Abstract
Group II chaperonins mediate protein folding in an ATP-dependent manner in eukaryotes and archaea. The binding of ATP and subsequent hydrolysis promotes the closure of the multi-subunit rings where protein folding occurs. The mechanism by which local changes in the nucleotide-binding site are communicated between individual subunits is unknown. The crystal structure of the archaeal chaperonin from Methanococcus maripaludis in several nucleotides bound states reveals the local conformational changes associated with ATP hydrolysis. Residue Lys-161, which is extremely conserved among group II chaperonins, forms interactions with the γ-phosphate of ATP but shows a different orientation in the presence of ADP. The loss of the ATP γ-phosphate interaction with Lys-161 in the ADP state promotes a significant rearrangement of a loop consisting of residues 160–169. We propose that Lys-161 functions as an ATP sensor and that 160–169 constitutes a nucleotide-sensing loop (NSL) that monitors the presence of the γ-phosphate. Functional analysis using NSL mutants shows a significant decrease in ATPase activity, suggesting that the NSL is involved in timing of the protein folding cycle.
Keywords: ATPase activity, chaperonins, nucleotide-sensing, protein folding, structure
Introduction
Correct folding of newly translated proteins is crucial for cellular function. A class of proteins called chaperonins plays a key role in the protein folding process, helping to minimize the formation of potentially toxic aggregates inside the cell (Frydman, 2001). Chaperonins consist of two back-to-back stacked rings of 7–9 subunits each, which create a protected environment for protein folding (Horwich et al, 2006; Reissmann et al, 2007). Although the subunit architecture, consisting of an equatorial, intermediate and apical domain, is preserved in all chaperonins, they are divided into two major groups. Group I chaperonins exist in bacteria and eukaryotic organelles and consist of a double homo-heptameric ring, such as GroEL from Escherichia coli (Horwich et al, 2006). Group II chaperonins are found in archaeal and eukaryotic cytosol and consist of a double hetero (or homo)-octameric or nonameric ring (Frydman et al, 1992; Shomura et al, 2004). The major structural difference between the two groups is that group I chaperonins have a ring-shaped cofactor (GroES-like) acting as a detachable lid that binds to the apical domains and closes the chamber. In contrast, the group II chaperonins lack the GroES-like cofactor and instead possess a built-in lid formed by protrusions at the tip of the apical domains that open and close in an ATP-dependent manner (Reissmann et al, 2007; Booth et al, 2008).
The peptide acceptor state (open) and protein folding state (closed) of the group II chaperonin from Methanococcus maripaludis (Cpn) has been described previously using cryo-EM (Clare et al, 2008; Zhang et al, 2010) and crystallographic (Pereira et al, 2010) methods. In contrast to group I chaperonins, in which the equatorial domains share a similar conformation between the open and closed states and the largest motions occurs at the intermediate and apical domains, the three domains of the group II chaperonin subunit reorient upon ATP hydrolysis in order to close the chamber (Pereira et al, 2010; Zhang et al, 2010). Recently, a crystal structure of an open state group II chaperonin in complex with ADP (Hou et al, 2010) showed a rotation of the apical domain by ∼30° compared with the open Cpn structure in complex with ATPγS nucleotide (Pereira et al, 2010). ATP binding alone causes a counter-clockwise rotation of the apical domain. Subsequent ATP hydrolysis drives the subunits to close the chamber completely (Zhang et al, 2011). A closed state structure of the yeast chaperonin in complex with substrate (actin) revealed an asymmetric configuration of the rings (Dekker et al, 2011). The ring movements involved in the folding cycle of group II chaperonins are coordinated in time and space via complex allosteric regulation (Kafri et al, 2001; Kafri and Horovitz, 2003; Bigotti et al, 2006; Reissmann et al, 2007). ATP binding to one subunit enhances ATP association with subunits in the same ring, whereas ATP binding to one ring inhibits ATP association with the subunits of the adjacent ring (Kafri et al, 2001; Kafri and Horovitz, 2003; Bigotti et al, 2006; Reissmann et al, 2007). The synchronized ring action depends on communication between individual subunits. A major question in understanding ATP-driven protein folding machines is how nucleotide binding and hydrolysis is sensed and communicated between the subunits to promote a large conformational change from the open to closed state. Though biochemical evidence suggests that the lid is an important determinant for cooperativity in the chaperonin, the structural components within the catalytic site that are responsible for communicating the nucleotide state around the ring remains a mystery (Reissmann et al, 2007).
Here, we obtain crystal structures of nucleotide states of Cpn at sufficient resolution to allow us to examine, in detail, the effects that different nucleotides states have on the group II Cpn structures. The crystal structures of the wild-type Cpn (Cpn-WT) and a previously described variant: Cpn-rls (Douglas et al, 2011) were solved in complex with the ATP analogue (AMP–PNP), ADP–AlFx and ADP. These structures reveal the significant conformational change associated with ATP hydrolysis. These changes highlight the importance of a region adjacent to the nucleotide-binding site that we identify as the nucleotide-sensing loop (NSL). Functional assays of ATPase activity and lid closure using NSL mutants corroborate the role of the NSL in ATP sensing and subsequent regulation of ATPase timing.
Results
Structural understanding of the nucleotide-binding site of group II chaperonins
Previously obtained open and the closed structures of Cpn were solved at 6.0 and 3.3 Å resolution, respectively (Pereira et al, 2010). Analysis of the detailed local conformational changes associated with nucleotide binding was not possible at these resolution limits. Fortuitously, in the crystals of the recently described substrate release mutant (Douglas et al, 2011) of the chaperonin (Cpn-rls) in complex with AMP–PNP, we observed a significantly higher resolution of X-ray diffraction (see Table I for details). This allowed us to identify the detailed effect of different nucleotide states on the Cpn structure. We also solved the structure of the Cpn-WT protein in complex with ADP in order to eliminate the possibility that the conformational changes were actually a result of the rls loop mutations. We observed that conformational changes in the nucleotide region are preserved between WT and the rls-mutant (Supplementary Figure S1), the only minor differences in the latter being in the region of the rls loop in the apical domain (Supplementary Figure S2). We therefore use the more general Cpn nomenclature in the remainder of this manuscript when discussing the structural changes, referring to both WT and rls-mutant structures.
Table 1. Statistics for data collection and refinement of Cpn-WT-ADP, Cpn-rls-ADP, Cpn-rls-AMP-PNP and Cpn-rls-ADP-AlFx crystal structures.
| Cpn-WT-ADP | Cpn-rls-ADP | Cpn-rls-AMP-PNP | Cpn-rls-ADP-AlFx | |
|---|---|---|---|---|
| A. Data collection | ||||
| Wavelength (Å) | 1.000 | 1.000 | 1.000 | 1.000 |
| Resolution range (Å) | 50–2.80 (2.90–2.80) | 50–2.34 (2.42–2.34) | 50–2.24 (2.32–2.24) | 50–2.70 (2.79–2.70) |
| Detector distance (mm) | 380 | 320 | 260 | 350 |
| Φ (deg) collected/ΔΦ (deg) | 245/1.0 | 360/0.5 | 240/1.0 | 240/1.0 |
| Exposure time (s) | 8 | 3 | 3 | 3 |
| Temperature of collect (K) | 100 | 100 | 100 | 100 |
| B. Data statistics | ||||
| Space group | I222 | I222 | I222 | I222 |
| Unit-cell parameters (Å) | a=162.16, b=185.00 and c=184.77 | a=161.56, b=185.58 and c=185.49 | a=161.02 b=184.47 and c=184.78 | a=162.55, b=184.46 and c=185.49 |
| Unique reflections | 63 540 (3207) | 108 907 (5438) | 117 431 (5932) | 69 455 (3501) |
| Multiplicity | 9.9 (8.6) | 5.4 (5.4) | 7.3 (7.1) | 9.9 (9.9) |
| Data completeness (%) | 100 (99.9) | 100 (99.9) | 100 (100) | 100 (100) |
| I/σ(I) | 14.8 (1.6) | 16.8 (1.6) | 20.3 (2.0) | 18.2 (1.9) |
| Rsyma (%) | 13.5 | 7.8 | 9.4 | 11.6 |
| C. Structure refinement | ||||
| Rfactorb (%) | 18.9 | 17.3 | 17.0 | 17.2 |
| Rfreec (%) | 21.7 | 21.1 | 22.0 | 22.6 |
| RMS from ideal geometry | ||||
| Bond lengths (Å) | 0.009 | 0.008 | 0.008 | 0.007 |
| Bond angles (deg) | 1.136 | 1.068 | 1.066 | 0.961 |
| Protein residues per AUd | 2064 (4 Subunits) | 2064 (4 Subunits) | 2064 (4 Subunits) | 2064 (4 Subunits) |
| ATP analogue (AMP-PNP) | — | — | 4 | — |
| ADP | 4 | 4 | — | 4 |
| AlFx | — | — | — | 4 |
| Mg2+ | 4 | 4 | 8 | 4 |
| Ramachandran plot | ||||
| Favoured region (%) | 97.7 | 98.1 | 98.5 | 97.4 |
| Outliers region (%) | 0 | 0.1 | 0.2 | 0.2 |
| aRsym=ΣhklΣiIi (hkl)–<I (hkl)>/ΣhklΣiIi (hkl), where Σhkl denotes the sum over all reflections and Σi is the sum over all equivalent and symmetry-related reflections. | ||||
| bRfactor=ΣFobs–Fcalc/Σ Fobs. | ||||
| cRfree=Rfactor for 5% of the data were not included during crystallographic refinement. | ||||
| dAU, asymmetric unit. | ||||
In order to address the effect of ATP hydrolysis on group II chaperonins, the Cpn-WT and Cpn-rls mutant were solved in complex with an ATP analogue (AMP–PNP), ADP–AlFx and ADP. All of the structures were solved in a closed conformation with resolutions ranging from 2.2 to 2.8 Å. The overall structure of closed state Cpn can be described as double homo-octameric rings stacked back-to-back, creating a spherically shaped complex, in contrast to the cylindrical form observed for the open state (Pereira et al, 2010). A brief description of the three domains follows: The equatorial domain is involved in contacts between the upper and lower rings and interactions of adjacent subunits in the same ring. This domain also contains the nucleotide-binding site (Ditzel et al, 1998). The equatorial domain is the largest subunit domain and an inner intra-ring contact in the equatorial region forms a β-sheet from β-strands of adjacent subunits (Pereira et al, 2010; Zhang et al, 2010). The intermediate and apical domains show only intra-ring contacts in the closed conformation. The intermediate domain is composed of two flexible hinges. Interestingly, the intermediate domain also interacts with the nucleotide-binding site located at the top of equatorial domain. ATP engages in a specific interaction with the carbonyl group of residue Gly-160 in the intermediate domain, which is present on a loop formed by the residues 160–169 (Pereira et al, 2010). Comparing the Cpn-WT and Cpn-rls structures with a non-hydrolyzable ATP analogue bound and ADP bound reveals that the loss of the γ-phosphate causes a large structural change in this loop of the intermediate domain. Since the intermediate domain links the equatorial domain, which contains the nucleotide-binding site, and the apical domain, which contains the substrate-binding site, conformational change at this region can potentially regulate communication between ATP hydrolysis and the conformational cycle. The apical domain also forms the entrance to the chamber via an ∼30 residue insertion at the tip of each domain. The 160–169-loop is a likely candidate for communicating the bond scission in the equatorial region, to the lid closure in the distal apical domain.
Local conformational change associated with ATP hydrolysis: NSL
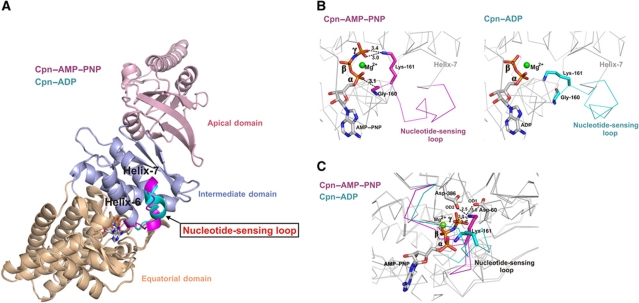
Comparison of the Cpn–AMP–PNP and Cpn–ADP states, which represent the states before and after ATP hydrolysis, respectively, shows conformational changes in residues 160–169 connecting helix-6 and helix-7 of the intermediate domain (Figure 1A) (Supplementary Movie 1). We designate this region the ‘NSL’, herein NSL, because of its role in sensing the state of nucleotide (i.e. pre- or post-hydrolysis) in the nucleotide-binding site. The change promoted by ATP hydrolysis is the conversion of a mainly random-coil structure for the NSL to an α-helical conformation generating an extension of helix-7 in the Cpn–ADP state. Similar structural transitions, as well as the coupling of these transitions to the nucleotide via an ATP sensor, have been described in other nucleotide-dependent molecular machines (Sprang, 1997; Meyer et al, 2003b; Corbett and Berger, 2005). Comparison of the C-α positions across the entire subunit of Cpn–AMP–PNP and Cpn–ADP shows an RMSD of 0.62 Å, whereas the RMSD between only the NSL residues is 3.14 Å, indicating a significant structural change in this region. The Cpn–AMP–PNP state shows two residues located in the NSL that interact directly with the phosphates of the bound nucleotide. The residues Gly-160 and Lys-161 make direct interactions with the α- and γ-phosphate, respectively. In the Cpn–ADP state, the main-chain carbonyl group of residue Gly-160 is no longer interacting with the α-phosphate and Lys-161 lacks the contact with the γ-phosphate (Figure 1B). The position assumed by Lys-161 in the Cpn–AMP–PNP state, due to the presence of the γ-phosphate, promotes an interaction of this residue via salt bridges with the two catalytic residues Asp-60 and Asp-386 (Figure 1C). The position of Lys-161 in the Cpn–AMP–PNP state may also be important for neutralizing the charges around the γ-phosphate group. The hydrolysis of the γ-phosphate group promotes the rearrangement of residue Lys-161; however, this conformational change does not affect the position of the core catalytic residues Asp-60 and Asp-386, which show similar locations in both the Cpn–AMP–PNP and Cpn–ADP states. The rearrangement of Lys-161, associated with the absence of the γ-phosphate, supports the theory that Lys-161 acts as an ATP sensor. We propose that this is a general mechanism for group II chaperonins, since this residue is strictly conserved within this class of chaperonins (Figure 2).
Figure 1.
(A) Rearrangement of the NSL between the Cpn–AMP–PNP (magenta) and Cpn–ADP (cyan) states. The loop assumes an α-helical structure in the Cpn–ADP state, extending helix-7. (B) Change in the interactions between residues Gly-160 and Lys-161, located in NSL, and the phosphate groups of the AMP–PNP and ADP states. In the Cpn–AMP–PNP structure, the residues Gly-160 and Lys-161 make direct contacts with the α- and γ-phosphates, respectively, whereas these interactions are not present in Cpn–ADP structure. (C) Additional electrostatic interactions of the residue Lys-161 in the AMP–PNP state. The position assumed by Lys-161 in the Cpn–AMP–PNP state caused by the presence of γ-phosphate promotes an interaction of this residue via salt bridges with the two catalytic residues Asp-60 and Asp-386.
Figure 2.
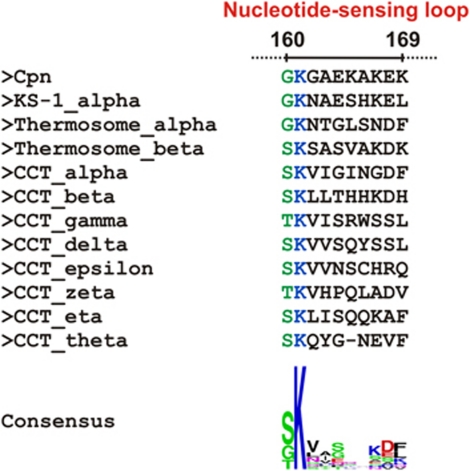
Sequence alignment of the NSL region of all group II chaperonins present in the Protein Data Bank (M. maripaludis; Thermococcus strain KS-1 subunit α; Thermoplasma acidophilum (Thermosome) subunits α and β; and the eight CCT subunits from Bos taurus). The two residues of the NSL of Cpn that interact directly with nucleotide are located at position 160 and 161 (regions coloured in green and blue, respectively). Gly-160 (Cpn sequence) is not highly conserved among the sequences; however, the interaction with nucleotide occurs via the main-chain carbonyl group and the side chain properties do not directly affect this contact. Interestingly, all of the sequences at position 160 have a residue with a short side chain (glycine, serine or threonine). A short side chain might confer the flexibility necessary for the NSL to assume the different conformations observed in the AMP–PNP and ADP states. In contrast with site 160, position 161 shows an extremely conserved lysine residue among the group II chaperonin sequences. The ATPase sensor mechanism proposed for Lys-161 appears to be a general mechanism for the entire class.
Facilitating ADP release after ATP hydrolysis
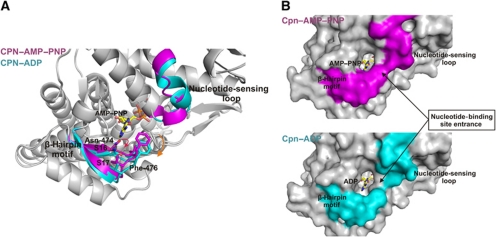
The Cpn–AMP–PNP and Cpn–ADP states display a distinct conformational change around the nucleotide-binding site in addition to the changes in the NSL. The large conformational changes in the NSL, arising from interactions with the nucleotide phosphates, generate a secondary structural rearrangement at the entrance to the nucleotide-binding site, near the nitrogenous adenine base. The entrance to the nucleotide-binding site has a β-hairpin motif consisting of the antiparallel strand-16 and strand-17 connected by three residues in a short loop (residue range 471–482 of the equatorial domain). This β-hairpin structural motif in Cpn–ADP moves away from the nucleotide when compared with the Cpn–AMP–PNP state (Figure 3A) (Supplementary Movie 1). The motion of this β-hairpin motif can be described as a rigid body motion, except for residue Asn-474, which undergoes a change in side chain direction. For the Cpn–AMP–PNP state, the OD1 atom of Asn-474 interacts with N1 and N6 in the adenine base. These interactions are not observed in the Cpn–ADP state and the side chain of Asn-474 displays a different orientation. These changes serve to modulate the size of the entrance portal to the nucleotide-binding site. An important residue in this regard is Phe-476, which makes hydrophobic contacts with NSL residue Ala-163 and the AMP–PNP nucleotide in the Cpn–AMP–PNP state. However, the position of Ala-163 is occupied by the charged residue Glu-164 in the Cpn–ADP state, disrupting the hydrophobic interactions observed in Cpn–AMP–PNP state and promoting a shift of Phe-476. Thus, the rearrangement in the β-hairpin region and NSL residues causes an opening of the nucleotide-binding site (Figure 3B) upon nucleotide hydrolysis. We postulate that the decrease in the number of contacts between the β-hairpin motif and the nucleotide, and the structural opening of the entrance observed in the Cpn–ADP state facilitates discharge of the ADP molecule to initiate a new cycle of protein folding activity.
Figure 3.
(A) Secondary structural rearrangements in the nucleotide-binding site related to NSL motion. The lower region of the nucleotide-binding site possesses a β-hairpin unit formatted from an antiparallel arrangement of strand-16 and strand-17. This β-hairpin structural motif undergoes a rigid body motion (indicated with the orange arrow) between the Cpn–AMP–PNP and Cpn–ADP states, except for the residue Asn-474, which shows a change in size chain direction (indicated with the red arrow). (B) A molecular surface representation of the Cpn–AMP–PNP (magenta) and Cpn–ADP (cyan) states. A rearrangement at the interface between the β-hairpin motif and the NSL promotes an opening of the nucleotide-binding site entrance in the Cpn–ADP state.
Functional analysis of the NSL
After identifying the NSL as the region responsible for nucleotide sensing, we investigated our structural findings by testing the biochemical consequences of mutations in the NSL region. To examine the influence of residue Lys-161 involved with γ-phosphate sensing, we constructed the mutant form Cpn-K161A. Interestingly, another residue in the NSL, Glu-164, makes contact at the interface of the ‘stem loop’, residues ∼45–57, and the C-terminus. This region has previously been hypothesized to act as a hinge point in the open to closed transition in Cpn (Zhang et al, 2010). We observe that Glu-164 participates in inter-subunits contact in the AMP–PNP state, but lacks any interactions with the neighbouring subunit in the ADP state, supporting a potential role in communicating the ATP hydrolysis signal around the ring. Therefore, we constructed the Cpn-E164A mutant to probe the contribution of this residue on the chaperonin cycle.
The last mutant form, Cpn-G160S, was created based on the analysis of primary sequences for the NSL region. This position is equivalent to residue 160 in the mammalian chaperonin TRIC/CCT from bovine and human, and is a serine residue in six subunits (α, β, δ, ε, η and θ) and a threonine residue in two subunits (γ and ζ) (Figure 4). Interestingly, bovine CCT has previously been shown to have a significantly slower ATP hydrolysis rate compared with Cpn (Reissmann et al, 2007). To confirm the impact of the residue at position 160 on the rate of ATP hydrolysis and thus the timing of the folding cycle, Gly-160 was mutated to a serine residue. Therefore, the NSL mutants were designed to probe the role of the residues involved in γ-phosphate sensing (Cpn-K161A), intra-ring contacts (Cpn-E164A), and timing of folding cycle (Cpn-G160S), and how they impact the dynamics of lid closure and ATPase activity.
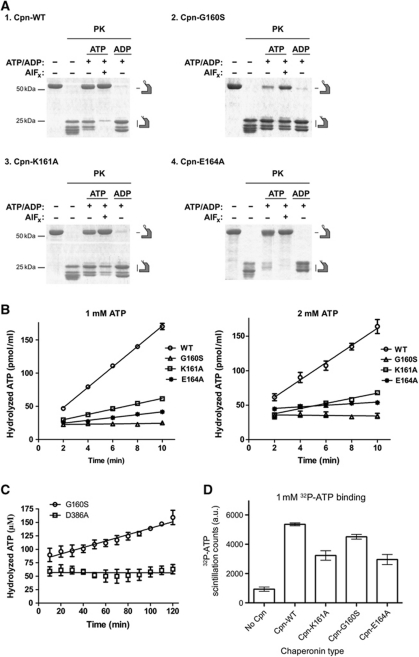
Figure 4.
(A) PK digestion of Cpn-WT, Cpn-G160S, Cpn-K161A and Cpn-E164A mutants, analysed by SDS–PAGE followed by Coomassie blue staining. PK leads to full digestion of the open Cpn lids. Incubation with ATP–AlFx locks the complex closed leading to complete PK protection of lids (compare ATP free and +ATP AlFx lanes). (B) ATP hydrolysis by Cpn-WT, Cpn-G160S, Cpn-K161A and Cpn-E164A measured at 1 and 2 mM of α-[32P]-ATP. (C) ATP hydrolysis by Cpn-G160S and the catalytic dead mutant Cpn-D386A measured at 1 mM α-[32P]-ATP. The mean with the error bars representing standard error of the mean (s.e.m.) is shown. (D) Filter-binding assays for Cpn-WT, Cpn-G160S, Cpn-K161A and Cpn-E164A. Shown is the mean, with the error bars representing s.e.m. Data shown in (C, D) are the result of three independent repeats.
Dynamics of lid closure
To assess the dynamics of the Cpn-WT and NSL mutants, we employed a proteinase-K (PK) assay that distinguishes the open, substrate binding, from the closed, substrate folding state in the Cpn. The PK digests the lid segments of Cpn when the complex is in the open state. However, if the Cpn is closed, the complex remains completely protease resistant (Reissmann et al, 2007; Douglas et al, 2011). In the absence of ATP, the complex is digested into roughly two pieces that are visualized as lower molecular weight bands on an SDS–PAGE gel at ∼29 kDa (Figure 4A, compare −ATP/ADP lanes and +ATP/+ATP and AlFx lanes). The Cpn-WT and Cpn mutants were incubated with either ADP, ATP or ATP–AlFx. Incubation with ADP (Figure 4A, compare +ADP lanes) did not protect the Cpn from PK digestion; however, the presence of ATP or ATP–AlFx promoted either partial or full protection (see Figure 4). Despite mutations in the NSL region, all mutants were still capable of changing conformation to the closed, PK protected, state. This analysis strongly suggests that residues Gly-160, Lys-161 and Glu-164 are not absolutely required for ATP hydrolysis, since the mutants (Cpn-G160S, Cpn-K161A and Cpn-E164A) are still capable of sampling the closed conformation when incubated with nucleotide or the hydrolysis mimic. However, we do observe variations in the levels of PK digestion of the mutants (Figure 4A, +ATP and +ATP–AlFx lanes) compared with Cpn-WT, suggesting that these residues are critical for linking the ATPase activity of the complex to the conformational cycling from the open to closed state.
Impact on ATPase activity
Since the dynamics, opening and closure, of the Cpn are tightly coupled to the ATPase activity, and the dynamics are perturbed in the NSL mutants, we characterized the ATPase activity of Cpn-WT as well as the NSL mutants. In order to compare the ATPase activity among Cpn-WT and the NSL mutants, we performed the assay at saturating ATP concentrations, 1 and 2 mM, values well above the Km for the chaperonin (Reissmann et al, 2007; Figure 4B; Table II). A decrease in the ATPase activity at higher concentration was observed for the Cpn-E164A mutant (Table II). This behaviour has been observed previously in the Cpn-WT (Reissmann et al, 2007), as well as the group I chaperonin GroEL (Yifrach and Horovitz, 1995), and is interpreted to be an indicator of negative cooperativity in the chaperonin. Disrupting the lateral contact at residue E164 may alter communication between adjacent subunits within a ring, but further investigation will be required to determine if there is a link to inter-ring negative cooperativity.
Table 2. ATPase activity data for Cpn-WT, Cpn-G160S, Cpn-K161A and Cpn-E164A.
| 1 mM ATP | 2 mM ATP | |
|---|---|---|
| Cpn-WT | 15.38±0.4031 | 12.49±1.018 |
| Cpn-G160S | 0.205±0.2522 | −0.189±0.5272 |
| Cpn-K161A | 4.018±0.2700 | 3.732±0.3664 |
| Cpn-E164A | 2.122±0.2837 | 1.137±0.2834 |
| ATPase activity measured as ATP hydrolyzed/min (μM/min), with standard error of the mean. | ||
| The data represent the results from three independent repeats. | ||
The replacement of the residue Gly-160 to serine, which is conserved in mammalian CCT, caused a severe reduction in the ATPase activity; however, the PK assay suggested this mutant was still capable of closing its chamber (Figure 4A and B). To further investigate the reduced ATP hydrolysis rate in Cpn-G160S, we monitored the ATPase activity over a longer time scale (Figure 4C). The ATP hydrolysis rate of Cpn-G160S was compared with the catalytically dead mutant Cpn-D386A, as negative control (Reissmann et al, 2007). These experiments suggested that the Cpn-G160S retains ATPase activity, albeit at a much slower rate than observed in the Cpn-WT. This result is consistent with the observation that the Cpn-G160S is partially PK resistant when incubated with either ATP or ATP–AlFx, indicative of a mixed population of closed and open chaperonins, which hydrolyze ATP much slower than the Cpn-WT (see above).
The mutation of the residue Lys-161, which is extremely conserved in all group II chaperonin sequences and directly contacts the γ-phosphate of ATP, generated a significant decrease in ATPase activity. The Cpn-K161A mutant showed an ATPase rate of 25% of the Cpn-WT activity. The Cpn-E164A mutant also shows a strong reduction in its ATPase activity compared with Cpn-WT. Intriguingly, Glu-164 is located ∼15 Å away from the nucleotide-binding site in either the AMP–PNP state or ADP state. Consequently, residue Glu-164 has no contact with nucleotide in either conformation and cannot be directly involved in nucleotide sensing or ATP hydrolysis. However, Glu-164 does make direct contact with the residues of the neighbouring subunits in the AMP–PNP state (Figure 5). Therefore, the reduction in ATPase activity for the Cpn-E164A mutant suggests that this residue plays a role in coupling lateral subunits in the ring to achieve optimal ATPase activity.
Figure 5.
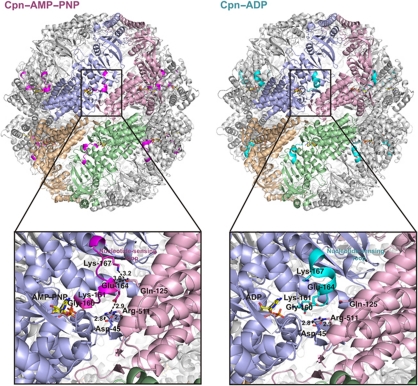
Overview of double ring Cpn–AMP–PNP and Cpn–ADP arrangements show that the NSL (coloured in magenta and cyan, respectively) is located between adjacent subunits. The interaction between Asp-45 and Arg-511 from adjacent subunit serves as hinge for intra-ring motion between the open and closed states of Cpn (Pereira et al, 2010; Zhang et al, 2010). The Cpn–AMP–PNP state demonstrates additional contacts between NSL residues and residues present on the equatorial domain of neighbouring subunits. The residue Glu-164 of NSL makes a salt bridge and a hydrogen bond with the residues Arg-511 and Gln-125. The residue Lys-167 makes an additional hydrogen bond with Gln-125. All these interactions are not present in Cpn–ADP state. Conformational changes in the NSL can be communicated to a lateral subunit by changes in intra-subunits contacts. ATPase assays using NSL residue mutant (Cpn-G160S, Cpn-K161A and Cpn-E164A) shows a significant difference on ATP hydrolysis activity compared with Cpn-WT demonstrating the influence of this region on enzyme activity. The hydrogen bonds are shown in broken lines and the distances are given in Angstroms.
ATP-binding affinity is linked to the NSL
The NSL mutants all show a reduced ATPase activity. To probe the influence of the NSL on the ATP binding we monitored the chaperonin capacity to bind radiolabelled nucleotide at a physiological ATP concentration (Figure 4D). It has previously been suggested that ATP binding to one subunit enhances ATP association with subunits in the same ring (Kafri and Horovitz, 2003; Bigotti et al, 2006; Reissmann et al, 2007). Therefore, ATP-binding affinity is dependent on communication between subunits. Both Cpn-K161A and Cpn-E164A mutants show a modest decrease in ATP binding compared with Cpn-WT (Figure 4D). Intriguingly, the Cpn-G160S mutant displayed comparable ATP binding to that seen in the Cpn-WT, despite a significant drop in ATPase activity (Figure 4C). This result, in conjunction with the PK assay, suggests that the binding of ATP by Cpn-G160S is similar as Cpn-WT but the timing of the conformation cycle is changed.
The reduced ATP binding observed in the Cpn-K161 mutant may result from a combination of disruption of communication of γ-phosphate state between subunits, and the loss of direct stabilizing interactions between the lysine nitrogen and the ATP γ-phosphate. Additionally, the Cpn-E164A mutant has a diminished capacity for ATP binding. The location of residue E164 some 15 Å from the active site argues that this effect cannot be a result of direct interaction with the ATP. This further supports a role for E164 in intra-ring communication and a potential impact on cooperativity within the chaperonin arising from the mutation. To examine the simultaneous influence of residues Lys-161 and Glu-164 involved with γ-phosphate sensing and the lateral subunit contacts, respectively, we created the double-mutant Cpn-K161A/E164A. This mutant did not exhibit additive effects, rather, it resembles the single mutants. A detailed biochemical analysis of this Cpn-K161A/E164A mutant can be found in Supplementary Figure S3.
The transition state Cpn–ADP–AlFx supports the role of Lys-161 as an ATP sensor
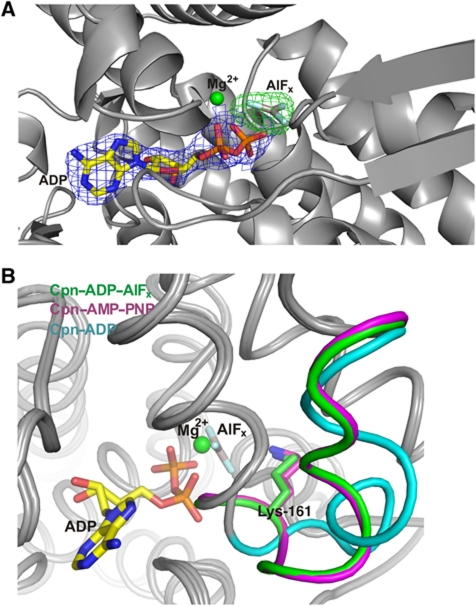
Aluminium fluoride (AlFx) acts as structural analogue of inorganic phosphate and has been widely utilized to analyse the functional cycle of numerous ATPases and chaperonins (Bigay et al, 1987; Menz et al, 2001; Chaudhry et al, 2003; Inobe et al, 2003; Meyer et al, 2003a; Iizuka et al, 2005). The transition state analogue Cpn–ADP–AlFx structure was solved in order to confirm the effect of the transition state on the NSL. The Cpn–ADP–AlFx complex crystallized in similar conditions used to solve the structure of Cpn–ADP state, except for the presence of 50 mM of AlFx in the crystallization buffer. Crystals of Cpn–ADP–AlFx diffracted to 2.7 Å resolution (Table I). The AlFx-binding site was identified using a cross-validated σA weighted difference electron density map (mFo–DFc) contoured at 7.0 σ (Figure 6A). The presence of AlFx at the γ-phosphate-binding site promotes a similar structural arrangement of the NSL observed for the Cpn–AMP–PNP state. The γ-phosphate oxygens of Cpn–AMP–PNP interact with equatorial domain residues Asn-59, Gly-61, Asp-91, Thr-93 and Thr-94 and with the Lys-161 from the NSL of the intermediate domain. One oxygen atom (O3G) of the γ-phosphate is involved in Mg2+ coordination. The interactions between the oxygen atoms of the γ-phosphate of AMP–PNP are replaced by fluorine contacts in the Cpn–ADP–AlFx state (Figure 6B; see Supplementary Figure S4 for a comparison with a previous AlFx group II chaperonin structure). The interaction between the NSL and the γ-phosphate, via Lys-161, is conserved in the Cpn–ADP–AlFx state. Therefore, the conformation of the NSL observed in the Cpn–ADP–AlFx state supports the role of Lys-161 in identifying the presence of the γ-phosphate and acting as the ATP sensor, also during the hydrolysis of nucleotide.
Figure 6.
(A) The AlFx-binding site was identified using a cross-validated σA weighted electron density map (mFo–DFc) contoured at 7.0 σ (coloured in green). A 2mFo–DFc electron density map around the ADP nucleotide contoured at 1.5 σ is shown in blue. (B) The presence of AlFx at the γ-phosphate-binding site promotes a similar structural arrangement in the NSL as observed for the Cpn–AMP–PNP state. The interactions between the oxygen atoms of the γ-phosphate of the AMP–PNP nucleotide are replaced by fluorine contacts in the Cpn–ADP–AlFx structure. The interaction between Lys-161 and the γ-phosphate is conserved in the Cpn–ADP–AlFx structure and allows the NSL to assume a similar conformation to Cpn–AMP–PNP.
Discussion
The structural and biochemical studies here show, for the first time, how the nucleotide state of the group II chaperonin is sensed and potentially communicated around the complex. We also see that the primary amino-acid sequence in a newly identified NSL directly impacts the rate of nucleotide hydrolysis and thus timing of the folding reaction. Finally, the structural changes associated with hydrolysis near the nucleotide-binding site open a portal that may favour product release, ADP, and entry of new ligand, ATP, into the active site.
The NSL is located at the inter-subunit interface. Structural changes in the NSL upon changes in nucleotide state modify the network of contacts involving neighbouring subunits, indicating a potential mechanism for intra-ring communication. A superposition of the open (substrate accepting) and closed (substrate folding) models shows an individual subunit rotation around 35° from open to closed state and the contact between Asp-45 and Arg-511 between adjacent subunits serves as hinge point for intra-ring motion (Pereira et al, 2010; Zhang et al, 2010). Since the NSL residues of the open Cpn model lacks any contact with neighbouring subunit residues, the existence of additional NSL residues interactions with neighbouring subunits associated with the presence of the γ-phosphate could be important in modulating the timing of Cpn ring opening and closing, which is central to protein folding function. Notably, the Cpn–AMP–PNP state shows several additional contacts between NSL residues (intermediate domain) and residues present in the equatorial domain of neighbouring subunits. Residue Glu-164 of the NSL makes an extra salt bridge and a hydrogen bond, respectively, with the residues Arg-511 and Gln-125 from the adjacent subunit. Residue Lys-167 makes an additional hydrogen bond with Gln-125. These interactions are not observed in the Cpn–ADP state (Figure 5). ATP hydrolysis and the conformational changes observed in the NSL create a weaker interaction between individual subunits allowing the ring to re-open in order to complete the refolding-reaction cycle. Therefore, the conformational changes observed in the NSL provide a regulatory mechanism for closing and opening the ring. Moreover, the location of the NSL permits signalling of ATP hydrolysis between subunits of the same ring, which underlies the intra-ring communication characteristic of the group II chaperonins (Figure 5). Consistent with all structures of group II chaperonins obtained to date, we observe symmetric complexes, and there are no differences at the inter-ring interface comparing the Cpn–ADP and Cpn–AMP–PNP states. Although we cannot discount the effects of crystal packing, to observe a difference between the upper and lower rings it is likely that the complex needs be trapped in a state with different nucleotide species in each ring.
Our studies also implicate a key NSL residue, 160 in the Cpn sequence, in determining the rate of nucleotide hydrolysis of chaperonin subunits. Mutation of this residue from a glycine to a serine dramatically reduces ATPase activity, which is very similar to the slower hydrolysis rate observed in bovine CCT/TRiC; in CCT/TRiC, the subunits in the complex possess either a serine or threonine at this position. The amino-acid identity at Cpn-160 provides a route for the modulation and fine-tuning of the nucleotide turnover rate, a function that is likely critical for setting the timer of the chaperonin to accommodate different client proteins and cellular environments. Chaperonins with different hydrolysis rates, and thus protein folding properties, could be engineered by modification of residues in the NSL, especially those that are equivalent to residue 160 in Cpn.
Investigations of the steps in ATP hydrolysis in group II chaperonins have shown that product release from the nucleotide pocket is the rate-limiting step (Bigotti et al, 2006). The structures of Cpn in complex with an ATP analogue and ADP demonstrate that the rearrangement in the NSL and β-hairpin promote an opening of the entrance of the nucleotide-binding site. Also, a decrease in the number of contacts between the β-hairpin motif and the nucleotide facilitates discharge of the ADP molecule. Thus, the diminished ATPase activity of the NSL mutant forms Cpn-G160S and Cpn-K161A could be partially a result of perturbed regulation of product release from the nucleotide pocket. Additionally, the NSL residues are located at an important interface between subunits. Changes in NSL contacts could alter intra-ring communication, modulating positive cooperativity, thus influencing the rates of ATP hydrolysis, as observed for the Cpn-E164A mutant. The structural and biochemical identification of the changes related to ATP hydrolysis provides important insights into the complex puzzle of protein folding by group II chaperonins and suggests routes for engineering to generate modified folding activities.
Materials and methods
Cloning, expression, purification and crystallization of Cpn-WT and Cpn-rls
Cloning, expression and purification of Cpn-WT have been reported elsewhere (Reissmann et al, 2007). The production of Cpn-rls was similar as described for Cpn-WT (Douglas et al, 2011). Both Cpn-WT and Cpn-rls proteins were concentrated and dialysed against 20 mM Hepes buffer pH 7.4 containing 50 mM NaCl, 5 mM MgCl2 and 10% glycerol. The final concentration of Cpn-WT and Cpn-rls used for crystallization trials were 10 and 12 mg/ml, respectively.
The complexes of Cpn-WT–ADP, Cpn-rls–AMP–PNP, Cpn-rls–ADP and Cpn-rls–ADP–AlFx were brought to 5.0 mM ADP (adenosine 5′-diphosphate), 5.0 mM of non-hydrolysable ATP analogue (adenosine 5′-[β,γ-imido]triphosphate, also called AMP–PNP) and 5.0 mM ADP+50 mM AlFx, respectively, prior to crystallization. Cpn-WT–ADP, Cpn-rls–AMP–PNP, Cpn-rls–ADP and Cpn-rls–ADP–AlFx were crystallized using a reservoir solution consisting of 0.2 M Li2SO4 and 20% PEG 3350. Crystals were obtained after 2 days by the sitting-drop vapour-diffusion method with the drops consisting of a mixture of 0.5 μl of protein solution and 0.5 μl of reservoir solution.
X-ray data collection and structure determination
Crystals of Cpn-WT–ADP, Cpn-rls–AMP–PNP, Cpn-rls–ADP and Cpn-rls–ADP–AlFx were sited in a reservoir solution containing 20% (v/v) of glycerol as cryo-protector solution. The X-ray datasets for all the Cpn nucleotides complexes were collected at the Berkeley Center for Structural Biology beamlines 8.2.1 and 8.2.2 of the Advanced Light Source at Lawrence Berkeley National Laboratory (LBNL). The diffraction data were recorded using ADSC-Q315r and ADSC-Q315 detectors, respectively. The datasets were processed using the program HKL-2000 (Otwinowski and Minor, 1997).
The crystal structures of Cpn-WT–ADP, Cpn-rls–AMP–PNP, Cpn-rls–ADP and Cpn-rls–ADP–AlFx were solved by the molecular-replacement method using as a search model the previously solved structure of Cpn-WT–AMP–PNP (PDB code 3KFB) (Pereira et al, 2010) with the program Phaser (McCoy et al, 2007) within the Phenix suite (Adams et al, 2010). Structure refinement was carried out using the phenix.refine program (Afonine et al, 2009). Translation-libration-screw (TLS) refinement was used, with each domain (equatorial, intermediate and apical) of each subunit identified as a TLS group. Manual rebuilding using COOT (Emsley and Cowtan, 2004) and addition of nucleotides, Mg2+ ions, AlFx and water molecules allowed construction of the final models. The ligands sites were interpreted in the various Cpn-WT and Cpn-rls structures using cross-validated sigmaA weighted electron density maps (mFo–DFc Fourier synthesis) contoured at 3.0 σ or higher. RMSD differences from ideal geometries for bond lengths, angles and dihedrals were calculated with Phenix (Adams et al, 2010). The overall stereochemical quality of the final models for Cpn-WT–ADP, Cpn-rls–AMP–PNP, Cpn-rls–ADP and Cpn-rls–ADP–AlFx were assessed using the program MOLPROBITY (Davis et al, 2007). The overall statistics for X-ray data collection and structures refinement are summarized in Table I.
Mutant construction for kinetic experiments
The Cpn-WT plasmid was used to create the Cpn-rls, Cpn-G160S, Cpn-K161A and Cpn-E164A mutants. The mutants of Cpn were generated using the QuickChange kit (Stratagene). Upon request, details of the specific primers used for construction of Cpn-rls, Cpn-G160S, Cpn-K161A and Cpn-E164A mutants will be provided.
PK protection assay
The purified Cpn-WT, Cpn-G160S, Cpn-K161A and Cpn-E164A proteins were incubated in ATPase buffer (100 mM KCl, 30 mM TRIS pH 7.4, 5 mM MgCl2, 10% glycerol, 1 mM DTT) at a final concentration of 0.25 μM. The reaction was supplemented with either ADP (1 mM), ATP (1 mM), 1 mM ATP–AlFx (1 mM ATP, 1 mM Al(NO3)3, 6 mM NaF) and incubated at 37°C for 10 min. The proteolysis was initiated by adding 20 μg/ml PK, and incubated for 10 min at 25°C then quenched by adding 5 mM PMSF to inhibit the protease activity. The reactions were incubated on ice for 10 min, and the digestion products were analysed by SDS–PAGE.
Kinetic experiments: ATPase assay
The ATPase activity of the Cpn-WT, Cpn-G160S, Cpn-K161A and Cpn-E164A were measured as previously described (Reissmann et al, 2007). In brief, after pre-incubation in ATPase buffer at 37°C, the chaperonin (0.25 μM) was supplemented with 1 mM and 2 mM α-[32P]-ATP. At times indicated, 2 μl of the reaction was transferred to a PEI-Cellulose TLC plate (Macherey-Nagel). The plates were developed in a TLC chamber containing 1 M LiCl and 0.5 M formic acid in H2O. After developing, the plates were dried and exposed with a phosphor screen (Kodak), then analysed using a Typhoon Scanner and Image Quant 5.2.
Filter-binding assays
In all, 1 μM Cpn (or Cpn mutant) was incubated in Cpn ATPase buffer (30 mM TRIS pH 7.4, 100 mM KCl, 5 mM MgCl2, 10% glycerol and 1 mM DTT) with 1 mM α-[32P]-ATP (0.1 μCi/μmol) for 5 min at 37°C. The reaction was then loaded on a 0.45-μm HA nitrocellulose filter (Millipore) connected to a vacuum system. The reaction was washed with 1 ml cold ATPase buffer. The filter was then dried and the amount of α-[32P]-ATP on the filter was assessed by scintillation counting. Each reaction was done in triplicate. Reported is the mean and s.e.m.
Accession numbers
The atomic coordinates and structure factors of Cpn-WT–ADP, Cpn-rls–ADP, Cpn-rls–AMP–PNP and Cpn-rls–ADP–AlFx have been deposited in the Protein Data Bank with accession codes 3RUQ, 3RUS, 3RUV and 3RUW, respectively.
Supplementary Material
Acknowledgments
We are grateful to the staff of the Berkeley Center for Structural Biology at the Advanced Light Source of Lawrence Berkeley National Laboratory. The Berkeley Center for Structural Biology was supported in part by the National Institutes of Health and National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source was supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract No. DE-AC02-05CH11231. This work was performed as part of the Center for Protein Folding Machinery; a NIH Roadmap-supported Nanomedicine Development Center, through Grant 2PN2EY016525.
Author contributions: PDA and JF designed and lead the project. JHP wrote the manuscript. JHP, CYR and RPM carried out the X-ray crystallography experiments. NRD, RK, KMK and JAK carried out the cloning, expression and purification of the proteins. NRD and TL carried out the PK digestion and ATPase activity assays. All the authors made contributions to the final manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Urzhumtsev A, Adams PD (2009) Automatic multiple-zone rigid-body refinement with a large convergence radius. J Appl Crystallogr 42: 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J, Deterre P, Pfister C, Chabre M (1987) Fluoride complexes of aluminium or beryllium act on G-proteins as reversibly bound analogues of the gamma phosphate of GTP. EMBO J 6: 2907–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigotti MG, Bellamy SR, Clarke AR (2006) The asymmetric ATPase cycle of the Thermosome: elucidation of the binding, hydrolysis and product-release steps. J Mol Biol 362: 835–843 [DOI] [PubMed] [Google Scholar]
- Booth CR, Meyer AS, Cong Y, Topt M, Sali A, Ludtke SJ, Chiu W, Frydman J (2008) Mechanism of lid closure in the eukaryotic chaperonin TRiC/CCT. Nat Struct Biol 15: 746–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry C, Farr GW, Todd MJ, Rye HS, Brunger AT, Adams PD, Horwich AL, Sigler PB (2003) Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics. EMBO J 22: 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare DK, Stagg S, Quispe J, Farr GW, Horwich AL, Saibil HR (2008) Multiple states of a nucleotide-bound group 2 chaperonin. Structure 16: 528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Berger JM (2005) Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI. Structure 13: 873–882 [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendal WB III, Soeyink J, Richardson JC, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35: 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Roe SM, McCormack EA, Beuron F, Pearl LH, Willison KR (2011) The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic chaperonins. EMBO J 30: 3078–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel L, Lowe J, Stock D, Stetter K-O, Huber H, Huber R, Steinbacher S (1998) Crystal structure of the thermosome, the archael chaperonin and homolog of CCT. Cell 93: 125–138 [DOI] [PubMed] [Google Scholar]
- Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J (2011) Dual action of ATP hydrolysis couples lid closure to substrate release into the group II chaperonin chamber. Cell 144: 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU (1992) Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J 11: 4767–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL, Farr GW, Fenton WA (2006) GroEL-GroES-mediated protein folding. Chem Rev 106: 1917–1930 [DOI] [PubMed] [Google Scholar]
- Hou Y, Hu Z, Zhang K, Wang L, Zhai Y, Zhou Q, Lander G, Zhu J, He Y, Pang X, Xu W, Bartlam M, Dong Z, Sun F (2010) Crystal structure of group II chaperonin in the open state. Structure 18: 1270–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka R, Yoshida T, Ishii N, Zako T, Takahashi K, Maki K, Inobe T, Kuwajima K, Yohda M (2005) Characterization of archaeal group II chaperonin-ADP-metal fluoride complexes: implications that group II chaperonins operate as a ‘two-stroke engine’. J Biol Chem 280: 40375–40383 [DOI] [PubMed] [Google Scholar]
- Inobe T, Kikushima K, Makio T, Arai M, Kuwajima K (2003) The allosteric transition of GroEL induced by metal fluoride-ADP complexes. J Mol Biol 329: 121–134 [DOI] [PubMed] [Google Scholar]
- Kafri G, Horovitz A (2003) Transient kinetic analysis of ATP-induced allosteric transitions in the eukaryotic chaperonin containing TCP-1. J Mol Biol 326: 981–987 [DOI] [PubMed] [Google Scholar]
- Kafri G, Willison KR, Horovitz A (2001) Nested allosteric interactions in the cytoplasmic chaperonin containing TCP-1. Protein Sci 10: 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz RI, Walker JE, Leslie A (2001) Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell 106: 331–341 [DOI] [PubMed] [Google Scholar]
- Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, Frydman J (2003a) Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell 113: 369–381 [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH (2003b) Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell 11: 647–658 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pereira JH, Ralston CY, Douglas N, Meyer D, Knee KM, Goulet DR, King JA, Frydman J, Adams PD (2010) Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle. J Biol Chem 285: 27958–27966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann S, Parnot C, Booth CR, Chiu W, Frydman J (2007) Essential function of the built-in lid in the allosteric regulation of eukaryotic and archaeal chaperonins. Nat Struct Biol 14: 432–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura Y, Yoshida T, Iizuka R, Maruyama T, Yohda M, Miki K (2004) Crystal structures of the group II chaperonin from Thermococcus strain KS-1: steric hindrance by the substituted amino acid, and inter-subunit rearrangement between two crystal forms. J Mol Biol 335: 1265–1278 [DOI] [PubMed] [Google Scholar]
- Sprang SR (1997) G protein mechanisms: insights from structural analysis. Ann Rev Biochem 66: 639–678 [DOI] [PubMed] [Google Scholar]
- Yifrach O, Horovitz A (1995) Nested cooperativity in the ATPase activity of the oligomeric chaperonin GroEL. Biochemistry 34: 5303–5308 [DOI] [PubMed] [Google Scholar]
- Zhang J, Baker ML, Schroder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, Frydman J, Chiu W (2010) Mechanism of folding chamber closure in a group II chaperonin. Nature 463: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ma B, DiMaio F, Douglas NR, Joachimiak LA, Baker D, Frydman J, Levitt M, Chiu W (2011) Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure. Structure 19: 633–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.