Abstract
Long non-coding RNAs (lncRNAs) are a numerous class of newly discovered genes in the human genome, which have been proposed to be key regulators of biological processes, including stem cell pluripotency and neurogenesis. However, at present very little functional characterization of lncRNAs in human differentiation has been carried out. In the present study, we address this using human embryonic stem cells (hESCs) as a paradigm for pluripotency and neuronal differentiation. With a newly developed method, hESCs were robustly and efficiently differentiated into neurons, and we profiled the expression of thousands of lncRNAs using a custom-designed microarray. Some hESC-specific lncRNAs involved in pluripotency maintenance were identified, and shown to physically interact with SOX2, and PRC2 complex component, SUZ12. Using a similar approach, we identified lncRNAs required for neurogenesis. Knockdown studies indicated that loss of any of these lncRNAs blocked neurogenesis, and immunoprecipitation studies revealed physical association with REST and SUZ12. This study indicates that lncRNAs are important regulators of pluripotency and neurogenesis, and represents important evidence for an indispensable role of lncRNAs in human brain development.
Keywords: long non-coding RNAs, neurogenesis, pluripotency, PRC2, SOX2
Introduction
The mammalian transcriptome comprises a vast number of long non-coding RNAs (lncRNAs), which are defined as transcripts >200 nucleotides with little or no protein-coding potential (Carninci et al, 2005). They participate in numerous biological processes that coordinate gene expression, through epigenetic modification (Khalil et al, 2009; Gupta et al, 2010; Mohammad et al, 2010; Tsai et al, 2010; Wang et al, 2011), mRNA splicing (Tripathi et al, 2010), control of transcription (Orom et al, 2010) or translation (Gong and Maquat, 2011) and genomic imprinting (Pandey et al, 2008; Redrup et al, 2009; Mohammad et al, 2010). Nevertheless, to date only a tiny fraction of lncRNAs have been functionally validated in biological or disease processes.
LncRNAs are emerging players in embryogenesis and in developmental processes (Amaral and Mattick, 2008; Dinger et al, 2008). Recent studies in embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) indicate that lncRNAs are integral members of the ESC self-renewal regulatory circuit (Sheik Mohamed et al, 2010; Guttman et al, 2011). In addition, Loewer et al (2010) showed that a large intergenic non-coding RNA (lincRNA), lincRNA-RoR, enhanced the reprogramming of fibroblasts into iPSCs. LncRNAs such as MALAT1, Evf-2 and Nkx2.2AS, have also been reported to specify neural cell fate and function (Tochitani and Hayashizaki, 2008; Bond et al, 2009; Bernard et al, 2010; Rapicavoli et al, 2010). LncRNAs are also dynamically expressed during neuronal–glia fate specification, and they appear to regulate the expression of protein-coding genes within the same genomic locus, suggesting lncRNA function (Mercer et al, 2010). Additional evidence suggesting functional roles of lncRNAs in the brain includes a computational analysis of in situ hybridization data from the Allen Brain Atlas, which identified 849 lncRNAs showing specific expression in the mouse brain (Mercer et al, 2008). Furthermore, neural lncRNAs have been shown to be regulated by transcription factors (Johnson et al, 2009) and epigenetic processes (Mercer et al, 2010). So far, most efforts aimed at understanding lncRNA functions in pluripotency and neural differentiation focussed on the mouse as a model system (Dinger et al, 2008; Tochitani and Hayashizaki, 2008; Mercer et al, 2010; Sheik Mohamed et al, 2010; Guttman et al, 2011). To date, the roles of lncRNAs in human embryonic and neural developmental gene networks have not been investigated. Given the generally poor evolutionary conservation of lncRNAs (Pang et al, 2006), there is a clear need to investigate whether lncRNAs are also important in human embryonic and neuronal developmental networks.
To address this, we established a highly efficient method to differentiate human ESCs (hESCs) into a homogeneous population of neural progenitor cells (NPCs), which then differentiate into mature neurons with 90% efficiency. In this study, we sought to identify human lncRNAs that are important in two key biological processes: pluripotency and neurogenesis. We present novel lncRNAs that are indispensable for both. These lncRNAs are likely to regulate many hundreds of mRNAs, possibly through interaction with histone-modifying complexes and transcriptional factors. These data highlight the importance of lncRNAs in fundamental human developmental processes.
Results
A homogeneous population of neural progenitors can be derived from human ES cells
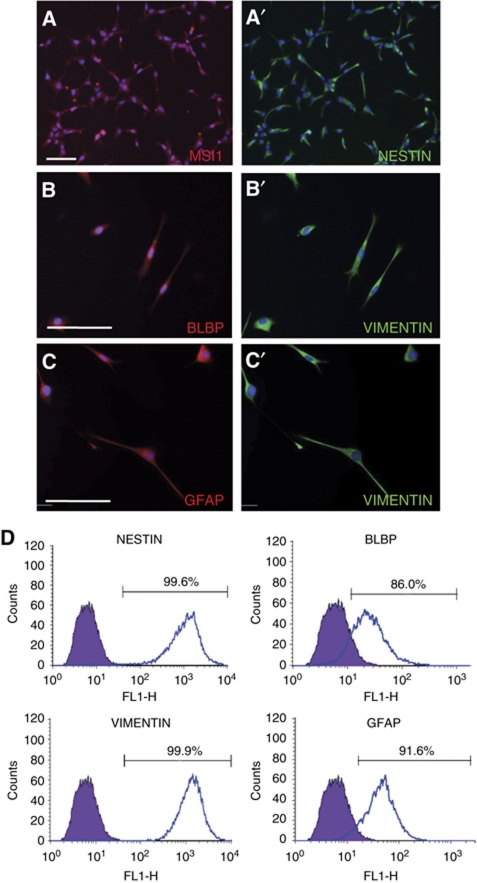
To investigate the roles of lncRNAs in neural development, we established a stepwise protocol to efficiently differentiate hESCs into neural progenitors and eventually into neurons. The co-culture technique of PA6 mouse stromal cells and hESCs, also termed stromal-derived induction activity (SDIA), has been previously reported by several groups to be able to generate numerous neural cell types including dopamine (DA) neurons (Kawasaki et al, 2000; Zeng et al, 2004). Using a modified SDIA differentiation protocol (Supplementary data; Supplementary Figure S1A), we derived a homogeneous population of NPCs from hESCs and human iPSCs, which expressed the neural progenitor markers NESTIN (NES), MUSASHI1 (MSI1) and the radial glia markers VIMENTIN (VIM), glial fibrillary acidic protein (GFAP), and brain lipid binding protein (BLBP) (Figure 1A and B′). This indicated that neural progenitors derived by the modified SDIA method were radial glia-like.
Figure 1.
Neural progenitors derived from H1 hESCs (H1-NPCs) homogenously expressed neural stem cell and radial glia markers. (A, A′) MUSASHI-1 (MSI1) and NESTIN (NES), both neural stem cell markers, were co-expressed in almost all the NPCs. (B–C′) The elongated H1-NPCs co-expressed radial glia markers BLBP, VIMENTIN (VIM) and GFAP. The scale bar indicates 100 μm. (D) Flow cytometry quantification of lineage markers in neural progenitors. Values show the percentage of immunopositive cells for the indicated antibodies.
The main advantage of this protocol was that a homogeneous population of radial glia-like neural progenitors expressing NES, VIM, BLBP and GFAP could be derived from undifferentiated hESCs (Figure 1D). These NPCs were expandable for at least 15 passages in the presence of mitogens bFGF and EGF to produce large numbers of cells for subsequent differentiation. In addition, these radial glia-like cells were karyotypically normal (Supplementary Figure S1B and C) and could be cryopreserved with high cell viability.
Human ESC-derived neural progenitors differentiate into functional DA neurons with high efficiency
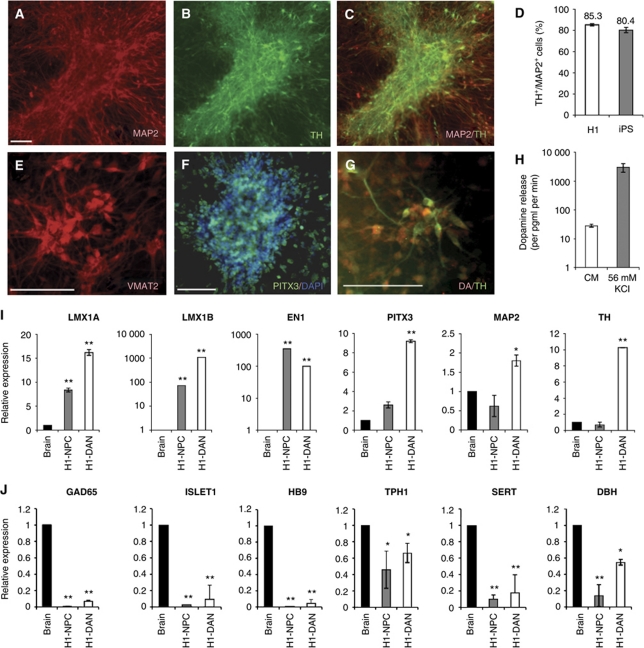
The NPCs derived from H1 hESCs (H1-NPCs) were differentiated into DA neurons by subjecting them to DA differentiation medium, consisting of SHH, FGF8 and ascorbic acid (see Supplementary data). At the end of the 14-day differentiation process, neurons immunopositive for both the mature neuron marker, MAP2, and the dopaminergic marker, tyrosine hydroxylase (TH), were abundant (Figure 2A–C), indicating that H1-NPCs were differentiated into DA neurons (H1-DANs). Further characterization revealed that other DA neuron markers such as VMAT2, PITX3 and DA were also expressed (Figure 2E–G). To further characterize the subtype of DA neurons derived, the gene expression profile of the derived neurons was compared against those of the whole brain and H1-NPC samples. The enrichment of mRNA expression of LMX1A, LMX1B, EN1, PITX3, MAP2 and TH confirmed that midbrain DA neurons were derived (Figure 2I), whereas the lack or decreased expression of GAD65, ISLET1, HB9, TPH1, SERT and DBH indicated that contaminating GABAergic, motor, sertonergic and noradrenergic neurons were absent (Figure 2J).
Figure 2.
H1-NPCs differentiate into TH+ midbrain DA neurons with high efficiency. (A) MAP2-expressing mature neurons were abundant at the end of the 14-day differentiation of H1-NPCs. (B, C) These neurons also expressed TH, indicative of a dopaminergic neuronal population. (D) In three independent experiments, the mean percentages of TH+/MAP2+ cells were 85.3% and 80.4% for H1- and iPSC-derived cultures, respectively. A panel of dopaminergic markers including (E) VMAT2, (F) PITX3 and (G) DA were also expressed by the H1-derived DA neurons. (H) In an in vitro test of function, cultured H1-derived DA neurons were induced to depolarize in 56 mM KCl. Using a DA ELISA assay, DA detected in the supernatant of the 56-mM KCl treatment was significantly 100-fold higher than dopaminer detected in the CM control—medium conditioned with the DA neurons under standard culture conditions for 48 h. (I) qPCR measurement of mRNAs encoding midbrain markers in cDNA from total brain, H1-derived neural precursors and dopaminergic neurons. (J) qPCR measurement of various neuronal subtype markers: GABAergic subtype (GAD65), motor neuron subtype (ISLET1 and HB9), serotonergic neurons (TPH1 and SERT) and noradrenergic neurons (DBH), indicative of an enriched dopaminergic population. The white scale bar indicates 100 μm. * and ** indicate P-values of <0.05 and <0.01, respectively.
Dopaminergic differentiation was very efficient, with 90% of the culture consisting of MAP2+ neurons, and 85% of TH+/MAP2+ cells in our cultures differentiated from hESCs, indicating that about 76% of the total cells in the culture were DA neurons (Figure 2D). We performed a gene ontology (GO) analysis of the genes that were upregulated in the neurons compared with undifferentiated hESCs, which indicated an enrichment of GO terms related to neuronal differentiation (Table I). The percentage of TH+/MAP2+ cells is one of the highest reported, and we report yields similar to a previous report (Cho et al, 2008), where they derived 86% of TH+/TUJ1+ cells (TUJ1 is a post-mitotic, early neuronal marker). A similar efficiency was also observed when human iPSCs were differentiated into DA neurons using the same technique, indicating the robustness of this differentiation method (Figure 2H).
Table 1. Genes expressed in H1-derived dopamine neurons were highly enriched for Gene Ontology terms relating to neuronal differentiation.
| Gene ontology biological process | Gene ontology term | Percentage of genes | |
|---|---|---|---|
| 1 | Neurogenesis | GO:0022008 | 13.35 |
| 2 | Generation of neurons | GO:0048699 | 12.28 |
| 3 | Neuron differentiation | GO:0030182 | 11.39 |
| 4 | Cell morphogenesis | GO:0000902 | 9.43 |
| 5 | Neuron development | GO:0048666 | 9.25 |
| 6 | Central nervous system development | GO:0007417 | 9.07 |
| 7 | Cell morphogenesis involved in differentiation | GO:0000904 | 8.54 |
| 8 | Brain development | GO:0007420 | 6.58 |
| 9 | Negative regulation of biosynthetic process | GO:0009890 | 6.41 |
| 10 | Positive regulation of gene expression | GO:0010628 | 6.23 |
| The top 10 terms are shown. | |||
| Gene clusters categorized into biological processes at levels 6–9 when analysed with FatiGO. P-value <0.01. | |||
In an in vitro test of the functionality of the hESC-derived DA neurons (H1-DANs), DA released by the neurons under normal culture condition and depolarizing condition (56 mM potassium chloride or KCl) was compared using an enzyme-linked immunosorbent assay (ELISA). Neurons incubated with 56 mM KCl released 105-fold more DA per minute (P<0.01) compared with the CM condition, indicating that H1-DANs were mature neurons responsive to depolarization by KCl in vitro (Figure 2H).
Microarray expression profiling identifies differentially expressed lncRNAs
The highly enriched cultures of human neural progenitors and neurons were then used for identification of lncRNAs that are necessary for neural development. We utilized a custom-designed microarray for long non-coding transcripts, as well as an Illumina beadchip microarray for protein-coding transcripts, to examine gene expression changes during the differentiation of hESCs into NPCs and subsequently into neurons. The lncRNA microarray design included 6671 transcripts identified in a number of published sources, and described in a previous publication (Jia et al, 2010). Importantly, the non-coding status of these transcripts was independently validated in that study. In total, the microarray contained 43 800 probes (Supplementary File 1), such that each lncRNA was represented by 6–8 probes, which achieved high sensitivity and specificity.
To summarize the microarray findings, comparing the NPC to hESC stages, we found 25% of protein-coding probes detected above background (6153 out of 24 526) and 4500 probes (18%) were significantly differentially detected (false discovery rate (FDR) <0.01; fold change >2). Of the lncRNA subset, 16% of probes were detected above background (7017 out of 43 800), with 9% (3885 probes) being differentially detected (P<0.05; fold change >2). When DA neuron stage was compared with the NPC stage, 24% of protein-coding probes were detected above background (5852 out of 24 526), with 13% of these (3076 probes) being differentially detected. Similarly, a smaller percentage (11.5%) of lncRNA probes (5058 probes) was expressed above background with 6% being differentially expressed (2622 probes). Altogether, we identified a total of 934 differentially regulated lncRNAs and 5051 differentially regulated mRNAs (Supplementary Figure S2D).
Identification of lncRNAs associated with pluripotency
We postulated that lncRNA transcripts important for hESC pluripotency maintenance would have an expression pattern similar to that of known pluripotency drivers such as OCT4, NANOG, and ZNF206, which are highly expressed in undifferentiated hESCs and downregulated upon differentiation (Supplementary Figure S2E). To identify lncRNAs that control pluripotency, we filtered for lncRNA transcripts that had at least four probes showing a greater than five-fold downregulation (P<0.05) when differentiated from hESCs to NPCs. In all, 36 lncRNAs were identified (Supplementary Figure S2F; Supplementary Table SV), including the telomerase RNA component TERC (Agarwal et al, 2010), indicating that our custom-designed array was able to identify pluripotency-associated lncRNAs.
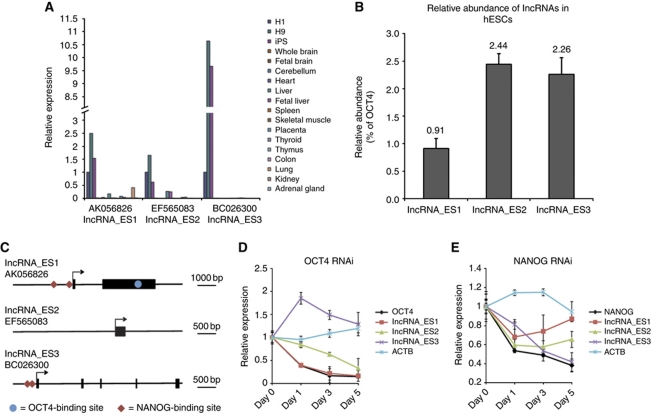
We next sought to determine if lncRNAs were essential for hESC pluripotency, using RNA interference (RNAi). Of the 36 pluripotency-associated lncRNAs, only 16 could have specific small interfering RNAs (siRNAs) designed to target them for knockdown, as the other 20 were substantially overlapping protein-coding genes, rendering it difficult to design specific siRNA sequences (Supplementary Table SIV). To select candidates for knockdown studies, we reasoned that if the identified lncRNAs were functional in maintaining pluripotency, their expression would be specific to pluripotent cells. Thus, we quantified the 16 lncRNAs’ expression in undifferentiated human pluripotent stem cells and a panel of somatic tissues. Three of the pluripotency lncRNAs were exclusively expressed in undifferentiated hESCs and iPSCs (Figure 3A), indicating that they were likely to play a role in pluripotency. Their expression was low (∼0.9–2.5%) compared with that of OCT4 mRNA level in undifferentiated hESCs (Figure 3B), suggesting that they might be playing a regulatory role. We named these transcripts lncRNA_ES1 (AK056826), lncRNA_ES2 (EF565083) and lncRNA_ES3 (BC026300). Inspection of histone marks covering these transcripts shows that all of them have epigenetic signatures indicative of active genes (Supplementary Figure S3).
Figure 3.
ES-specific lncRNAs are low abundance transcripts, and are probably regulated by pluripotent factors OCT4 and NANOG. (A) The expression levels of pluripotency lncRNAs were determined by qPCR in a panel of cell lines and somatic tissues. Three lncRNAs, which we named lncRNA_ES1, lncRNA_ES2 and lncRNA_ES3, were exclusively expressed in hESCs and iPSCs. (B) Comparison of lncRNA expression, as determined by qPCR, compared with the mRNA encoding OCT4. (C) Schematic showing OCT4- and NANOG-binding sites in the vicinity of the lncRNAs. Black arrows indicate transcription start sites, and thick black bars indicate exons. Binding site locations were obtained from ChIP-seq data in Chia et al (2010). (D, E) qPCR measurement of lncRNA expression changes in response to knockdown by RNAi of OCT4 and NANOG, respectively. ACTB encodes a housekeeping gene.
To validate that the pluripotency lncRNAs are bona fide non-coding transcripts, we chose to employ the Coding Potential Calculator (CPC) tool to predict protein-coding potential of the transcripts, as it combines a variety of parameters in conjunction with a support vector machine, and the accuracy of prediction was >95% (Kong et al, 2007). CPC indicated that lncRNA_ES1 and lncRNA_ES2 are very likely non-coding while lncRNA_ES3 could be a ‘weakly coding’ transcript, and the putative 40 amino-acid peptide has neither BLAST hits nor protein domains (Table II). The transcription start and end sites were also confirmed by deep sequencing of the hESC transcriptome (RNA-seq) and are presented in Supplementary Figure S4.
Table 2. Pluripotency lncRNAs in this study.
| lncRNA name | lncRNA ID | Genomic location (hg18/NCBI36) | Transcript length (bp) | Class of lncRNA | CPC scorea |
|---|---|---|---|---|---|
| lncRNA_ES1 | AK056826 | chr6:14,388,338-14,393,355 | 3150 | Intergenic | −1.15338 |
| lncRNA_ES2 | EF565083 | chr1:198,709,840-198,710,182 | 343 | Intergenic | −0.922722 |
| lncRNA_ES3 | BC026300 | chr13:53,593,076-53,605,002 | 1053 | Intergenic | 0.777772 |
| aA negative score assigned by the Coding Potential Calculator (CPC) indicates a non-coding transcript while a value between 0 and 1 indicates a ‘weakly coding’ transcript. | |||||
Pluripotency lncRNAs are regulated by transcription factors
Next, we investigated whether the pluripotency lncRNAs are regulated by transcription factors known to regulate pluripotency. We interrogated data available from the deep sequencing of chromatin immunoprecipitation (ChIP-seq) libraries in hESCs (Chia et al, 2010), which revealed that there are OCT4- and NANOG-binding sites located near the transcription start sites of three of the lncRNAs (Figure 3C). The proximity of these binding sites suggests that the lncRNAs may be direct, downstream targets of pluripotency factors OCT4 and NANOG. To test this, we monitored expression of the pluripotency lncRNAs over a period of 5 days following either OCT4 RNAi or NANOG RNAi. lncRNA_ES1 has an OCT4-binding site in its vicinity, and its expression decreased in response to OCT4 RNAi (Figure 3D). Pluripotency lncRNAs with a neighbouring NANOG-binding site (namely lncRNA_ES1 and lncRNA_ES3) also showed decreased expression upon NANOG RNAi (Figure 3E). Together, these results suggest that pluripotency lncRNAs are integrated into known pluripotency transcriptional networks.
Knockdown of lncRNAs result in hESC differentiation
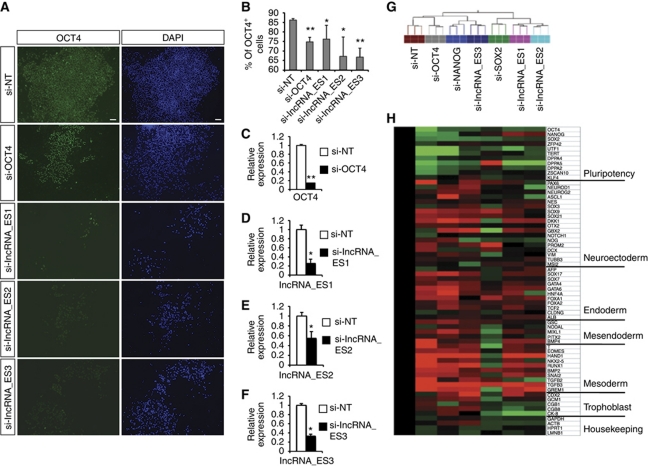
To determine if lncRNAs affect the pluripotent status of hESCs, we transfected siRNAs against the lncRNAs into hESCs. Two siRNAs were designed for each lncRNA and the more effective siRNA was subsequently used (Figure 4C to F; Supplementary Figure S5). Seven days later, pluripotency was assessed by OCT4 immunofluorescence, and RNA was also isolated for global gene expression by microarray profiling. Knockdown of any of the three pluripotency lncRNAs resulted in a loss of OCT4 protein (Figure 4A and B) and mRNA (Figure 4H). In addition, knockdown of lncRNAs resulted in downregulation of a panel of pluripotency markers and simultaneous upregulation of lineage markers corresponding to the neuroectoderm, endoderm and mesoderm germ layers (Figure 4H).
Figure 4.
Knockdown of pluripotency lncRNAs results in hESC differentiation. (A) H1 hESCs were transfected with the indicated siRNAs, and OCT4 protein level was assayed by immunofluorescence 7 days later. (B) The percentage of OCT4+ cells from (A) was quantified by flow cytometry. (C) Assessing the efficiency of siRNA knockdown by qPCR for OCT4. (D–F) Knockdown of lncRNA_ES1, lncRNA_ES2 and lncRNA_ES3, respectively. Two siRNA duplexes were designed against each transcript, and the most effective siRNA was used. * and ** indicate P-values of <0.05 and <0.01, respectively. (G) Hierarchical clustering of the siRNA-treated hESCs. (H) From the microarray, a panel of pluripotency and lineage markers were analysed and fold change values, relative to si-NT control, were presented in a heatmap. Green indicates downregulation and red indicates upregulation.
From the microarray data, hierarchical clustering revealed that lncRNA_ES3 RNAi expression patterns clustered closely with those from the NANOG RNAi, in accordance with the regulation of the lncRNA by pluripotency transcription factors (Figure 4G). However, lncRNA_ES1 and lncRNA_ES2 knockdown showed a global transcriptome profile most similar to SOX2 RNAi, suggesting that lncRNA_ES1 and lncRNA_ES2 could be maintaining pluripotency in a SOX2-dependent manner.
LncRNAs interact with SUZ12 and transcription factor SOX2
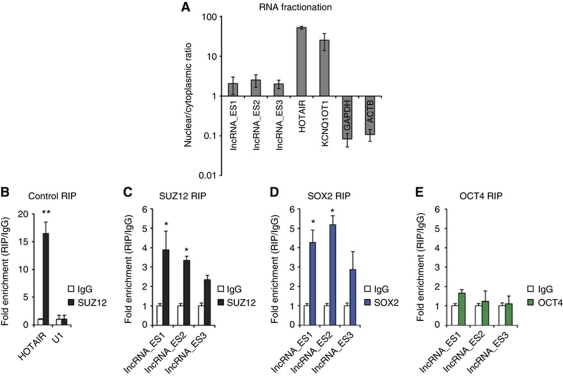
We sought to gain mechanistic insight into lncRNA involvement in hESC pluripotency. We first asked where lncRNAs were localized in the cell, with the idea that nuclear localization provides evidence for a role in epigenetic or transcriptional regulation. By means of RNA fractionation followed by quantitative PCR (qPCR), we found that lncRNA_ES1, lncRNA_ES2 and lncRNA_ES3 were preferentially retained in the nucleus (Figure 5A). Recent reports have linked nuclear lncRNAs to chromatin-modifying complexes (Khalil et al, 2009; Tsai et al, 2010; Guttman et al, 2011) and transcription factors (Bond et al, 2009). Hence, we asked if the lncRNAs could physically associate with nuclear factors to carry out their regulatory role in hESCs. We performed RNA immunoprecipitation (RIP) experiments in which RNA–protein complexes were crosslinked with formaldehyde, and immunoprecipitated with antibodies specific to SUZ12, a component of the PRC2 complex, and pluripotency transcription factors SOX2 and OCT4. We found that lncRNA_ES1 and lncRNA_ES2 were physically associated with SUZ12 and SOX2, but not OCT4 (Figure 5C–E). This is consistent with the clustering of the si-lncRNA_ES1 and si-lncRNA_ES2 samples with si-SOX2 (Figure 4G).
Figure 5.
Nuclear pluripotency lncRNAs are physically associated with PRC2 component SUZ12 and pluripotent transcription factor SOX2. (A) Cellular RNA was separated into the nuclear and cytoplasmic fraction, and qPCR was performed to compute nuclear/cytoplasmic ratio. HOTAIR and KCNQ1OT1 are known, nuclear-localized RNAs, while GAPDH and ACTB are protein-coding mRNAs expected to cytoplasmically localize. (B) RIP of HOTAIR using anti-SUZ12, a previously reported interaction (Gupta et al, 2010). RIP enrichment was measured by qRT–PCR, and values were normalized to background immunoprecipitation measured by isotype IgG. (C–E) RIP of lncRNAs using anti-SUZ12, anti-SOX2 and anti-OCT4, respectively. * and ** indicate P-values of <0.05 and <0.01, respectively.
Identification of lncRNAs associated with neuronal differentiation
Apart from roles in maintenance of pluripotency, we also asked whether any lncRNAs were necessary for differentiation in our system. Thus, we analysed our microarray data to identify lncRNAs with expression profiles suggestive of important roles in neuronal differentiation. We identified a group of 35 lncRNAs, which were highly expressed in mature neurons (more than three-fold) compared with hESCs and NPCs (Supplementary Figure S2J; Supplementary Table SVI). Of the 35 lncRNAs, 25 occupied a genomic location that did not overlap protein-coding genes, and could have siRNAs designed against them. As a proof of concept, we focussed on four neuronal lncRNAs for functional studies, namely, RMST (AK056164, AF429305 and AF429306), lncRNA_N1 (AK124684), lncRNA_N2 (AK091713) and lncRNA_N3 (AK055040). All of these transcripts were previously characterized in a comprehensive study of human transcripts (Imanishi et al, 2004). Similarly, protein-coding potential of these transcripts was determined using CPC, which indicated that these neuronal lncRNAs are most likely non-coding (Table III). Transcription start and end sites of the lncRNAs were validated by deep RNA sequencing of H1-derived neurons (Supplementary Figure S6).
Table 3. Properties of the neuronal lncRNAs in this study.
| lncRNA name | lncRNA ID | Genomic location (hg18/NCBI36) | Transcript length (bp) | Class of lncRNA | CPC scorea |
|---|---|---|---|---|---|
| RMST | AK056164 | chr12:96,382,930-96,451,675 | 2099 | Intergenic | −0.34905 |
| lncRNA_N1 | AK124684 | chr8:77,478,848-77,481,928 | 3081 | Intergenic | −1.22643 |
| lncRNA_N2 | AK091713 | chr11:121,465,023-121,556,316 | 1931 | Overlapping | 0.172903 |
| lncRNA_N3 | AK055040 | chr7:81,413,696-81,415,731 | 2035 | Proximal | −1.21246 |
| aA negative score assigned by the Coding Potential Calculator (CPC) indicates a non-coding transcript while a value between 0 and 1 indicates a ‘weakly coding’ transcript. | |||||
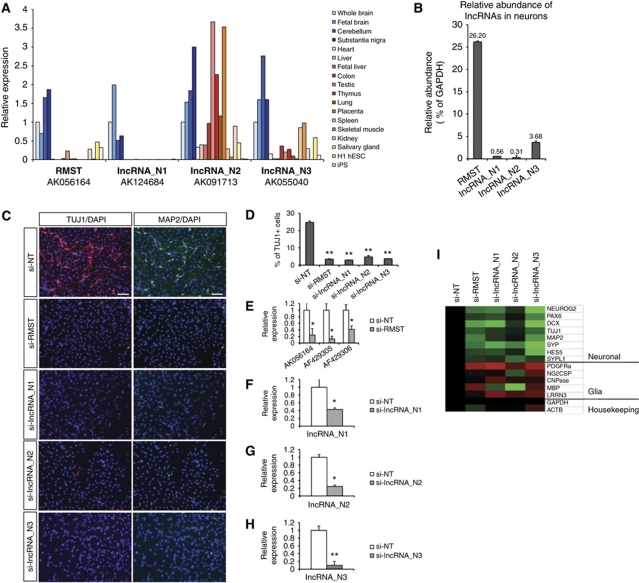
We reasoned that if the neuronal lncRNAs were functional, they should be expressed in the brain. Quantitative analysis of transcript expression (Figure 6A) revealed that RMST and lncRNAs_N1–3 were all expressed in brain structures (whole brain, fetal brain, substantia nigra and cerebellum). While expression of RMST and lncRNA_N1 were more confined to brain regions, lncRNA_N2 and lncRNA_N3 were also present in other somatic tissues. As with the case of pluripotency lncRNAs, neuronal lncRNAs were not abundant (∼0.3–26% relative to GAPDH mRNA levels), consistent with their proposed regulatory roles (Figure 6B).
Figure 6.
Neuronal lncRNAs are expressed in brain tissues, and are required for neurogenesis. (A) Expression of neuronal lncRNAs RMST, lncRNA_N1, lncRNA_N2 and lncRNA_N3 were analysed by qPCR in brain tissues (whole brain, fetal brain, cerebellum and substantia nigra), as well as a panel of other somatic tissues and pluripotent stem cells. RMST and lncRNA_N1 were more specific to brain tissues than lncRNA_N2 and lncRNA_N3. (B) The abundance of the neuronal lncRNAs was shown relative to GAPDH mRNA levels in H1-derived neurons. (C) Immunostaining of ReN-VM neural stem cells treated with the respective siRNAs, and induced to differentiate for 7 days. TUJ1 is a marker of early post-mitotic neurons, and MAP2 is a marker of mature neurons. (D) The percentage of TUJ1-expressing cells was quantified by flow cytometry. (E–H) We validated the efficiency of siRNAs against lncRNAs by qPCR. Shown in each case is the most efficient siRNA that was used for subsequent experiments. For RMST, all three isoforms were targeted. (I) Following knockdown and differentiation, a panel of neuronal and glia markers were analysed by qPCR, and the fold changes, relative to si-NT control, were represented in a heatmap. Green indicates downregulation and red indicates upregulation. * and ** indicate P-values of <0.05 and <0.01, respectively.
Neuronal lncRNAs are required for neuronal differentiation
To determine if the neuronal lncRNAs were required for neurogenesis, we investigated the effect of their knockdown on neuronal differentiation. We transfected siRNAs against each of the neuronal lncRNAs, and induced differentiation of the ReN-VM neural stem cells in N2B27 medium. Seven days later, neuronal differentiation was assayed at the protein level, by immunostaining of TUJ1+ early post-mitotic neurons and/or MAP2+ late mature neurons, as well as at the mRNA level. We tested two siRNA duplexes per lncRNA, and the most efficient siRNA was subsequently used (Figure 6E–H; Supplementary Figure S7). While the non-target siRNA (si-NT) control yielded TUJ1+ and MAP2+ neurons, very few stained cells were observed where the neuronal lncRNAs were knocked down (Figure 6C). This was confirmed by FACS analysis of TUJ1+ cells transfected with the respective siRNAs. The si-NT control yielded ∼25% TUJ1+ neurons, while knockdown of the neuronal lncRNAs resulted in <5% TUJ1+ neurons (Figure 6D). Together, these data indicate that the neuronal lncRNAs we tested were required for efficient neuronal differentiation.
qPCR at day 7 further showed that reduced lncRNA levels in neural progenitors resulted in decreased expression of neurogenic markers including NEUROG2, PAX6, DCX, TUJ1, MAP2, SYP, HES5 and SYPL1, and a simultaneous increase in glia markers PDGFRα, NG2CSP, CNPase, MBP and LRRN3 (Figure 6I). This indicates that loss of the neuronal lncRNAs alters cellular differentiation fate from a neurogenic to a gliogenic programme, and suggests that the lncRNAs play a key role in neural cell fate specification.
Neuronal lncRNAs support neurogenesis by association with nuclear proteins
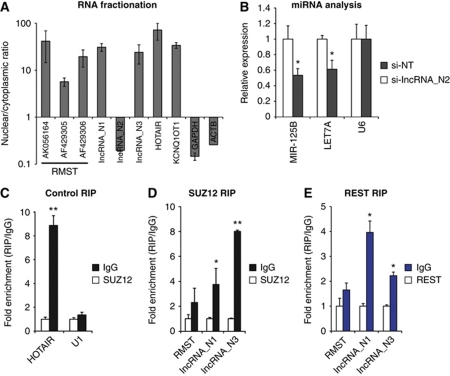
We next investigated the molecular mechanism of action of the neuronal lncRNAs. First, we sought to determine the subcellular localization of the neuronal lncRNAs, by means of RNA fractionation followed by qPCR. With the exception of lncRNA_N2, the neuronal lncRNAs were preferentially nuclear retained (Figure 7A). Thus RMST, lncRNA_N1 and lncRNA_N3 might be interacting with nuclear factors and/or chromatin to support neurogenesis, while lncRNA_N2 possibly has a role in the cytoplasm.
Figure 7.
Neuronal lncRNAs act via diverse mechanisms. (A) Quantification of relative expression of lncRNAs in nuclear and cytoplasmic cell fractions. (B) Quantification of changes in hosted miRNAs in response to lncRNA_N2 knockdown. MiRNAs were quantified using Taqman miRNA qPCR. (C–E) RIP of lncRNAs with SUZ12 and REST antibodies. The interaction of HOTAIR with SUZ12 is a known interaction that serves as a positive control (Gupta et al, 2010). * and ** indicate P-values of <0.05 and <0.01, respectively.
Next, we sought to identify physical interactions of nuclear lncRNAs with nuclear proteins, specifically with SUZ12 and REST, as the former has been reported to be associated with many lncRNAs (Khalil et al, 2009), and the latter is an important transcription factor that represses neurogenesis and is part of the complex bound by the lncRNA HOTAIR (Naruse et al, 1999; Su et al, 2004; Tsai et al, 2010). Three individual RIP experiments confirmed that lncRNA_N3 was significantly enriched in the SUZ12 IP over isotype IgG IP control (Figure 7D), suggesting that lncRNA_N3 was involved in the epigenetic silencing of genes. On the other hand, lncRNA_N1 was enriched in the REST IP compared with isotype IgG IP (Figure 7E), suggesting that lncRNA_N1 associates with the REST/coREST complex to regulate gene expression and neural cell fate specification.
Cytoplasmic lncRNA_N2 affects microRNA expression
We noticed that lncRNA_N2 was cytoplasmic and contains the microRNAs (miRNAs) MIR-125B and LET7 within its introns (Supplementary Figure S6C). These miRNAs are known to be important for neurogenesis (Rybak et al, 2008; Le et al, 2009). This suggests that lncRNA_N2 represents the processing product of the miRNA host transcript, and that knockdown of this transcript could repress neuronal lineage commitment. To this end, we performed a knockdown of lncRNA_N2 in neural stem cells, isolated total RNA 48 h later, and compared MIR-125B and LET7A levels with that of the non-target siRNA control. qPCR revealed that lncRNA_N2 was knocked down by about 75% (Figure 7B), and MIR-125B and LET7A levels were reduced significantly by about 50%. This indicated that the lncRNA_N2 is likely to promote neurogenesis by maintaining MIR-125B and LET7A levels in neural progenitors.
Discussion
It is now evident that large numbers of lncRNAs exist in the mammalian transcriptome (Carninci et al, 2005; Guttman et al, 2009), and they function via diverse mechanisms (Wilusz et al, 2009). In this study, we identified lncRNAs essential for the maintenance of pluripotency and neuronal differentiation in human cells and established that they physically interact with key nuclear proteins to execute their biological functions.
While it has been observed that some lncRNAs act in cis (Ponjavic et al, 2009), a recent report indicated that a unique class of lncRNAs termed large intergenic non-coding RNAs or lincRNAs primarily affect gene expression in trans (Guttman et al, 2011). For the lncRNAs in the present study, we found evidence for a regulatory function in trans. With the exception of cytoplasmic lncRNA_N2, which may represent the miRNA host transcript that gives rise to miRNAs in that region, we did not observe a significant change of gene expression within a 600-kb window (300 kb upstream and 300 kb downstream) following knockdown of the other six lncRNAs (Supplementary Figure S8). Therefore, while some lncRNAs may work in cis, it appears that the lncRNAs in this study are trans-acting.
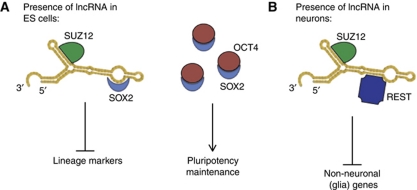
Although lncRNAs have been previously linked to stem cell pluripotency (Dinger et al, 2008; Sheik Mohamed et al, 2010; Guttman et al, 2011), we report for the first time that these ‘pluripotent lncRNAs’ could complex with SOX2 to control hESC pluripotency. Endogenous RIP experiments in hESCs indicated that lncRNA_ES1 and lncRNA_ES2 were physically associated with the transcription factor SOX2 and PRC2 component SUZ12, suggesting that lncRNAs function as a modular scaffold for different proteins or protein complexes to assemble onto (Tsai et al, 2010). Our results indicated that both lncRNA_ES1 and lncRNA_ES2 associated with SUZ12 and SOX2, but not OCT4, and were involved in pluripotency. Therefore, we propose a model whereby pluripotent lncRNAs may act as a scaffold in which SUZ12 and the repressive PRC2 complex is recruited to silence SOX2 neural targets in pluripotent hESCs (Figure 8A). Bioinformatic predictions (Bellucci et al, 2011) suggest that SOX2 preferentially binds to the 5′ end of lncRNA_ES1 while SUZ12 preferentially associates with the 3′ end of the lncRNA (Supplementary Figure S9), in accordance with the cell-type-specific ‘flexible scaffold’ function of lncRNAs proposed by Guttman et al (2011). This proposed scaffolding role of lncRNAs is, however, still subject to experimental validation. In addition, analysis of H3K27 trimethylation marks at promoters of SOX2 target genes in hESCs would shed light on how the lncRNA/protein complex regulates pluripotency.
Figure 8.
Proposed mechanism for role of lncRNA in hESC pluripotency and neuronal differentiation. (A) In undifferentiated hESCs, we propose a mechanism whereby lncRNAs serve as a modular scaffold for SUZ12 (or PRC2) and SOX2. SOX2 may recruit the chromatin-modifying complex to silence neuroectoderm lineage markers by H3K27 methylation. Binding of lncRNA to SOX2 may prevent OCT4 or other core ES transcription factors from binding. Since lncRNAs are present in low abundance, that would not affect the core ES transcriptional network by sequestering SOX2. (B) In a mechanism similar to (A), we envisioned that neuronal lncRNAs may link chromatin modifiers and transcription factors, in order to silence glia lineage genes while promoting neurogenesis.
Our data also indicate an indispensable role of lncRNAs in neurogenesis. While cytoplasmic lncRNA_N2 may be the miRNA host gene responsible for neurogenic miRNAs MIR-125B and LET7A in the same genomic locus, we identified physical association of other neuronal lncRNAs with SUZ12 and REST, and envision a similar mechanism for these lncRNAs (Figure 8B). While it is likely that lncRNAs regulate biological processes through epigenetic modifications, elucidation of molecular mechanisms require more studies, including a genome-wide assessment of histone marks in native and perturbed lncRNA conditions. It has been proposed that lncRNAs may represent a key undiscovered genetic component in the evolution of the human brain (Mattick and Mehler, 2008), but little evidence has been presented for functional roles of lncRNAs in the human nervous system. The data presented in this study represent the first direct demonstration that lncRNAs are necessary components of neural developmental gene networks in human, and imply that deregulation of lncRNA expression may contribute to developmental and neurological disorders.
Materials and methods
Cell culture and neural differentiation
H1 hESCs (passage number 20–35) was grown feeder-free on Matrigel (BD Biosciences) in conditioned medium. Neural differentiation was initiated using a modified SDIA method, in which neural progenitors were enriched and cultured as a monolayer in NPC medium consisting of mitogens bFGF and EGF. Dopaminergic neuronal differentiation was achieved by culture of neural progenitors in N2B27 medium previously described (Ying et al, 2003) supplemented with 200 ng/ml SHH, 100 ng/ml FGF8 and 200 μM ascorbic acid (see Supplementary data for details). A human fetal mesencephalon-derived neural stem cell line, ReN-VM (ReNeuron, Millipore), was cultured as previously described (Donato et al, 2007). Differentiation of ReN-VM cells was achieved by culture in N2B27 medium.
Immunofluorescence
Cells were fixed, permeabilized, and blocked as with standard immunostaining procedures (see Supplementary data). Primary antibodies were diluted in blocking buffer and incubated overnight at 4°C. The list of antibodies and dilutions used is provided in Supplementary Table SI. Secondary antibodies conjugated with AlexaFluor-488 or AlexaFluor-594 (Molecular Probes, Invitrogen) were diluted 1:2500 in blocking buffer and incubated for 2 h at room temperature. DAPI (0.5 μg/ml) was used to visualize cell nuclei.
RNA extraction
Total RNA was extracted in TriZol (Invitrogen), and purified using the RNeasy Mini Kit with DNase I treatment (Qiagen), following the manufacturers’ instructions. Total RNA for miRNA expression analysis was extracted in TriZol and was not column purified. For RNA fractionation, we used the PARIS Kit to isolate RNA from the nuclear and cytoplasmic compartments, following the manufacturer's instructions.
Custom lncRNA array design
We designed a custom microarray to interrogate human lncRNA expression. Potential lncRNAs were gathered from a variety of sources (see Supplementary Table SVII and Supplementary File S1 for details). These lncRNAs essentially comprise the same lncRNA catalogue described in Jia et al (2010). Altogether this set comprised 6673 transcripts. Using Agilent eArray tool, we designed six distinct 60-mer microarray probes against each transcript, and printed these on a custom array slides, along with standard control probes.
Microarray hybridization and data analysis
Sentrix® Human Ref-8 Expression BeadChip microarrays (Illumina) were used for genome-wide expression analysis of coding genes. For hybridization on the Illumina arrays, cRNA was synthesized and labelled using TotalPrep RNA Amplification Kit (Ambion), following the manufacturer's instructions. We utilized a custom-designed microarray to analyse genome-wide lncRNA expression. For this purpose, total RNA was amplified and labelled using Agilent's One-Color Quick Amp Labeling Kit, according to the manufacturer's recommendations.
Scanned data from the BeadChip raw files for all samples were retrieved and background corrected using BeadStudio (Illumina), and subsequent analyses were completed in GeneSpring GX (Agilent). Data were normalized both within and between arrays, and corrected for multiple testing using Benjamini–Hochberg analysis. We defined genes as significantly differentially expressed when FDR is <0.05.
RNA interference
siRNAs targeting lncRNAs for knockdown were designed using Invitrogen's Block-It RNAi Designer (https://rnaidesigner.invitrogen.com/rnaiexpress). Two duplexes were designed for each lncRNA, and only the most effective siRNAs were used for subsequent studies. The sequences of the duplexes are provided in Supplementary Table SII. For transfection, hESCs were seeded in 12-well plates at about 100 clumps per well in MTeSR medium. In all, 100 pmol of siRNAs was complexed with 5 μl of Lipofectamine RNAiMAX reagent (Invitrogen), according to the manufacturer's protocol. Following transfection, the medium was replaced with fresh MTeSR medium and re-transfection was performed at days 2 and 4 after the initial siRNA transfection as previously described.
For transfection of neural stem cells, cells were seeded at 0.2 × 106 per well of a 12-well plate. In all, 50 pmol of siRNAs was complexed with 2.5 μl of Lipofectamine RNAiMAX reagent. Fresh medium was replaced 24 h after transfection. Re-transfection was performed once more 48 h after the initial transfection.
Quantitative real-time PCR
Total RNA was extracted as described above. Reverse transcription was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). For qPCR, primers that span splice junctions were used wherever possible. The lists of primers used are found in Supplementary Tables SIII and SIV). The set-up of qPCR reactions is described in Supplementary data. In all qPCR experiments, a minimum of three technical replicates and three biological replicates were performed. Fold change was normalized to GAPDH mRNA expression unless otherwise specified.
For miRNA expression analysis, reverse transcription was performed using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) following the manufacturer's instructions. TaqMan probes and primers were used for qPCR. Fold change was normalized to U6 snRNA expression. To compute significance, Student's t-test was performed, and a P-value of <0.05 was deemed statistically significant.
RNA immunoprecipitation
RIP was performed as previously described (Niranjanakumari et al, 2002). Briefly, cells were detached with Accutase (Millipore), crosslinked in 1% formaldehyde for 15 min and quenched with 2.5 M glycine for 5 min. The cell pellet was resuspended in modified RIPA buffer (150 mM NaCl, 50 mM Tris, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP-40) supplemented with RNase inhibitor Superase.In (Ambion) and Complete protease inhibitor (Roche). The cell suspension was briefly sonicated at low amplitude for 5 × 30 s cycles using a Bioruptor sonicator to lyse nuclei. Cell debris was removed by centrifugation at 4°C, precleared with Protein G dynal beads (Invitrogen) before adding to respective antibodies pre-bound with Protein G dynal beads for 3 h at room temperature. In all, 5 μg of antibodies was used for each RNA-IP. The following antibodies raised in rabbit were used anti-SUZ12 (ab12073), anti-SOX2 (ab59776), both from Abcam, and, anti-OCT4 (H-134), anti-REST (H-290), both from Santa Cruz Biotechnology. Beads were then washed three times in modified RIPA buffer, and twice in high salt RIPA buffer (1 M NaCl, 50 mM Tris, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP-40). Crosslinks were reversed and proteins were digested with Proteinase K (Invitrogen) at 65°C for 2 h. RNA was extracted in Trizol and precipitated in isopropanol.
Supplementary Material
Acknowledgments
We thank Irene Aksoy for the human iPS cell line. We express our gratitude to Linda Lim and Akshay Bhinge for critical reading of this manuscript. We also thank Kee-Yew Wong for technical assistance and Gireesh Bogu for RNA-seq analysis. We are also thankful to Leonard Lipovich and Hui Jia (Wayne State University) for helping with the lncRNA microarray design. SYN was supported by the NUS Graduate School Scholarship (NGSS), and this research was funded by the Biomedical Research Council and Agency for Science, Technology and Research (A*STAR).
Author contributions: SYN conceived the project, designed and performed the experiments, interpreted the results and wrote the manuscript. RJ helped design the human lncRNA microarray, analysed the data, contributed ideas and wrote the manuscript. LWS contributed ideas, interpreted the results and edited the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agarwal S, Loh YH, McLoughlin EM, Huang J, Park IH, Miller JD, Huo H, Okuka M, Dos Reis RM, Loewer S, Ng HH, Keefe DL, Goldman FD, Klingelhutz AJ, Liu L, Daley GQ (2010) Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature 464: 292–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Mattick JS (2008) Noncoding RNA in development. Mamm Genome 19: 454–492 [DOI] [PubMed] [Google Scholar]
- Bellucci M, Agostini F, Masin M, Tartaglia GG (2011) Predicting protein associations with long noncoding RNAs. Nat Methods 8: 444–445 [DOI] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29: 3082–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD (2009) Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci 12: 1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V et al. (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, Huss M, Soh BS, Kraus P, Li P, Lufkin T, Lim B, Clarke ND, Bard F, Ng HH (2010) A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468: 316–320 [DOI] [PubMed] [Google Scholar]
- Cho MS, Lee YE, Kim JY, Chung S, Cho YH, Kim DS, Kang SM, Lee H, Kim MH, Kim JH, Leem JW, Oh SK, Choi YM, Hwang DY, Chang JW, Kim DW (2008) Highly efficient and large-scale generation of functional dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA 105: 3392–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS (2008) Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato R, Miljan EA, Hines SJ, Aouabdi S, Pollock K, Patel S, Edwards FA, Sinden JD (2007) Differential development of neuronal physiological responsiveness in two human neural stem cell lines. BMC Neurosci 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi T, Itoh T, Suzuki Y, O’Donovan C, Fukuchi S, Koyanagi KO, Barrero RA, Tamura T, Yamaguchi-Kabata Y, Tanino M, Yura K, Miyazaki S, Ikeo K, Homma K, Kasprzyk A, Nishikawa T, Hirakawa M, Thierry-Mieg J, Thierry-Mieg D, Ashurst J et al. (2004) Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol 2: e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L (2010) Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 16: 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Teh CH, Jia H, Vanisri RR, Pandey T, Lu ZH, Buckley NJ, Stanton LW, Lipovich L (2009) Regulation of neural macroRNAs by the transcriptional repressor REST. RNA 15: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H, Mizuseki K, Nishikawa S, Kaneko S, Kuwana Y, Nakanishi S, Nishikawa SI, Sasai Y (2000) Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron 28: 31–40 [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: W345–W349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H, Lim B, Lodish HF (2009) MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol 29: 5290–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL (2010) Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42: 1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Mehler MF (2008) RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci 31: 227–233 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA 105: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF (2010) Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci 11: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C (2010) Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 137: 2493–2499 [DOI] [PubMed] [Google Scholar]
- Naruse Y, Aoki T, Kojima T, Mori N (1999) Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc Natl Acad Sci USA 96: 13691–13696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA (2002) Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26: 182–190 [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C (2008) Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32: 232–246 [DOI] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS (2006) Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet 22: 1–5 [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP (2009) Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet 5: e1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Blackshaw S (2010) The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol 10: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, Nagano T, Cobb BS, Fraser P, Reik W (2009) The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 136: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG (2008) A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10: 987–993 [DOI] [PubMed] [Google Scholar]
- Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L (2010) Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 16: 324–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Kameoka S, Lentz S, Majumder S (2004) Activation of REST/NRSF target genes in neural stem cells is sufficient to cause neuronal differentiation. Mol Cell Biol 24: 8018–8025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochitani S, Hayashizaki Y (2008) Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem Biophys Res Commun 372: 691–696 [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A (2003) Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21: 183–186 [DOI] [PubMed] [Google Scholar]
- Zeng X, Cai J, Chen J, Luo Y, You ZB, Fotter E, Wang Y, Harvey B, Miura T, Backman C, Chen GJ, Rao MS, Freed WJ (2004) Dopaminergic differentiation of human embryonic stem cells. Stem Cells 22: 925–940 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.