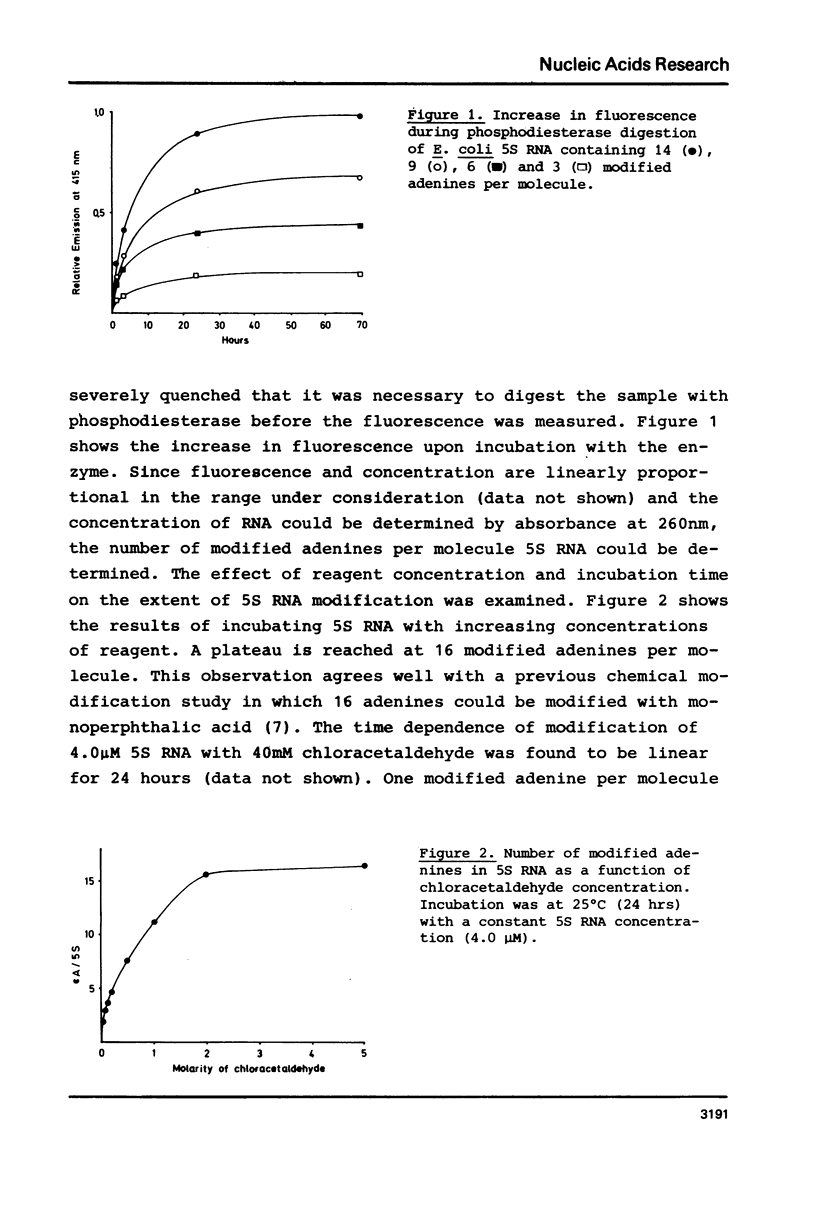
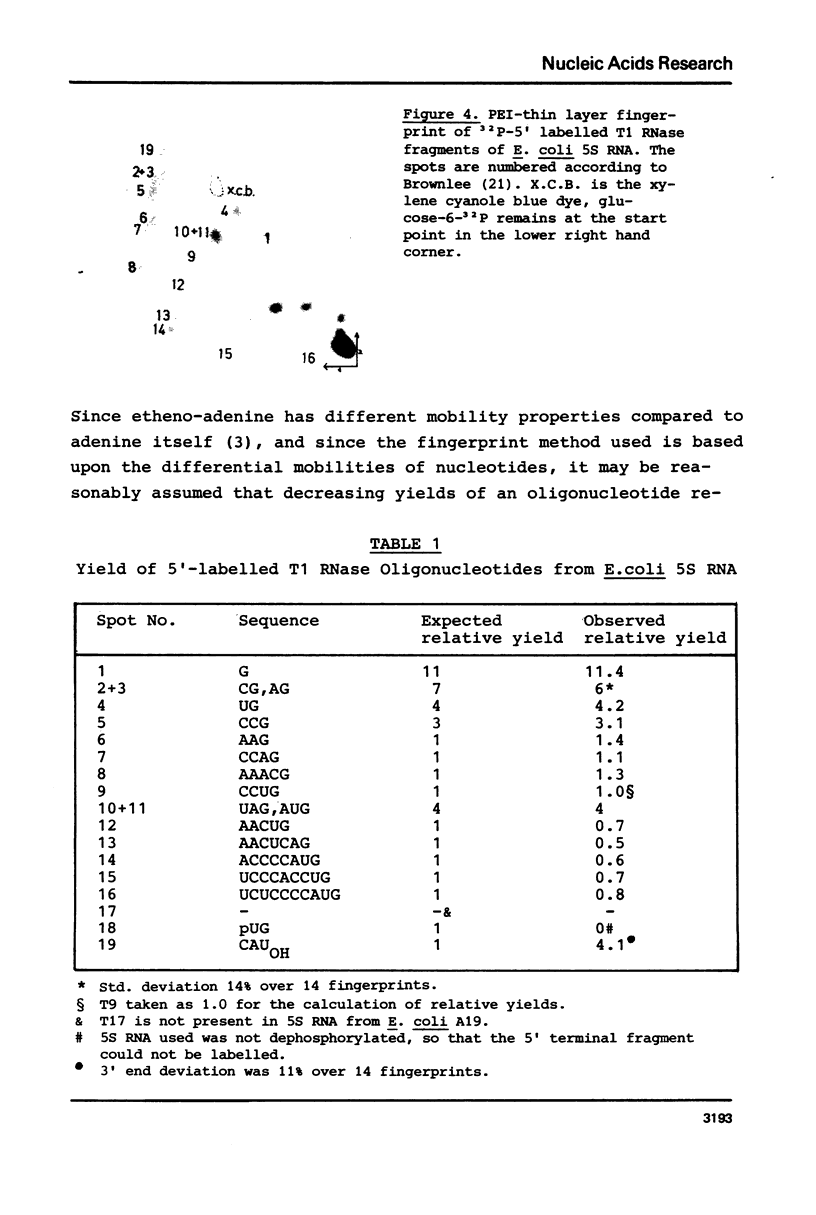
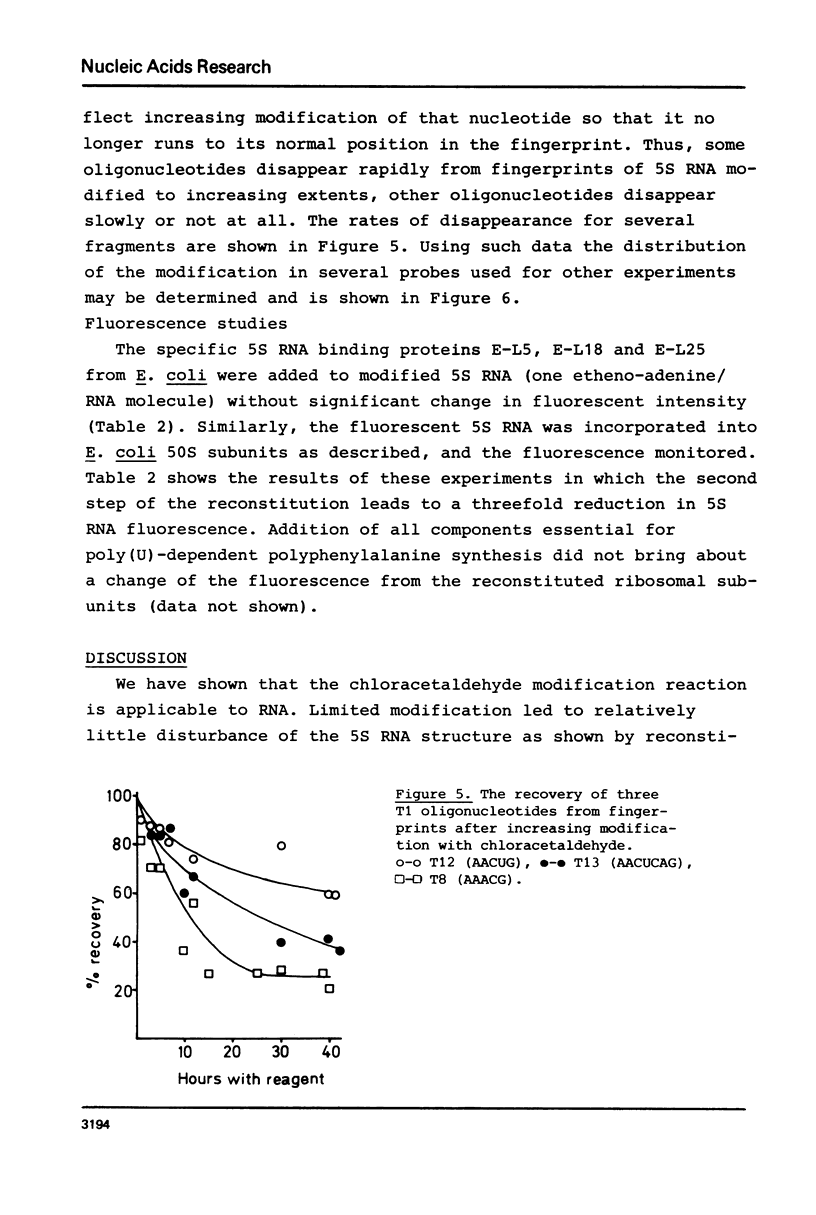
Abstract
Reaction of 5S RNA with chlorocetaldehyde leads to the conversion of unpaired adenines to the fluorescent 1,N6-etheno-adenine derivatives. Up to 16 of the 23 adenines in free 5S RNA can be modified, the fastest reacting are A29, A34, A57-59. Partial modification of adenines in this area results in a 20% reduction in the efficiency of 5S RNA incorporation into 50S subunits during reconstitution and a 15% reduction in the activity of these subunits in peptide synthesis. Fluorescence from 1,N6-etheno-adenine is quenched in free 5S RNA and is not detectably further influenced by the binding of proteins E-L5, E-L18 and E-L25, nor by the first stage of the two step E. coli 50S subunit reconstitution procedure. However, the fluorescence is further reduced to near zero after the second step of the reconstitution. Thus, 5S RNS free in solution contains 16 unpaired adenines, those in the region between A29 and A59 particularly accessible to modification by chlorocetaldehyde. This portion of the 5S RNA molecule appears to undergo either a conformational change or interacts with other ribosomal components in the last stage of subunit reassembly.
Full text
PDF




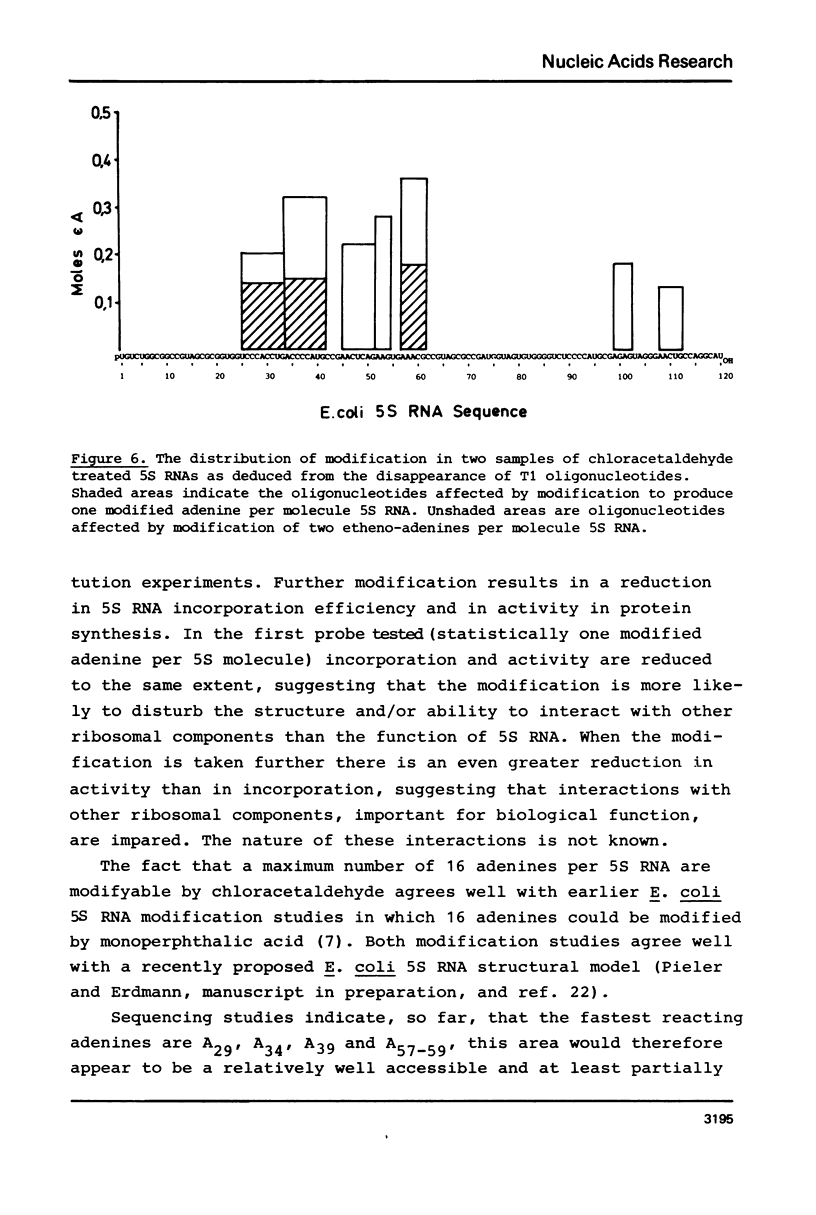
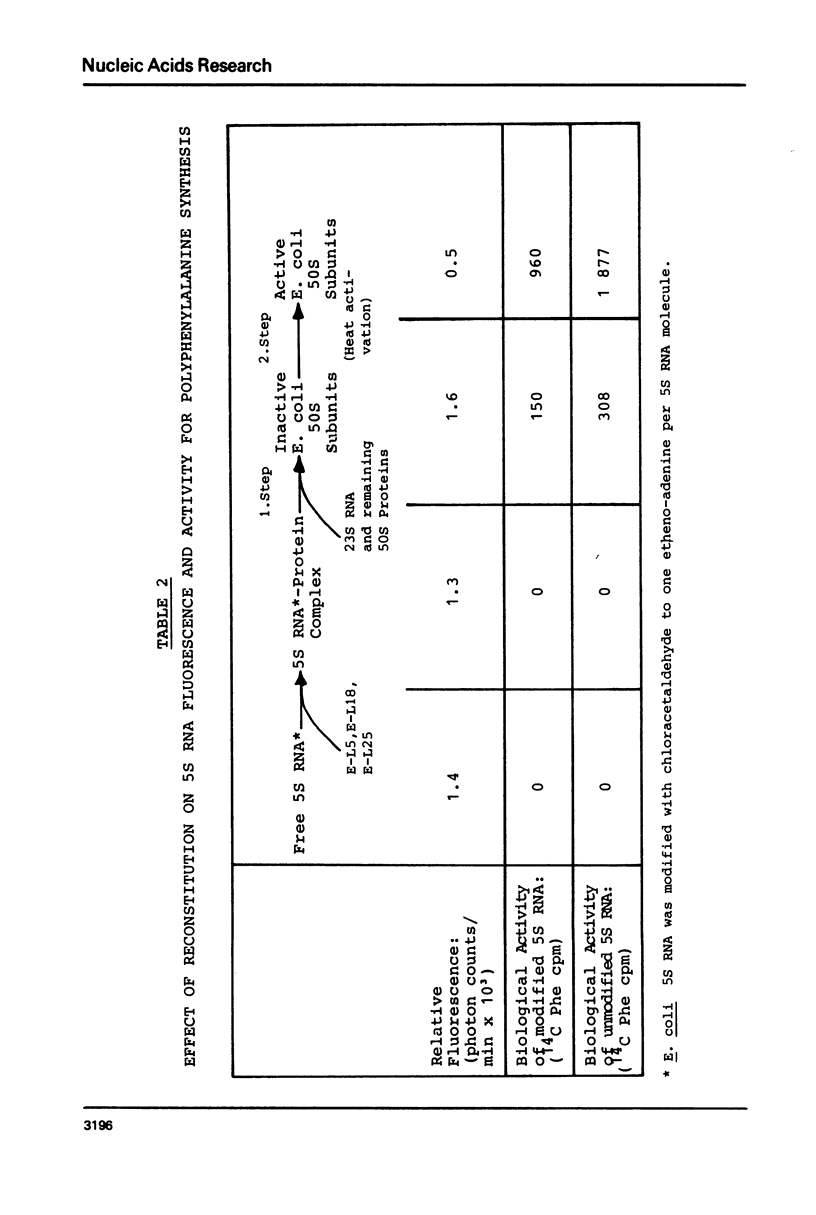


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrio J. R., Secrist J. A., 3rd, Leonard N. J. Fluorescent adenosine and cytidine derivatives. Biochem Biophys Res Commun. 1972 Jan 31;46(2):597–604. doi: 10.1016/s0006-291x(72)80181-5. [DOI] [PubMed] [Google Scholar]
- Bellemare G., Jordan B. R., Rocca-Serra J., Monier R. Accessibility of Escherichia coli 5S RNA base residues to chemical reagents. Influence of chemical alterations on the affinity of 5S RNA for the 50S subunit structure. Biochimie. 1972;54(11):1453–1466. doi: 10.1016/s0300-9084(72)80087-7. [DOI] [PubMed] [Google Scholar]
- Burrell H. R., Horowitz J. Binding of ribosomal proteins to RNA covalently coupled to agarose. Eur J Biochem. 1977 May 16;75(2):533–544. doi: 10.1111/j.1432-1033.1977.tb11554.x. [DOI] [PubMed] [Google Scholar]
- Cramer F., Erdmann V. A. Amount of adenine and uracil base pairs in E. coli 23S, 16S and 5S ribosomal RNA. Nature. 1968 Apr 6;218(5136):92–93. doi: 10.1038/218092a0. [DOI] [PubMed] [Google Scholar]
- Cronenberger J. H., Erdmann V. A. Stimulation of polypeptide polymerization by blocking of free sulphydryl groups in Escherichia coli ribosomal proteins. J Mol Biol. 1975 Jun 15;95(1):125–137. doi: 10.1016/0022-2836(75)90340-x. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A., Doberer H. G. Structure and function of 5S RNA: the role of the 3' terminus in 5S RNA function. Mol Gen Genet. 1972;114(2):89–94. doi: 10.1007/BF00332779. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A., Fahnestock S., Higo K., Nomura M. Role of 5S RNA in the functions of 50S ribosomal subunits. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2932–2936. doi: 10.1073/pnas.68.12.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A. Structure and function of 5S and 5.8 S RNA. Prog Nucleic Acid Res Mol Biol. 1976;18:45–90. [PubMed] [Google Scholar]
- Larrinua I., Delihas N. Accessibility of guanine at position 44 in the invariant sequence 5'CCG44AAC3' of Escherichia coli 5S RNA to reaction with kethoxal. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4400–4404. doi: 10.1073/pnas.76.9.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus K. H., Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F., Chaires J. B. Functional modification of 16S ribosomal RNA by kethoxal. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3115–3118. doi: 10.1073/pnas.69.11.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom O. W., Craig B. B., Hardesty B. A. The conformation of the anticodon loop of yeast tRNAPhe in solution and on ribosomes. Biopolymers. 1978 Dec;17(12):2909–2931. doi: 10.1002/bip.1978.360171212. [DOI] [PubMed] [Google Scholar]
- Secrist J. A., 3rd, Barrio J. R., Leonard N. J., Weber G. Fluorescent modification of adenosine-containing coenzymes. Biological activities and spectroscopic properties. Biochemistry. 1972 Sep 12;11(19):3499–3506. doi: 10.1021/bi00769a001. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Jou W. M., Fiers W. Analysis of 32P-labeled bacteriophage MS2 RNA by a mini-fingerprinting procedure. Anal Biochem. 1976 May 7;72:433–446. doi: 10.1016/0003-2697(76)90551-0. [DOI] [PubMed] [Google Scholar]
- Wagner R., Garrett R. A. Chemical evidence for a codon-induced allosteric change in tRNALys involving the 7-methylguanosine residue 46. Eur J Biochem. 1979 Jul;97(2):615–621. doi: 10.1111/j.1432-1033.1979.tb13151.x. [DOI] [PubMed] [Google Scholar]
- Wrede P., Erdmann V. A. Activities of B. stearothermophilus 50 S ribosomes reconstituted with prokaryotic and eukaryotic 5 S RNA. FEBS Lett. 1973 Jul 15;33(3):315–319. doi: 10.1016/0014-5793(73)80219-4. [DOI] [PubMed] [Google Scholar]
- Wrede P., Pongs O., Erdmann V. A. Binding oligonucleotides to Escherichia coli and Bacillus stearothermophilus 5 S RNA. J Mol Biol. 1978 Mar 25;120(1):83–96. doi: 10.1016/0022-2836(78)90296-6. [DOI] [PubMed] [Google Scholar]
- Zimmermann J., Erdmann V. A. Identification of Escherichia coli and Bacillus stearothermophilus ribosomal protein binding sites on Escherichia coli 5S RNA. Mol Gen Genet. 1978 Apr 17;160(3):247–257. doi: 10.1007/BF00332968. [DOI] [PubMed] [Google Scholar]