Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16: 2272–2280 [DOI] [PubMed] [Google Scholar]
- Koide T, Moriwaki K, Uchida K, Mita A, Sagai T, Yonekawa H, Katoh H, Miyashita N, Tsuchiya K, Nielsen TJ, Shiroishi T (1998) A new inbred strain JF1 established from Japanese fancy mouse carrying the classic piebald allele. Mamm Genome 9: 15–19 [DOI] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W (2003) Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35: 88–93 [DOI] [PubMed] [Google Scholar]
- Lee D-H, Singh P, Tsai SY, Oates N, Spalla A, Spalla C, Brown L, Rivas G, Larson G, Rauch TA, Pfeifer GP, Szabó PE (2010) CTCF-dependent chromatin bias constitutes transient epigenetic memory of the mother at the H19-Igf2 imprinting control region in prospermatogonia. PLOS Genet 6: e1001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BZ, Huang Z, Cui QY, Song XH, Du L, Jeltsch A, Chen P, Li G, Li E, Xu GL (2011) Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res 21: 1172–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC (2003) Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet 35: 97–102 [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Okamura E, Shimotsuma M, Fukamizu A, Tanimoto K (2009) A randomly integrated transgenic H19 imprinting control region acquires methylation imprinting independently of its establishment in germ cells. Mol Cell Biol 29: 4595–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O′Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LP, VerMilyea MD, Turner BM (2006) Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet 38: 835–841 [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH (2007) DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M (2009) Structural basis for recognition of H4K4 methylation status by DNA methyltransferase 3A ATRX-DNMT3-DNMTL domain. EMBO Rep 10: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannetier M, Julien E, Schotta G, Tardat M, Sardet C, Jenuwein T, Feil R (2008) PR-SET7 and SUV4-20H regulate H4 lysine-20 methylation at imprinting control regions in the mouse. EMBO Rep 9: 998–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Smits G, Fraser P, Reik W, Paro R (2007) Non-coding transcripts in the H19 imprinting control region mediate gene silencing in transgenic Drosophila. EMBO Rep 8: 1068–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24: 2264–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y (2005) Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278: 440–458 [DOI] [PubMed] [Google Scholar]
- Shemer R, Birger Y, Riggs AD, Razin A (1997) Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA 94: 10267–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, Hata K, Martin SL, Noce T, Kuramochi-Miyagawa S, Nakano T, Sasaki H, Pillai RS, Nakatsuji N, Chuma S (2009) The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 17: 775–787 [DOI] [PubMed] [Google Scholar]
- Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU (2003) Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet 34: 75–79 [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS (1995) A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet 9: 407–413 [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R (2004) Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet 36: 1296–1300 [DOI] [PubMed] [Google Scholar]
- Verona RI, Thorvaldsen JT, Reese KJ, Bartolomei MS (2008) The transcriptional status but not the imprinting control region determines allele-specific histone modifications at the imprinted H19 Locus. Mol Cell Biol 28: 71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H (2006) Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 20: 1732–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, Gotoh K, Hiura H, Arima T, Fujiyama A, Sado T, Shibata T, Nakano T, Lin H, Ichiyanagi K, Soloway PD et al. (2011) Role of piRNA and non-coding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332: 848–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE (2010) Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329: 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu T, Sugiyama N, De Felice M, Yeom YI, Ohbo K, Masuko K, Obinata M, Abe K, Scholer HR, Matsui Y (1999) Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev Growth Differ 41: 675–684 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, Jeltsch A (2010) Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res 38: 4246–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM (2009) PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol 16: 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng HH, Wei CL (2007) Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1: 286–298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.