Abstract
The four serotypes of dengue virus (DENV-1 to -4) cause the most important emerging viral disease. Protein E, the principal viral envelope glycoprotein, mediates fusion of the viral and endosomal membranes during virus entry and is the target of neutralizing antibodies. However, the epitopes of strongly neutralizing human antibodies have not been described despite their importance to vaccine development. The chimpanzee Mab 5H2 potently neutralizes DENV-4 by binding to domain I of E. The crystal structure of Fab 5H2 bound to E from DENV-4 shows that antibody binding prevents formation of the fusogenic hairpin conformation of E, which together with in-vitro assays, demonstrates that 5H2 neutralizes by blocking membrane fusion in the endosome. Furthermore, we show that human sera from patients recovering from DENV-4 infection contain antibodies that bind to the 5H2 epitope region on domain I. This study, thus, provides new information and tools for effective vaccine design to prevent dengue disease.
Keywords: antibody, dengue, structure
Introduction
The four serotypes of the mosquito-transmitted dengue virus (DENV) constitute the largest vector-borne viral disease burden on the planet (Monath, 1994). There is neither an approved vaccine nor specific therapy against these pathogens. DENV belongs to the flavivirus genus of the Flaviviridae family, which includes other important human pathogens such as Japanese encephalitis (JE), West Nile (WN), yellow fever (YF), and tick borne encephalitis (TBE) viruses (Lindenbach and Rice, 2001). The virus particles measure around 500 Å in diameter and possess a lipid bilayer that harbours 180 copies of the membrane and envelope (E) glycoproteins. Entry into host cells occurs via receptor-mediated endocytosis followed by low pH induced fusion of the viral and endosomal membranes (van der Schaar et al, 2008). The E protein is required for both steps of the entry pathway.
Protein E displays about 30% sequence variability across DENV serotypes. Crystal structures are available for the pre- and/or post-fusion forms for the soluble ectodomain of E (sE) from several flaviviruses, including DENV serotypes 1–3 (Rey et al, 1995; Modis et al, 2003, 2004, 2005; Bressanelli et al, 2004; Zhang et al, 2004; Kanai et al, 2006; Nybakken et al, 2006; Nayak et al, 2009). The E protein fold and virion architecture are conserved in all flaviviruses (Lindenbach and Rice, 2001). sE contains three domains (DI, DII and DIII). DI consists of a 9-stranded β-barrel with strands labelled A0 through I0. DII is formed by insertions in loops D0E0 and H0I0 and carries a hydrophobic fusion loop at its tip. DIII has an immunoglobulin superfamily fold and is thought to bind to cell surface receptors (Crill and Roehrig, 2001). The ectodomain is attached to the viral membrane by a downstream amphipathic stem region followed by a double trans-membrane helix at the C-terminus of the protein. In the mature virion, 90 E dimers form a closed shell around the viral membrane, defining the outer surface of the mature virus particle (Kuhn et al, 2002). In the acidic conditions of the endosome, the E subunits dissociate into monomers and insert their fusion loops into the endosomal membrane, which induces the co-axial trimerization of the protein via DI and DII. The simultaneous or subsequent folding back of DIII and stem regions against the lateral surface of the trimer then brings the viral and target membranes together, in the first step of membrane fusion (Bressanelli et al, 2004; Modis et al, 2004; Nayak et al, 2009). However, information about intermediate states of E along the fusion pathway is lacking, and as such the molecular details of how the assembly of trimers is coupled to DIII relocation remain obscure. Likewise, how this enormous reorganization of the E protein subunits is accomplished within the context of the flavivirus virion is unknown.
The E protein is the target of potently neutralizing antibodies against DENV and other flaviviruses. Several flavivirus immunocomplexes have been studied by cryo-electron microscopy (EM) in combination with X-ray crystallography (Nybakken et al, 2005; Kaufmann et al, 2006; Lok et al, 2008; Cherrier et al, 2009). A murine monoclonal antibody (Mab) binding to DIII from DENV-1 through DENV-3 was shown to prevent cell attachment by disrupting the virion architecture (Lok et al, 2008), while two WN virus-specific Mabs of murine and human origin were found to neutralize post-attachment by interfering with the acid induced disassembly of the E glycoprotein shell during the initial stages of the membrane fusion pathway (Kaufmann et al, 2009, 2010; Thompson et al, 2009).
Dengue infection confers life-long immunity to the infecting serotype only (Sabin, 1952). Subsequent infections by different serotypes carry an elevated risk of life-threatening disease (Halstead, 2003). Weakly neutralizing, cross-reactive antibodies that bind to virus particles from multiple serotypes dominate the human immune response to dengue (Stiasny et al, 2006; Lai et al, 2008; Crill et al, 2009). Severe disease associated with heterotypic DENV infections is thought to involve enhancement of DENV infection of Fcγ receptor-bearing cells by these antibodies, a phenomenon called antibody-dependent enhancement (ADE; Halstead, 2003). Consequently, a safe vaccine would need to protect against all four DENV serotypes. The epitopes recognized by strongly neutralizing, serotype-specific human antibodies are of particular interest, although none have been described. DIII carries the epitopes of potent, serotype-specific murine antibodies (Gromowski and Barrett, 2007; Sukupolvi-Petty et al, 2007; Shrestha et al, 2010; Wahala et al, 2010), but the role of anti-DIII antibodies in the human immune response to DENV is unclear (Crill et al, 2009; Wahala et al, 2009). Analysis of memory B cells from DENV-infected individuals identified potently neutralizing, serotype-specific IgGs binding to unknown epitopes on either DI or DII (Beltramello et al, 2010). Repertoire cloning studies on bone marrow samples from DENV-infected chimpanzees yielded similar results, leading to the isolation of the potently neutralizing Mab 5H2 (Men et al, 2004), which is specific for DENV-4 and binds to an epitope on DI (Lai et al, 2007), a portion of E whose immunogenic properties are poorly characterized. Here, we performed a structural and functional characterization of Mab 5H2.
Results
Crystal structures of DENV 4 sE in complex with Fab 5H2
We determined the crystal structure of DENV-4 sE in complex with Fab 5H2 to 3.2 Å resolution. The asymmetric unit contains two copies of sE, organized as in the flavivirus pre-fusion head-to-tail dimer (Rey et al, 1995), with each E subunit bound by a Fab molecule (Figure 1A and B). Analysis of this structure showed that the 5H2 epitope resides on DI. Therefore, in an attempt to improve the resolution, we then engineered a construct that allowed the production of the isolated recombinant DI (rDI; see Figure 1C and Materials and methods). We obtained crystals of rDI in complex with the Fab that diffracted to 2.7 Å resolution and contained two copies of the complex in the asymmetric unit. The statistics of the crystallographic analyses and refinement of the atomic models are provided in Table I. The structure of DI from the rDI/Fab complex can be superposed on its counterpart from the sE/Fab structure with a root mean square deviation (r.m.s.d.) of 0.8 Å for 88 equivalent Cα atoms (Supplementary Figure S1A), showing that DI can fold independently of the presence of the other domains of E. In particular, the conformation of the rDI C-terminal region is identical to that of DI within sE, up to and including residue 295, such that the N-terminal portion of the DI/DIII linker is in the correct configuration in spite of the absence of DIII. Superposition of the rDI/Fab complexes onto the sE/Fab complexes, omitting the Fab constant domains from the calculation, gives r.m.s.d of 0.6–0.8 Å for 314–318 equivalent Cα atoms (Supplementary Figure S1B). Comparison of the structures of the rDI/Fab and sE/Fab complexes showed that the Fab recognizes essentially identical sets of epitope residues, which maintain an identical conformation as far as we can tell, given the difference in resolution of the two structures. The antibody/antigen complex buries ∼850 Å2 of accessible surface area per molecule, with a shape correlation factor of 0.61 (Lawrence and Colman, 1993), which are values typical of antibody–antigen complexes.
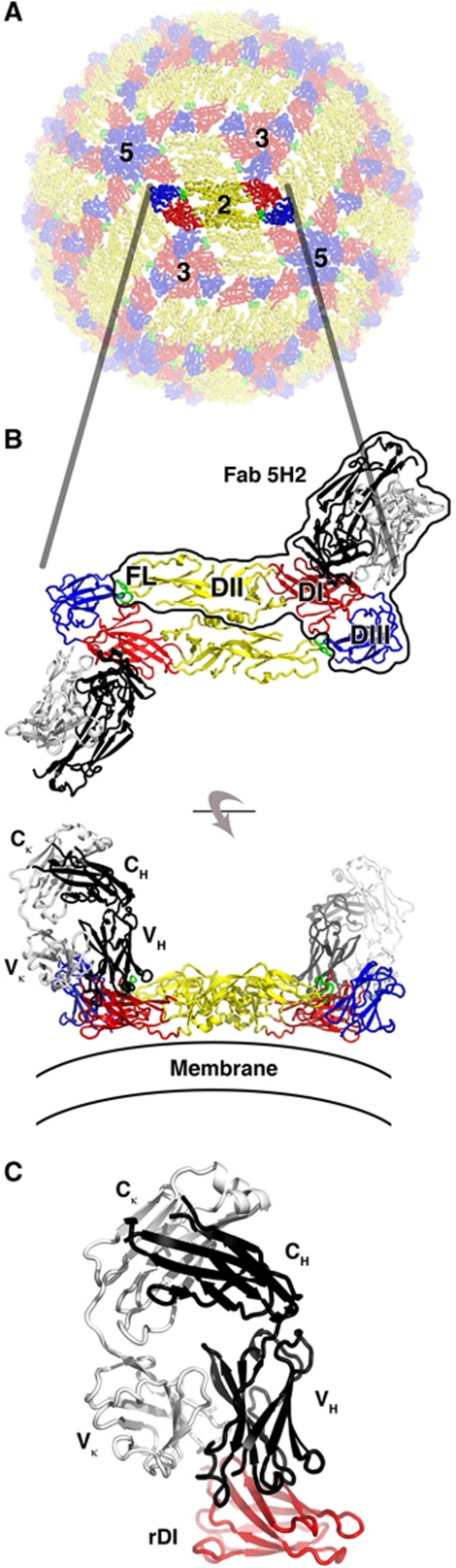
Figure 1.
Crystal structures of Fab 5H2 in complex with DENV-4 sE and rDI. (A) Structure of the E glycoprotein shell from the mature DENV-2 virion (PDB accession code 1THD). Domain I (residues 1–52, 132–192 and 281–295), domain II (residues 53–131 and 193–280) and domain III (residues 296–395) are shown in red, yellow and blue, respectively. The fusion loop (residues 100–108) is shown in green. One E dimer has been emphasized for clarity. The adjacent icosahedral five-, three- and two-fold symmetry axes are indicated. (B) Two views of the DENV-4 sE dimer in complex with Fab 5H2. The heavy and light chains of the Fabs are shown in dark and light grey, respectively. The viewing direction in the upper panel corresponds roughly to that in (A). One copy of the sE/Fab complex has been outlined. The lower panel shows the structure viewed 90° away, roughly in the plane of the E glycoprotein shell in the virus. The variable and constant domains of the heavy chain and κ light chain are labelled. The viral membrane is indicated. (C) One copy of the rDI/Fab crystal structure with rDI shown in red.
Table 1. Data collection and refinement statistics.
| Crystal features | sE: Fab 5H2 | DI: Fab 5H2 |
|---|---|---|
| Space group | P21 | P212121 |
| No. of copies per asymmetric unit | 2 | 2 |
| Unit cell parameters | ||
| a, b, c (Å) | 95.11, 134.75, 106.08 | 78.94, 113.86, 169.58 |
| α, β, γ (deg) | 90, 106.70, 90 | 90, 90, 90 |
| Data quality | ||
| Resolution (Å)a | 47.74–3.23 (3.41–3.23) | 47.27–2.71 (2.86–2.71) |
| No. of observations | 135 123 (16475) | 167 940 (11882) |
| Number of unique reflections | 39 919 (5445) | 41 316 (5211) |
| Completeness (%) | 97.3 (91.7) | 97.7 (86.4) |
| Redundancy | 3.4 (3.0) | 4.1 (2.3) |
| I/σ(I) | 10.0 (2.0) | 11.3 (2.0) |
| Rsym (%)a,b | 10.2 (46.5) | 9.0 (42.9) |
| Model quality | ||
| Resolution range of refinement (Å) | 47.74–3.23 | 15.00–2.71 |
| R/Rfree (%)c | 24.1/24.6 | 22.2/25.2 |
| No. of atoms (protein/carbohydrate/solvent) | 12 319 (12291/28/0) | 8529 (8120/0/409) |
| Average atomic B-factor | 69.5 | 58.5 |
| R.m.s. deviation in bond lengths/angles (Å, deg) | 0.007/0.90 | 0.007/0.97 |
| Ramachandran plotd | ||
| Favoured (%) | 95.65 | 95.85 |
| Allowed (%) | 4.03 | 3.49 |
| Disallowed (%) | 0.32 | 0.66 |
| aValues in parentheses correspond to the highest resolution shell. | ||
| bRsym=100 × [∑h∑i∣Ii(h)−〈I(h)〉∣]/∑h∑iIi(h) where 〈I(h)〉 is the mean of all observations Ii(h) of reflection h. | ||
| cR=∑h∣Fo(h)−Fc(h)∣/∑h Fo(h) where Fo(h) and Fc(h) are the observed and calculated structure factor amplitudes of reflection h. Rfree corresponds to a randomly selected 5% of reflections excluded from the refinement. | ||
| dCalculated using the molprobity webserver (http://molprobity.biochem.duke.edu/). | ||
The conformation rDI N-terminal region
The DENV-4 rDI structure differs significantly, however, from DI in sE in the first 20 residues of the molecule, comprising the N-terminus, the A0 strand and the A0B0 loop (Supplementary Figure S1C). In the pre-fusion dimer, the N-terminal end of sE is located at the DI/DIII interface, with residues 8–13 forming the A0 strand and residues 14–19 in the A0B0 loop (Figure 2A), as observed previously for DENV-2 and DENV-3 (Modis et al, 2003, 2005; Zhang et al, 2004). By contrast, in rDI, the A0 strand consists of residues 4–6, so that rDI A0B0 loop (residues 7–19) is longer than in sE (Figure 2B). Residues 1–20 do not interact with the bound Fab. Furthermore, the observed conformation is the same in the two copies of the rDI/Fab complex in the crystal asymmetric unit, indicating that it does not result from crystal-packing interactions. The conformation of these residues is, therefore, likely to correspond to the lowest free energy state of DI when DIII is absent.
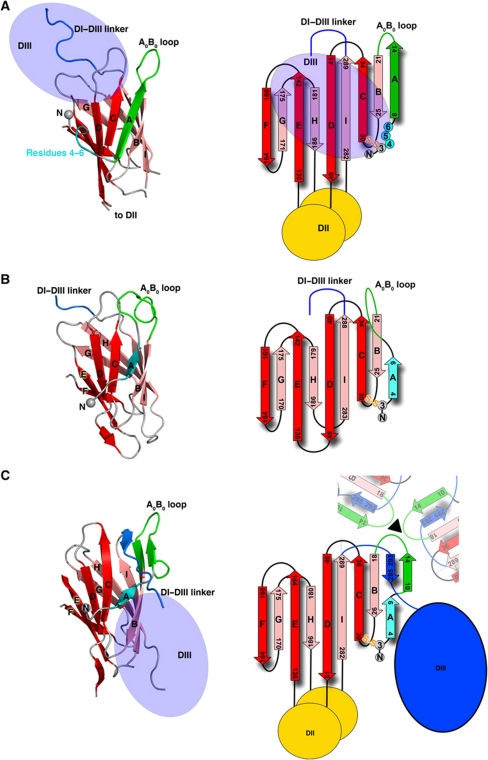
Figure 2.
Structural analysis of the rDI N-terminal region. (A–C) DI from DENV-4 sE (A), DENV-4 rDI (B) and the post-fusion form of the DENV-2 sE (C) are shown. For each part of the figure, the panel on the left shows the structure of DI from the relevant crystal structure while the right panel shows a schematic representation of the DI secondary structure. The colour scheme is the same in both panels. Beta-strands on the ACDEF face of the DI β-barrel are shown in red, while those on the opposing BIHG face are shown in salmon pink. The A0–I0 strands are labelled A–I. The N-terminal residue is represented by a grey sphere. Residues 4–6, 8–20 and 293–300 are coloured cyan, green and blue, respectively. Domains II and III are represented by yellow and blue ovals where appropriate. In the secondary structure schematics, ‘S-S’ represents the disulphide bridge between residues Cys3 and Cys30. The schematic in (C) also indicates the interactions with the A0-B0 loops of neighbouring subunits around the three-fold axis of the trimer.
The fusogenic conformational change of sE from dimer to trimer is accompanied by a remodelling of the sE N-terminal region and the DI/DIII linker (Modis et al, 2004). In the post-fusion sE trimer, the DI A0 strand is also formed by residues 4–6, as in rDI (Figure 2C). In the trimer, residues 10–14 in the A0B0 loop form an additional β-strand that clips onto the B0 strand, directing the polypeptide chain towards the trimer three-fold axis, where it interacts with its counterparts in the adjacent E subunits. This rearrangement makes room for the DI–DIII linker in its post-fusion conformation, which inserts as a short β-strand between the A0 and C0 strands. In the structure of the rDI/Fab complex, the conformation of residues 1–6 is very similar to that observed in the post-fusion form of sE (Supplementary Figure S1C; Figure 2B and C). However, the rDI A0B0 loop is instead turned towards the C0 strand, thus occluding the post-fusion binding site for the DI–DIII linker (the latter being in its pre-fusion conformation, accordingly; Figure 2B). These results show that the N-terminal region of DI spontaneously adopts a conformation that is mid-way between those found in the pre- and post-fusion forms when DIII is absent. Hence, the N-terminal region of E could adopt this conformation when the pre-fusion DI–DIII interface is broken during the fusogenic conformational change.
The 5H2 epitope
Mab 5H2 was raised against DENV-4 strain 814669, which is 100% identical in the E protein gene to strain Myanmar 1976 used in our constructs. The 5H2 epitope is located on the exposed, F0G0 edge of the DI β-barrel. The Fab contacts β-strands F0, G0 and H0 and the loops connecting them, as well as the polypeptide segment downstream of β-strand I0, in the DI–DIII linker (Figure 3A). The antibody–antigen interactions are listed in Supplementary Tables 1 and 2 (see Supplementary data). The side chains of E residues Glu172 and Lys174 (G0 strand), Asp177 (G0H0 loop), Glu180 (H0 strand), and Arg293 hydrogen bond to residues in the Fab (Figure 3B). Of particular note are residues Lys174 and Arg293 (coloured green in Figure 3A and B). Residue Lys174 forms the only salt bridge across the interface, with residue AspH100e in the complementarity determining region (CDR) H3 loop of the Fab. Residue Arg293 is part of the linker between DI and DIII, which undergoes a major relocation during the fusogenic conformational change of E. This residue stacks beneath ArgL32 (L2 loop) and donates a hydrogen bond to AsnL92 (L3 loop). Despite the absence of DIII in the rDI/Fab complex, the electron density maps of both the rDI/Fab and sE/Fab complexes are very clear in this region and show identical interactions.
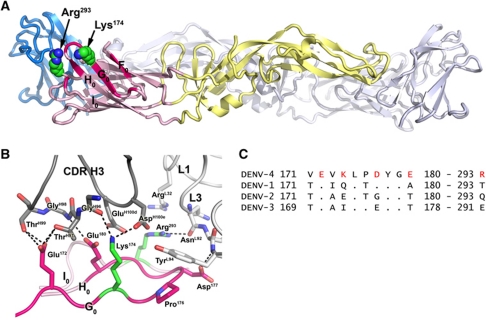
Figure 3.
The 5H2 epitope. (A) Side view of the DENV-4 sE dimer. Domains I-III of the near subunit are coloured light pink, pale yellow and blue, respectively. The 5H2 epitope residues on DI are coloured magenta. Strands F0-I0 are labelled. The side chains of residues Lys174 and Arg293 are shown as spheres (green—carbon and blue—nitrogen). (B) Close-up of the antibody–antigen interface from the rDI/Fab structure. Residues in E follow the colour scheme in (A). The heavy and light chains of the Fab are in dark and light grey. Carbon atoms follow the main chain colour scheme, while nitrogen and oxygen atoms are blue and red, respectively. Hydrogen bonds and salt bridges are shown as dashed lines. The CDR loops H3, L1 and L3 of the antibody are indicated. (C) Amino-acid sequence comparison between the 5H2 epitope region on DENV-4 strain Myanmar 1976, and the corresponding portion of E taken the from consensus sequences for DENV-1 to -3. DENV-4 residues that use their side chains to form salt bridges or hydrogen bonds with the antibody are written in red. A dot denotes conservation with DENV-4.
Aside from the conserved residue Glu172, the epitope residues that engage the antibody using their side chains (Lys174, Asp177, Glu180, and Arg293) are variable across serotypes (Figure 3C). In particular, the consensus residue 174 is glutamine, glutamate, or isoleucine in DENV-1, -2, and -3, respectively. DENV-4 neutralization escape variants generated against 5H2 most commonly harbour a single mutation, Lys174Glu, which completely abrogates 5H2 binding (Lai et al, 2007). The unique identity and interactions of DENV-4 residue Lys174 are thus a critical determinant of Mab 5H2 binding affinity and specificity. Additionally, the interactions made by Asp177, Glu180, and Arg293 described above would also be generally incompatible with the identity of the corresponding residues in the other DENV serotypes (Figure 3B and C), explaining the specificity of 5H2 for DENV-4 strains.
The epitopes of serotype-specific murine antibodies binding to DIII have been shown to harbour sequence variations between genotypes that can have major effects on Mab binding and neutralization efficacy (Balsitis et al, 2010; Brien et al, 2010; Sukupolvi-Petty et al, 2010; Wahala et al, 2010). The key residues of the 5H2 epitope (Glu172, Lys174, Pro176, Asp177, Glu180, and Arg293) are conserved in almost all (28/30) DENV-4 strains present in the database. Accordingly, Mab 5H2 was previously shown to neutralize three geographically diverse DENV-4 strains (814669, H241, and 341750) with equally high efficiency (Men et al, 2004). Thus, we anticipate that 5H2 will efficiently neutralize the vast majority of DENV-4 strains currently circulating worldwide.
5H2 inhibits fusogenic conformational changes in E
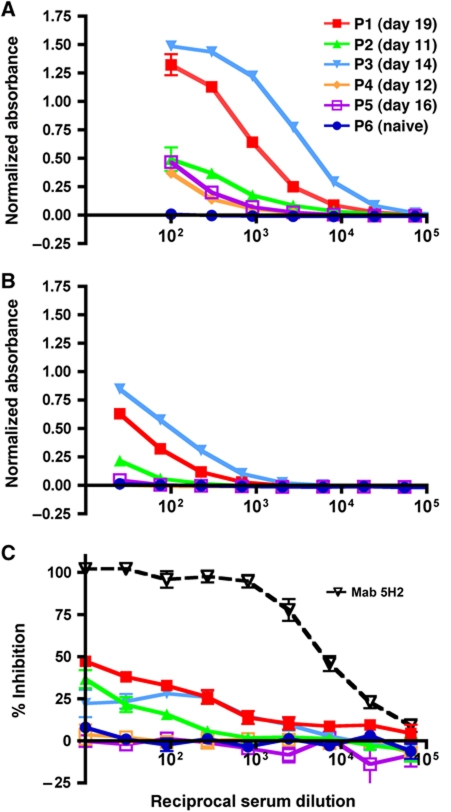
Mab 5H2 was previously shown to block a post-attachment step of the DENV-4 entry pathway, since treatment of DENV-4 virions with the antibody either before or after virus binding to cultured cells is equally effective in blocking infection (Lai et al, 2007). This suggests that 5H2 neutralizes by preventing fusion of the viral and endosomal membranes, as found for other anti-flavivirus Mabs (Crill and Roehrig, 2001; Nybakken et al, 2005; Kaufmann et al, 2006, 2009, 2010; Thompson et al, 2009; Vogt et al, 2009). Indeed, two Mabs binding to the 5H2 epitope region on the TBE virus E protein were shown to block viral membrane fusion with liposomes (Stiasny et al, 2007). We tested the effect of Mab 5H2 on viral membrane fusion with liposomes using a previously reported assay (Zaitseva et al, 2010; see Materials and methods). For this purpose, DENV-4 virions whose membranes had been pre-labelled with a self-quenching concentration of the fluorescent lipid DiD, were incubated with various concentrations of Mab 5H2 and mixed with liposomes that mimic the phospholipid composition of late endosomes. Fusion of the viral membranes with the liposomes was then induced by acidification to pH 5.5, resulting in dilution of DiD in the liposomes and an increase in DiD fluorescence due to de-quenching. Mab 5H2 inhibited lipid mixing in a dose-dependent manner (Figure 4A and B), to levels comparable to those observed for two fusion-blocking human Mabs against WN virus, CR4354 and CR4348 (Vogt et al, 2009). Indirect ELISAs using Mab 5H2 and immobilized rDI, performed at pH 5.5 and 7.0, yielded Kd values of 2.9 nM (95% confidence interval 2.6–3.3 nM) and 1.3 nM (1.2–1.5 nM), respectively (see Supplementary Figure S2), indicating that endosomal acidification would not significantly reduce the affinity of the antibody for the virus. These observations strongly suggest that neutralization of DENV-4 by Mab 5H2 involves blocking membrane fusion within the endosome.
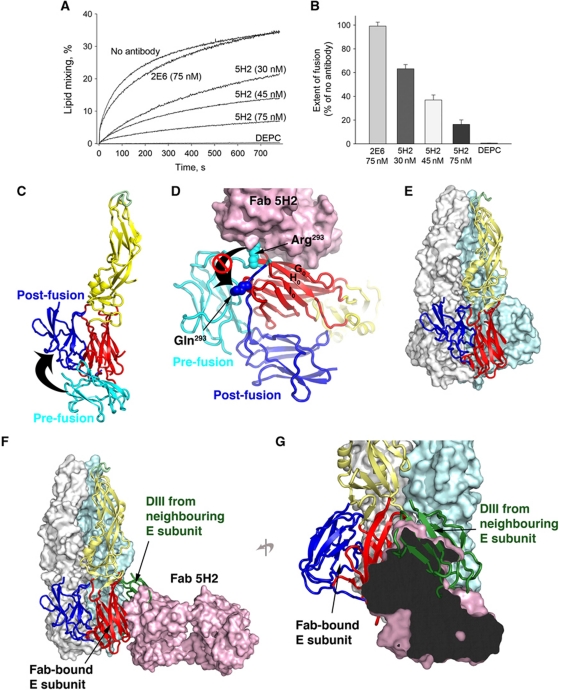
Figure 4.
Inhibition of E-mediated membrane fusion by Mab 5H2. (A, B) Inhibition of fusion between DiD-labelled DENV-4 virions and liposomes by Mab 5H2. The graph in (A) shows membrane fusion measured online (see Materials and methods) following acidification to pH 5.5, for DENV-4 virions pre-incubated with the indicated concentration of Mab 5H2, a non-binding control Mab 2E6, no antibody, or diethylpyrocarbonate (DEPC). (B) The extent of fusion, measured at 700 s post acidification as a percentage of the DiD signal with no antibody. Data for at least three independent experiments are shown. (C) The fusogenic conformational change of a single E subunit. The DENV-4 sE subunit, as imaged in our crystal structure of the sE/Fab complex, is shown with DI–DIII coloured red, yellow and cyan, respectively, with the fusion loop in green. The position of DIII in the post-fusion conformation is shown in blue. This was obtained by superposing the structure of the post-fusion form of the DENV-2 sE onto the DENV-4 sE subunit using residues in DI. (D) 5H2 binding stabilizes the pre-fusion form of the E subunit to which it is bound. The pre-fusion DENV-4 sE in complex with Fab 5H2 is coloured as in (C) with the Fab shown in pink space-fill. The position of DIII and the DI–DIII linker in the post-fusion state, based on the post-fusion DENV-2 sE structure as described above, are shown in blue. The side chains of residues Arg293 and Gln293, which are located in the DI–DIII linkers of DENV-4 and DENV-2, respectively, are shown as spheres. (E) The DENV-2 post-fusion sE trimer (PDB accession code 1OK8). The near subunit is shown in cartoon representation with DI–DIII coloured red, yellow and blue and the fusion loop in green. The adjacent subunits in the trimer are shown in white and pale-cyan space-fill. (F, G) Fab 5H2 would block DIII relocation of the adjacent E subunit in the trimer. Fab 5H2 (pink space-fill) was docked onto DI of the near sE subunit (see Materials and methods). (F) A close-up view with part of the molecular surface of the Fab cut away, indicating the steric overlap between the Fab and the DIII from a neighbouring E subunit (dark green cartoon).
A key feature of the fusogenic conformational change of E is the relocation of DIII (Bressanelli et al, 2004; Modis et al, 2004; Nayak et al, 2009; Figure 4C). This is accompanied by a substantial swing-around of the residues in the DI–DIII linker, which would require residue Arg293 to shift ∼10 Å away from its position in the pre-fusion state (Figure 4D). The interactions of Mab 5H2 with this residue (Figure 3B) are thus likely to prevent this movement (Figure 4D). To further investigate the effects of 5H2 on the membrane fusion transition, we docked our DENV-4 rDI/Fab 5H2 crystal structure onto one subunit in the DENV-2 sE post-fusion trimer (see Figure 4E and F and Materials and methods). In this location, the Fab clashes severely with DIII from the neighbouring subunit in the trimer (Figure 4F and G): thus, the bound Fab would prevent the fusogenic conformational change in the adjacent subunit by occluding the lateral binding site for its DIII. Thus, 5H2 appears to directly prohibit the fusogenic conformational change simultaneously in two E subunits in the pre-hairpin trimer. A recent study showed that stable association of the DENV-2 sE trimer with liposomes requires at least two out of the three subunits to adopt the post-fusion hairpin conformation (Liao et al, 2010). These observations strongly indicate that pre-hairpin trimers containing one Fab-bound subunit would be incapable of pursuing their transition into the fusogenic hairpin conformation.
The 5H2 epitope region is immunogenic in humans
Comparison of the 5H2 sequence with the chimpanzee and human antibody germ-line genes (see Materials and methods and Supplementary Figure S3) demonstrates that a human equivalent to 5H2 (hu5H2) sharing 98% amino-acid sequence identity with 5H2 over its variable domains can be constructed from the gene segments listed in Table II. It is, therefore, possible that hu5H2 would be raised in humans in response to infection by DENV-4. To investigate this, we analysed five serum samples from patients convalescing from secondary DENV-4 infections, to see their IgG binding responses to purified DENV-4 rDI and sE by indirect ELISA (Figure 5A and B). The absorbance values were standardized using Mab 5H2 (see Materials and methods), allowing the binding profiles of each serum to rDI and sE to be quantitatively compared. All (5/5) patient sera tested bound to sE (Figure 5A), while those from patients 1–3 showed a lower but nonetheless clear response to rDI whose magnitude varied between samples in-step with the corresponding variations in the sE response (Figure 5B). Naive serum (patient 6) showed essentially no binding to either sE or rDI. An analysis of the ELISA binding profiles estimated that 3.6, 2.3 and 3.0% of the sE-specific IgG from patients 1–3 binds to rDI (see Materials and methods). We then performed competition ELISA experiments that quantified the ability of antibodies in the sera to inhibit the binding of rDI to immobilized Mab 5H2. Patient sera 1–3 inhibited rDI binding to the plate by 20–50% at the lowest dilution (1/10), while sera 4 and 5 showed essentially no inhibition (Figure 5C), consistent with the low levels of anti-rDI antibodies in these samples. These results suggest that at least a portion of the anti-rDI antibodies in patient sera bind to epitopes on DI that overlap with that of Mab 5H2.
Table 2. The chimpanzee and human antibody germ-line genes closest in sequence to 5H2a.
| Geneb | Chimpanzee |
Human |
|||
|---|---|---|---|---|---|
| Genome locationc | ID 5H2 (%)d | IMGT ref.e | ID |
||
| 5H2 (%)d | Chimp (%)f | ||||
| V H | 14/-/107109845-107110132 | 89 | IGHV4-59*04 | 87 | 98 |
| D | NI | — | IGHD1-7*01 | 64 | — |
| J H | 14/-/106345815-106345864 | 79 | IGHJ3*01 | 79 | 98 |
| V κ | 2a_random/-/2274595-2274877 | 96 | IGKV-13*02 | 95 | 97 |
| J κ | 2a/-/89920744-89920782 | 100 | IGKJ4*01 | 97 | 100 |
| aSee also Supplementary Figure S3 and Materials and Methods. | |||||
| bV, variable; D, diversity; J, junction. The subscript denotes the heavy (H) or κ-light (κ) chain. | |||||
| cChromosome/strand/nucleotide_start-nucleotide_end. NI—not identified. | |||||
| dPercent nucleotide sequence identity with 5H2. | |||||
| eNamed according to the IMGT convention (http://www.imgt.org/). | |||||
| fNucleotide sequence identity with the corresponding chimpanzee genes. | |||||
Figure 5.
Sera from patients recovering from DENV-4 infection contain antibodies that bind to the 5H2 epitope region on DI. (A, B) Indirect ELISAs using immobilized DENV-4 sE (A) or rDI (B) and human serum samples taken from five patients (P1–5) recovering from DENV-4 infection, and naive serum (P6). The day post-onset of symptoms on which the sample was taken is given in the key. (C) Inhibition of rDI binding to immobilized Mab 5H2 by the same serum samples, evaluated by competition ELISA. Reciprocal serum dilution values on the x-axis are on a log scale. Error bars show the standard error of the mean. When they are not visible they are smaller than the marker.
Discussion
A major reorganization of the flavivirus E protein takes place during virus entry into target cells. In response to the acidic environment of the endosome, the E dimers in the mature virion dissociate into monomers, which then re-assemble into fusogenic trimers. Crystal structures are available for the pre- and post-fusion forms of sE, but a detailed molecular understanding of the fusion transition is lacking due to the absence of information about intermediate states. The observed conformation of rDI provides a snapshot of an intermediate conformation of this domain during the transition from dimer to trimer. In this intermediate, the A0B0 loop adopts a conformation that occludes the post-fusion binding site for the DI–DIII linker, preventing DIII relocation. Once the E subunits have formed the pre-fusion trimer, the A0B0 loops would change into the post-fusion conformation, forming inter-subunit contacts with each other and concomitantly exposing the post-fusion binding sites for the DI–DIII linkers. Thus, the A0B0 loop might act as a molecular switch that permits DIII relocation to proceed only within the context of the correctly assembled pre-hairpin trimer, and ensures that this occurs simultaneously in all three subunits.
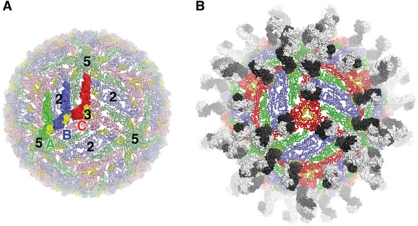
To investigate how 5H2 would bind to the mature DENV-4 virion, we docked our crystal structures onto the available quasi-atomic model of the mature DENV-2 virion (Zhang et al, 2004; see Materials and methods), the accepted model for flavivirus organization in general. The icosahedral asymmetric unit of the mature flavivirus virion contains three independent E protein subunits (referred to as A, B and C—Figure 6A) that inhabit chemically distinct environments (Kuhn et al, 2002). This exercise suggests that 5H2 can bind unencumbered to subunits A and B, but not to subunit C, the latter burying its epitope in contacts with three-fold symmetry-related E molecules (Figure 6B; Supplementary Figure S5). Thus, we predict that the mature DENV-4 virion has 120 epitopes that are accessible for 5H2 binding, as found for every flavivirus/Fab complex imaged experimentally to date (Kaufmann et al, 2006, 2010; Lok et al, 2008; Cherrier et al, 2009). The C subunits in our model may provide a source of fusion-competent E protomers, requiring a greater proportion of the accessible epitopes on the virion to be bound by the antibody to bring about neutralization. Although our crystal structures suggest that 5H2 would prohibit the formation of functional trimers, it is also possible that the antibody also interferes with earlier stages of the fusion pathway, as seen for two antibodies against WN virus (Kaufmann et al, 2006, 2009, 2010). Further studies, including a cryo-EM reconstruction of the DENV-4/Fab 5H2 immunocomplex, will be required to explore fully how 5H2 disables the viral membrane fusion apparatus. 5H2 may prove to be useful tool to provide further new insights into how the membrane fusion pathway is implemented in the context of the virion, and how the architecture of the mature virion relates to its function as a membrane fusion machine.
Figure 6.
Model of Fab 5H2 binding to the mature DENV-4 virion. (A) Overview of the quasi-atomic model of the DENV-4 E protein shell, constructed as described in Materials and methods. The three unique E protein subunits in the icosahedral asymmetric unit (subunits A–C) are shown in space-filling representation and coloured green, blue and red, respectively. The symmetry copies of these subunits are coloured light green, light blue and pink, respectively, and are shown as loop trace. The Mab 5H2 epitopes are coloured yellow. The icosahedral five-, three- and two-fold symmetry operators bordering on or existing within one icosahedral facet are labelled. (B) Predicted decoration of the mature DENV-4 virion by Fab 5H2. Here, all of the A, B and C subunits are coloured green, blue and red, respectively. The heavy/light chains of the Fab molecules are coloured dark grey/white.
The strongly neutralizing DENV epitopes targeted by the human immune system are unknown, despite their obvious importance to vaccine development. Analysis of immune sera and memory B cells recovered from infected individuals indicates that long-term, serotype-specific immunity to DENV involves antibodies that bind to undescribed epitopes located on either DI or DII of the E protein (Crill et al, 2009; Wahala et al, 2009; Beltramello et al, 2010). Here, we have shown that convalescent phase patient sera can contain antibodies that bind to DI and that the epitopes of at least some of these antibodies overlap with the 5H2 epitope. Interestingly, a small number of DENV-4 strains, which belong to genotype III, possess glutamate in plate of lysine at position 174 of E (Klungthong et al, 2004), which abrogates 5H2 binding (Lai et al, 2007) as discussed above. Perhaps, this mutation has arisen from selective pressure from 5H2-like antibodies in humans. Further studies will be required to further characterize DI-specific human antibodies. We have demonstrated here for the first time that correctly folded rDI can be produced in the absence of the other E protein domains. Recombinant DIs could be used as new tools to further probe the antigenic structure of E in humans. Furthermore, tetravalent formulations of DENV rDIs offer an unexplored vaccination strategy with the potential to elicit strongly neutralizing serotype-specific antibodies simultaneously against all four serotypes.
Materials and methods
Protein production, purification and crystallization
A DNA fragment containing the DENV-4 (strain Myanmar 1976; unpublished) genomic region coding for the prM-sE (1680 nt in total including all of prM and the E ectodomain, ending at codon 394 of E) was amplified by PCR using specific primers and inserted into the plasmid pT351. This is a shuttle vector containing selection markers for yeast and E. coli, as well as a metallothionein-inducible expression cassette for Drosophila cells. In the construct, called pT351/DENV-4 sE, the prM-sE sequence is in frame with the Drosophila BiP signal peptide, which directs the recombinant protein to the secretory pathway, and with a StrepTag (http://www.iba.com) for affinity purification at its C-terminus, preceded by an enterokinase cleavage site for specific proteolytic removal of the tag. The presence of the prM sequence ensures folding of sE in the ER in the presence of its natural chaperone. Upon prM cleavage by Drosophila Furin in the TGN, the sE dimer dissociates from the complex and is found secreted in the medium. Drosophila S2 cells (Invitrogen, Carlsbad, CA) were co-transfected with pT351/DENV-4 sE and a vector conferring resistance to puromycin, using the effectene transfection reagent (Qiagen). The selected cells were adapted to serum-free growth medium and grown to high density before induction with 500 μM CuSO4. The supernatant was collected 10 days later, concentrated using a flow concentration system with a 10-kDa cutoff membrane (Vivascience), and sE purified by affinity chromatography using a streptactin column. The eluate was concentrated and further purified by size-exclusion chromatography, using a superdex 200 10/300 column (GE healthcare) with 0.5 M NaCl and 50 mM Tris (pH 8.0). For rDI, we used a synthetic gene (Genscript) corresponding to DENV-4 E residues 1–50, 135–190 and 281–298. The D0 and E0 loops were joined by inserting a Gly-Gly linker between residues 50 and 135, while a Thr residue was inserted in-between residues Gly190 and Gly281 to link the G0 and H0 strands. This was cloned into the pT351 vector, expressed in Drosophila S2 cells and purified from supernatants exactly as described for the prM-sE construct above. The purified proteins were concentrated to 8 mg/ml (sE) and 10 mg/ml (rDI) in 150 mM NaCl, 50 mM TRIS pH 8.0.
The recombinant, His-tagged 5H2 Fab fragment was produced in the periplasm of E. coli essentially as described previously (Men et al, 2004). DNA fragments encoding the Fab 5H2 heavy- and light-chain sequences were cloned into a pCOMB3H phage display vector that had been modified to encode a six-residue histidine tag at the 3′-end of the heavy-chain insert (Glamann et al, 1998). This plasmid was transformed into E. coli strain XL-1 Blue. Selected colonies were grown to an early exponential phase (optical density at 600 nm of ∼0.2) in 1 L of L-broth containing 1% glucose, 100 mg/l ampicillin and 10 mg/l tetracycline at 30°C. Fab production was induced by transferring the bacteria to 2 L of L-broth containing 100 mg/l ampicillin, 10 mg/l tetracycline and 0.1 mM isopropyl-β-D-thio-galactopyranoside (IPTG) and growing for 5 h at 30°C. The bacteria were pelleted and re-suspended in 50 mM sodium phosphate pH 8, 10 mM Tris–HCl pH 8, 100 mM NaCl, 5 mM MgCl2 and 20 mM imidazole. The soluble Fab product was released from the bacterial periplasm by three cycles of freeze thaw, and the preparation clarified by centrifugation. The recombinant Fab was purified from the supernatant by immobilized metal affinity chromatography followed by cation exchange chromatography. Fab 5H2 containing fractions were pooled and concentrated to 11 mg/ml in 20 mM HEPES, pH 7.3, 100 mM NaCl.
The sE/Fab and rDI/Fab complexes were formed by mixing the purified proteins at 1:1 stoichiometry and an overall concentration of ∼10 mg/ml. Vapour diffusion crystallization trials were performed immediately, without further purification or incubation of the sample, at 19°C. Drops were formed by mixing equal volumes of the protein and reservoir solution. Crystals of the sE/Fab complex grew from 8% polyethylene glycol 8000, 8% 4-methyl pentane diol (MPD) and 0.1 M HEPES, pH 7.5 as the reservoir solution, while the rDI/Fab complex was equilibrated against 2.0 M ammonium sulphate. The crystals were cryo-cooled in liquid nitrogen after they were first transferred into a cryo-protectant solution consisting of the reservoir solution condition supplemented with 25% MPD (sE/Fab) or 25% glycerol (rDI/Fab).
Structure determination and analysis
The X-ray data were collected at the Swiss Light Source beamline PX1 using the high-resolution diffractometer set-up and the Pilatus 6M detector (Broennimann et al, 2006). Data were processed using XDS (Kabsch, 1988). Scaling and reduction were performed using SCALA (Evans, 2005) and programs from the CCP4 suite (Collaborative Computing Project No. 4, 1994). Starting phases for crystals of the sE/Fab complex were obtained by molecular replacement using MOLREP (Vagin and Teplyakov, 1997). Here, the crystal structures of a Fab fragment from an anti-uranyl antibody (E Stura, unpublished) and the DENV-2 sE (PDB accession code 1OKE) were used as search models. The structure was rebuilt in COOT (Emsley and Cowtan, 2004) and refined using a pre-release version of BUSTER (Bricogne, 1993), with restrained all-atom B-factor refinement and local structure similarity non-crystallographic symmetry (LSSNCS) restraints. The structure of the rDI/Fab complex was solved by molecular replacement using the corresponding portion of the sE/Fab 5H2 complex. The structure of the rDI/Fab was refined in BUSTER using restrained all-atom B-factor refinement, LSSNCS restraints and TLS refinement with each domain defined as a separate group. Water molecules were added automatically in the later stages of refinement and verified manually. Structural superpositions, buried surface area and surface complementarity coefficient calculations, and inter-molecular contacts were performed using programs from the CCP4 suite (Collaborative Computing Project No. 4, 1994). Figures were prepared in PYMOL (http://pymol.sourceforge.net).
The model of Fab 5H2 bound to the post-fusion form of the E protein was generated by superposing the rDI/Fab complex (chains A, H and L) onto the post-fusion structure of DENV-2 sE (PDB accession code 1OK8; Modis et al, 2004) via superposition of the corresponding DIs (r.m.s.d.=1.2 Å between equivalent DI Cα atoms). Binding of Fab 5H2 to the mature DENV-4 virion was modelled by superposing the DI–DIII/Fab and DII from our 3.2 Å resolution sE/Fab 5H2 crystal structure (chains A, H and L) separately onto the DI–DIII and DII, respectively, of each E protein subunit in the icosahedral asymmetric unit of the quasi-atomic model of the DENV-2 E protein shell (PDB accession code 1THD). Icosahedral symmetry was then applied. This gave r.m.s.d. in Cα coordinates of 0.9 and 1.1 Å over DI–DIII and DII, respectively. This method mimicked the original fitting procedure that was used to position the DENV-2 sE protein crystal structure into the 9.5 Å resolution cryo-EM reconstruction of the mature DENV-2 virion (Zhang et al, 2004).
DENV-4 and cultured cells
Mosquito C6/36 cells (American Type Culture Collection, Manassas, VA) were grown in minimum essential medium (MEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 0.05 mg/ml gentamicin and 2.5 units/ml amphotericin B (Fungizone). Production and purification of DENV-4 was performed essentially as described in Kuhn et al (2002). Confluent C6/36 cells were infected with DENV-4 strain 814669 at multiplicity 0.2. After 4 days, the virus was harvested and cleared from cell debris. Subsequently, virions were precipitated with 8% polyethylene glycol 8000 overnight, and re-suspended in NTE buffer (20 mM Tris–HCl, 120 mM NaCl, 1 mM EDTA). Virions were purified on 10–35% potassium tartrate step gradients by ultracentrifugation at 4°C for 2 h at 35 000 r.p.m. The virus fraction was concentrated and cleared using Amicon centrifugal filter devices (Millipore, Billerica, MA, USA). The DENV-4 purity was analysed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE). The viral titre was determined by focus assay in Vero cells.
Antibodies
Large-scale production and purification of full-length humanized Mab 5H2 (Men et al, 2004) following transient transfection of 293T cells was performed by Kemp Biotechnology (Gaithersburg, MD). The humanized chimpanzee MAb 2E6, which was used as a non-binding control in the fusion inhibition assays (described below), was derived by repertoire cloning from a chimpanzee initially immunized with Langat virus (LGTV) and then boosted with attenuated TBE virus/DENV-4 chimera. The TBE virus/DENV-4 chimera is a chimeric flavivirus in which the prM and E genes of DENV-4 have been substituted for their counterparts from LGTV. Mab 2E6 is specific for the TBE virus complex and neutralizes LGTV and the TBE virus/DENV-4 chimera at a high titre in vitro (manuscript in preparation).
Fusion inhibition assay
We characterized fusogenic activity of DENV-4 virions towards liposomes as described in Zaitseva et al (2010). Briefly, viral particles were labelled with a self-quenching concentration of the fluorescent lipid DiD from a Vybrant cell-labeling kit (Molecular Probes, Eugene, OR). Large unilamellar liposomes of 100 nm diameter were formed by extrusion from the mixture of 1,2-dioleoyl-sn-glycero-3-phosphocholine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, soy L-α-phosphatidylinositol, bis(monooleoylglycero) phosphate, S,R Isomer (all from Avanti Polar Lipids, Alabaster, AL) in a molar ratio of 5:2:1:2. This lipid mixture mimics the phospholipid composition of late endosomal membranes (Kobayashi et al, 2002). Mixtures of purified DENV-4 virions (∼105 focus forming units in 100 μl) with various concentrations of Mab 5H2 or control (Mab 2E6) were pre-incubated for 30 min at 37°C and then for 2 h at 4°C, prior to addition to liposomes (in 20 mM Tricine-HCl and 140 mM NaCl, pH 7.8) to give a final lipid concentration of 30 μM in a volume of 2 ml. The fusion reaction was triggered by adding a pre-titrated amount of MES/acetic acid buffer to reach pH 5.5. We recorded fluorescence at excitation and emission wavelengths of 620 and 665 nm, respectively, using an Aminco Bowman Series 2 luminescence spectrometer (Rochester, NY). At the end of each recording, we added Triton X-100 to a final concentration of 0.1% to fully de-quench DiD (‘100% lipid mixing’). In control experiments, we abolished the fusogenic activity of DENV-4 by a 15-min incubation with 2 mM diethylpyrocarbonate (DEPC, Sigma, St Louis, MO), a histidine-modifying reagent (Zaitseva et al, 2010).
Sequence analysis of the 5H2 variable domains
In order to establish the chimpanzee antibody germ-line genes from which 5H2 derives, we performed a BLAT search (http://genome.ucsc.edu/) of the 5H2 nucleotide sequence against the chimpanzee genome (March 2006 CGSC 2.1/panTro2 assembly—Chimpanzee Sequencing and Analysis Consortium, 2005). This gave the VH, Vκ and Jκ genes listed in Table II as the matches with the greatest nucleotide sequence identity with 5H2. The JH gene could not be retrieved using this method, probably because of the large number of somatic hypermutations in the fourth framework region of the 5H2 heavy chain (see Supplementary Figure S3). A BLAT search against the chimpanzee genome using the human JH sequence (identified below) as the query returned the chimpanzee JH gene listed in Table II. We were unable to identify the chimpanzee D segment, which was not surprising since D segment nucleotide sequences are typically too short to be retrieved by these methods. Comparison of the 5H2 nucleotide sequence with these chimpanzee gene segments revealed the somatic hypermutations that had occurred during affinity maturation.
The human germ-line gene segments listed in Table II, and the model for VH-D-JH recombination given in Supplementary Figure S3, were obtained by analysis of the 5H2 nucleotide sequence with the IMGT/V-QUEST program (Brochet et al, 2008). As expected, the human gene segments were >97% identical with their chimpanzee counterparts (Table II). Affinity maturation of hu5H2 was simulated by inserting the somatically hypermutated bases from the 5H2 nucleotide sequence into the corresponding positions of the human gene segments, unless those substitutions were already present or would be silent. This gave an amino-acid sequence ∼98% identical to that of 5H2 over the variable domains.
Human serum samples
The human sera used in this study had been deposited with the National Reference Center for Arboviruses, Institut Pasteur de la Guyane, French Guiana, following the patients’ non-opposition to the reuse of their samples for research purposes. These sera had been collected from six patients who displayed clinical symptoms of dengue (temperature ⩾38.5°C, headache, myalgia and/or arthralgia). Seropositivity for dengue was established using an IgM capture ELISA that used mouse-brain extracts as antigens (Dussart et al, 2008). The infecting serotype was identified by RT–PCR (Lanciotti et al, 1992) on acute phase blood samples. The serum samples used in this study were collected from the same patients 11–19 days post-onset of symptoms, and characterized as primary or secondary infections using IgG and IgM antibody capture ELISA kits (Panbio, Brisbane, Australia) according to the manufacturers instructions. These procedures established that patients 1–5 had presented with secondary DENV-4 infections, while patient 6 had not been exposed to dengue.
DENV-4 rDI and sE indirect ELISA with human sera
The wells of 96-well microtitre plates were coated overnight at 4°C with 10 ng of purified DENV-4 rDI or 300 ng of purified DENV-4 sE in 100 μl of PBS. The wells were washed three times with PBS/0.1% Tween-20 (PBS-T) and blocked for 2 h at 37°C with 200 μl of PBS-T/5% non-fat dried milk (NDM). In the following sections, the plates were washed three times with PBS-T after all incubation steps. The plates were incubated for 1 h at 37°C with 100 μl/well of each human serum sample in a three-fold dilution series (starting at 1/25 for rDI and 1/100 for sE) in dilution buffer (PBS-T/1% NDM). Positive and negative controls consisted of substituting the sera for a saturating concentration (50 nM) of Mab 5H2 in dilution buffer, or dilution buffer alone, respectively. The plates were then incubated for 1 h at 37°C with 100 μl/well of a horseradish peroxidase (HRP)-conjugated mouse anti-human IgG secondary antibody (Jackson, West Grove, PA, USA) diluted 1:1000 in dilution buffer, followed by 100 μl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution (KPL, Gaithersburg, MD, USA) for 10 min at room temperature (RT). The reaction was stopped by the addition of 50 μl/well of 0.25 N H2SO4 solution and absorbance values at 450 nm recorded using a plate reader. Normalized, background-corrected absorbance values, A′, from the wells of each plate were obtained using the absorbance values A and the equation A′=(A−A0)/(A1−A0), where A1 and A0 are the mean absorbance values of the positive and negative controls, respectively, from the same plate. This accounted for the differing amounts of each antigen (rDI or sE) coated onto the ELISA plates and allowed the absorbance values between different plates to be standardized. One experiment was performed for rDI and sE, with each serum sample dilution series tested once on each of three plates.
The percentage of the sE-specific IgG binding to rDI in sera from patients 1–3 was estimated as follows. Inspection of the normalized, background-corrected binding profiles for each serum sample with rDI and sE showed that they were approximately linear as a function of serum dilution for absorbance values <0.5. The absorbance values as a function of reciprocal serum dilution factor (1/n) were thus fitted over this range by linear regression using Prism (GraphPad Software). The gradient is given by the high-dilution limit of the fixed-slope dose–response function as (A′1−A′0) × (C/K), where A′0 and A′1 are the normalized, corrected absorbance values with no antibody and at saturation of antibody binding, respectively, C is the concentration of sE- or rDI-specific antibody (as appropriate) in the undiluted serum sample, and K is the concentration of sE- or rDI-specific antibody (as appropriate) that would correspond to half-maximal binding. The binding profiles show that the ratio of the values of (A′1–A′0) for rDI and sE will not differ substantially from unity. Furthermore, assuming that the overall affinity K of the polyclonal response to DI does not differ significantly from that of the polyclonal response to the full-length ectodomain, the ratio of the gradients for rDI and sE will correspond approximately to the fraction of the total sE-specific IgG that binds to rDI. The linear fitting to the rDI and sE binding data for patient sera 1–3 is shown in Supplementary Figure S4. The values of the resulting gradients from patients 1–3 were 25±0.42, 5.7±0.12 and 72±1.5 for rDI, and 690±8.2, 250±9.8 and 2400±26 for sE. These values predict that rDI-specific IgG accounts for ∼3.6, 2.3 and 3.0% of the total sE-specific IgG titres of patient sera 1–3, respectively.
DENV-4 rDI indirect ELISA with Mab 5H2 at pH 5.5 and 7.0
This was performed using rDI as the capture antigen exactly as described above, except that the serum samples were replaced by three-fold serial dilutions of Mab 5H2 (starting at 333 nM) in 150 mM NaCl, 0.25% bovine serum albumin and 100 mM sodium phosphate/50 mM citric acid buffer at pH 5.5 or 7.0. One experiment was performed in which binding at each pH value was tested in triplicate. The data were fitted with fixed-slope dose–response curves in Prism.
Competition ELISA
The wells of 96-well microtitre plates were coated for 48 h at 4°C with 100 ng of purified Mab 5H2 in 100 μl of PBS. Purified, strep-tagged rDI at 0.75 ng/ml was pre-incubated with three-fold serial dilutions of each serum sample (starting at 1/10) in dilution buffer, in 96-well round-bottomed plates at 37°C for 90 min. Positive controls for rDI binding consisted of replacing the patient sera with naive serum or dilution buffer, while in the negative control both serum and rDI were substituted for dilution buffer. The positive control for competition for rDI binding consisted of replacing serum with a three-fold dilution series of Mab 5H2. During this incubation step, the wells of the ELISA plate were washed three times with PBS-T and blocked for 1 h at 37°C with 200 μl of blocking buffer. In all, 100 μl of each rDI/serum pre-incubate was transferred to the wells of the ELISA plate, which was then incubated at 37°C for 1 h. Recombinant domain I bound onto the immobilized Mab 5H2 was detected by incubating with 100 μl/well of a mouse anti-strep tag antibody (Qiagen) diluted 1:1000 in dilution buffer for 1 h at 37°C, followed by 100 μl/well of an HRP-conjugated goat anti-mouse secondary antibody (Sigma) diluted 1:1000 in dilution buffer for 1 h at 37°C. The rest of the assay was performed as described for the indirect ELISAs above. Values of percent inhibition of rDI binding to the plate, relative to naive serum, were calculated from the absorbance values A as 100 × (Anaive−A)/(Anaive−A0), where Anaive is the mean absorbance value obtained using naive serum in place of patient serum, and A0 is the mean background (calculated from the negative control). Two independent experiments were performed in which each serum sample was tested in single and triplicate measurements, respectively. The data from both experiments were combined to give the results shown in Figure 5C.
Accession codes
The coordinates and structure factors of the DENV-4 sE and rDI in complex with Fab 5H2 were deposited with the Protein Data Bank (accession numbers 3UAJ and 3UC0, respectively).
Supplementary Material
Acknowledgments
We thank Scott Halstead and Susie Kliks for their important support through the PDVI; Ahmed Haouz and Patrick Weber from the Protein Crystallization platform at the Pasteur Institute, Paris, and the staff of the PX1 beamline at the Swiss Light Source for beamline support; Clemens Vonhrein, Oliver Smart and Gerard Bricogne (Global phasing limited) for assistance in data processing and structure refinement; Dr S-T Yang (National Institutes of Health) for advice on liposome-based assays; Dr W Gillette and his colleagues at the Protein Expression Laboratory, SAIC-Frederick/NCI (Frederick, MD) for assistance in the production of recombinant Fab 5H2. This work was supported by a PDVI grant to FAR, the Intramural Research Program of the National Institute of Allergy and Infectious Diseases to CJL, the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development and a National Institutes of Health Intramural Biodefense Research grant (both to LVC). JJBC was supported by an EMBO long-term fellowship (ALTF-194-2005) and a Marie Curie Intra-European Fellowship (EIF-25456-DENLIG). FAR also acknowledges support from Merck-Serono and from the ANR grant ‘DENTRY’.
Author contributions: MENS and CMK performed the cloning, expression and purification of the rDI and sE constructs. MENS and EAS co-crystallized rDI and sE with Fab 5H2. JJBC and FAR collected synchrotron X-ray data. JJBC, EAS and SD processed the data and solved the crystal structures. JJBC performed the rebuilding, refinement and analysis of the structures and, in conjunction with PD, performed the ELISAs. APG produced and purified dengue virus for the liposome fusion assays. EZ performed liposome fusion assays. FAR and CJL devised the study. JJBC and FAR wrote the paper in conjunction with CJL and LVC.
Footnotes
The authors declare that they have no conflict of interest.
References
- Balsitis SJ, Williams KL, Lachica R, Flores D, Kyle JL, Mehlhop E, Johnson S, Diamond MS, Beatty PR, Harris E (2010) Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog 6: e1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, Rey FA, Varani L, Whitehead SS, Diamond MS, Harris E, Lanzavecchia A, Sallusto F (2010) The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8: 271–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA (2004) Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J 23: 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G (1993) Direct phase determination by entropy maximization and likelihood ranking: status report and perspectives. Acta Crystallogr D Biol Crystallogr 49: 37–60 [DOI] [PubMed] [Google Scholar]
- Brien JD, Austin SK, Sukupolvi-Petty S, O’Brien KM, Johnson S, Fremont DH, Diamond MS (2010) Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J Virol 84: 10630–10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, Giudicelli V (2008) IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36: W503–W508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broennimann C, Eikenberry EF, Henrich B, Horisberger R, Huelsen G, Pohl E, Schmitt B, Schulze-Briese C, Suzuki M, Tomizaki T, Toyokawa H, Wagner A (2006) The PILATUS 1M detector. J Synchrotron Radiat 13: 120–130 [DOI] [PubMed] [Google Scholar]
- Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Chen BR, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG, Fremont DH (2009) Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. EMBO J 28: 3269–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 [DOI] [PubMed] [Google Scholar]
- Crill WD, Hughes HR, Delorey MJ, Chang GJ (2009) Humoral immune responses of dengue fever patients using epitope-specific serotype-2 virus-like particle antigens. PLoS One 4: e4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill WD, Roehrig JT (2001) Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol 75: 7769–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussart P, Petit L, Labeau B, Bremand L, Leduc A, Moua D, Matheus S, Baril L (2008) Evaluation of two new commercial tests for the diagnosis of acute dengue virus infection using NS1 antigen detection in human serum. PLoS Negl Trop Dis 2: e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Evans PR (2005) Scaling and assessment of data quality. Acta Crystallogr D 62: 72–85 [DOI] [PubMed] [Google Scholar]
- Glamann J, Burton DR, Parren PW, Ditzel HJ, Kent KA, Arnold C, Montefiori D, Hirsch VM (1998) Simian immunodeficiency virus (SIV) envelope-specific Fabs with high-level homologous neutralizing activity: recovery from a long-term-nonprogressor SIV-infected macaque. J Virol 72: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromowski GD, Barrett AD (2007) Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology 366: 349–360 [DOI] [PubMed] [Google Scholar]
- Halstead SB (2003) Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res 60: 421–467 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1988) Automatic indexing of rotation diffraction patterns. J App Cryst 21: 67–72 [Google Scholar]
- Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, Fikrig E, Marasco WA, Koski RA, Modis Y (2006) Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J Virol 80: 11000–11008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Chipman PR, Holdaway HA, Johnson S, Fremont DH, Kuhn RJ, Diamond MS, Rossmann MG (2009) Capturing a flavivirus pre-fusion intermediate. PLoS Pathog 5: e1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Nybakken GE, Chipman PR, Zhang W, Diamond MS, Fremont DH, Kuhn RJ, Rossmann MG (2006) West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc Natl Acad Sci USA 103: 12400–12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG (2010) Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proc Natl Acad Sci USA 107: 18950–18955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungthong C, Zhang C, Mammen MP Jr, Ubol S, Holmes EC (2004) The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology 329: 168–179 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Beuchat MH, Chevallier J, Makino A, Mayran N, Escola JM, Lebrand C, Cosson P, Gruenberg J (2002) Separation and characterization of late endosomal membrane domains. J Biol Chem 277: 32157–32164 [DOI] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH (2002) Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CJ, Goncalvez AP, Men R, Wernly C, Donau O, Engle RE, Purcell RH (2007) Epitope determinants of a chimpanzee dengue virus type 4 (DENV-4)-neutralizing antibody and protection against DENV-4 challenge in mice and rhesus monkeys by passively transferred humanized antibody. J Virol 81: 12766–12774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CY, Tsai WY, Lin SR, Kao CL, Hu HP, King CC, Wu HC, Chang GJ, Wang WK (2008) Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J Virol 82: 6631–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV (1992) Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol 30: 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM (1993) Shape complementarity at protein/protein interfaces. J Mol Biol 234: 946–950 [DOI] [PubMed] [Google Scholar]
- Liao M, Sanchez-San Martin C, Zheng A, Kielian M (2010) In vitro reconstitution reveals key intermediate states of trimer formation by the dengue virus membrane fusion protein. J Virol 84: 5730–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM (2001) The viruses and their replication. In Fields Virology, Knipe DM, Howley PM (eds), 4th edn, pp 991–1041. Philadelphia: Lippincott Williams and Wilkins
- Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG (2008) Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol 15: 312–317 [DOI] [PubMed] [Google Scholar]
- Men R, Yamashiro T, Goncalvez AP, Wernly C, Schofield DJ, Emerson SU, Purcell RH, Lai CJ (2004) Identification of chimpanzee Fab fragments by repertoire cloning and production of a full-length humanized immunoglobulin G1 antibody that is highly efficient for neutralization of dengue type 4 virus. J Virol 78: 4665–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC (2003) A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci USA 100: 6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC (2004) Structure of the dengue virus envelope protein after membrane fusion. Nature 427: 313–319 [DOI] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC (2005) Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol 79: 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath TP (1994) Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA 91: 2395–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak V, Dessau M, Kucera K, Anthony K, Ledizet M, Modis Y (2009) Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J Virol 83: 4338–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH (2006) Crystal structure of the West Nile virus envelope glycoprotein. J Virol 80: 11467–11474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH (2005) Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature 437: 764–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC (1995) The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375: 291–298 [DOI] [PubMed] [Google Scholar]
- Sabin AB (1952) Research on dengue during World War II. Am J Trop Med Hyg 1: 30–50 [DOI] [PubMed] [Google Scholar]
- Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, Kim T, O’Brien KM, Nelson CA, Johnson S, Fremont DH, Diamond MS (2010) The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog 6: e1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Brandler S, Kossl C, Heinz FX (2007) Probing the flavivirus membrane fusion mechanism by using monoclonal antibodies. J Virol 81: 11526–11531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Kiermayr S, Holzmann H, Heinz FX (2006) Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J Virol 80: 9557–9568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, Fremont DH, Diamond MS (2010) Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol 84: 9227–9239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH, Diamond MS (2007) Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J Virol 81: 12816–12826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BS, Moesker B, Smit JM, Wilschut J, Diamond MS, Fremont DH (2009) A therapeutic antibody against west nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog 5: e1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin AA, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J App Cryst 30: 1022–1025 [Google Scholar]
- van der Schaar HM, Rust MJ, Chen C, van der Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM (2008) Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog 4: e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt MR, Moesker B, Goudsmit J, Jongeneelen M, Austin SK, Oliphant T, Nelson S, Pierson TC, Wilschut J, Throsby M, Diamond MS (2009) Human monoclonal antibodies against West Nile virus induced by natural infection neutralize at a postattachment step. J Virol 83: 6494–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala WM, Donaldson EF, de Alwis R, Accavitti-Loper MA, Baric RS, de Silva AM (2010) Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog 6: e1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala WM, Kraus AA, Haymore LB, Accavitti-Loper MA, de Silva AM (2009) Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology 392: 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva E, Yang ST, Melikov K, Pourmal S, Chernomordik LV (2010) Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog 6: e1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG (2004) Conformational changes of the flavivirus E glycoprotein. Structure 12: 1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.