Abstract
The CCA-adding enzyme [ATP(CTP):tRNA nucleotidyltransferase] adds CCA to the 3′ ends of transfer RNAs (tRNAs), a critical step in tRNA biogenesis that generates the amino acid attachment site. Here, we show that the CCA-adding enzyme plays a key role in tRNA quality control as it selectively marks structurally unstable tRNAs and tRNA-like small RNAs for degradation. Instead of adding CCA to the 3′ ends of these transcripts, CCA-adding enzymes from all three kingdoms of life add CCACCA. In addition, hypomodified mature tRNAs are subjected to CCACCA addition as part of a rapid tRNA decay (RTD) pathway in vivo. We conjecture that CCACCA addition is a universal mechanism for controlling tRNA levels and preventing errors in translation.
Although tRNAs require CCA at their 3′ ends for amino acid attachment and correct positioning in the ribosome, CCA is not encoded in nearly all eukaryotic tRNA genes or in many archaeal and bacterial tRNA genes (1-4). Instead, the CCA-adding enzyme post-transcriptionally adds CCA to the 3′ ends of tRNAs and tRNA-like transcripts (5-8). Recent work has identified two tRNA-like small RNAs, mascRNA (MALAT1-associated small cytoplasmic RNA) and the MEN β tRNA-like small RNA, that are generated by 3′ end processing of long nuclear-retained noncoding RNAs in human and mouse cells (9-11) (Fig. 1A). These RNAs are ∼70% similar in sequence and are generated by enzymes involved in canonical tRNA biogenesis, including RNases P and Z. Although the long noncoding RNAs from which they are processed are expressed at roughly equal levels, the MEN β tRNA-like small RNA is below the threshold of detection by Northern blot analysis (Fig. 1B) (12). This suggests that these tRNA-like transcripts are differentially regulated post-transcriptionally. Using a more sensitive 3′ rapid amplification of cDNA ends (RACE) PCR approach, we detected expression of the MEN β tRNA-like small RNA in vivo (Fig. S1). Surprisingly, nearly all sequenced transcripts ended in CCACCA or CCACC, which is not encoded in the genome. This 3′ end modification was not detected on mascRNA (9) and thus would be consistent with CCACCA addition signaling RNA degradation.
Fig. 1. CCACC(A) is added to the MEN β tRNA-like small RNA.
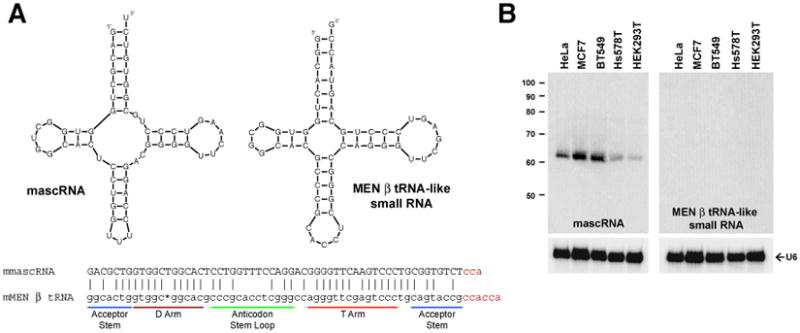
(A) mascRNA and the MEN β tRNA-like small RNA are predicted to fold into cloverleaf secondary structures. The mouse homologs are shown. 3′ RACE revealed that different 3′ terminal sequences are post-transcriptionally added to the RNAs (shown in red). (B) Northern blot analysis using 15 μg of total RNA from five cell lines.
Although the CCA-adding enzyme is thought to terminate after CCA synthesis (7, 8), it was the most likely candidate to catalyze CCACCA addition. We thus expressed and purified His-tagged versions of the CCA-adding enzyme from human (13), Escherichia coli (14), and Sulfolobus shibatae (15) (Fig. S2, A to C) and confirmed that all three enzymes terminate polymerization once CCA has been added to canonical tRNAs (Fig. S2, D to F). In contrast, CCACCA or CCACC was added to the MEN β tRNA-like small RNA in vitro (Fig. S3), recapitulating the in vivo 3′ RACE results. This indicates that the CCA-adding enzyme catalyzes CCA or CCACCA addition depending on the characteristics of the RNA substrate.
To determine the sequence elements required for CCACCA addition, we investigated converting mouse mascRNA from a CCA to a CCACCA target by generating chimeric mascRNA-MEN β tRNA-like transcripts (Fig. 2A and fig. S4A). Swapping the acceptor stem (Mut 1) was sufficient to convert mascRNA from a CCA to a CCACCA target in vitro with near 100% efficiency, whereas the other arms had no effect on CCA addition (Fig. 2B and fig. S4). CCA-adding enzymes from all three kingdoms of life added CCACCA to the Mut 1 transcript, despite the eukaryotic (Fig. 2B) and eubacterial enzymes (Fig. S4C) having a different catalytic mechanism from archaeal enzymes (Fig. 2C) (16-20). Substituting the acceptor stems of canonical tRNAs with that of the mouse MEN β tRNA-like small RNA similarly converted those transcripts to CCACCA targets in vitro (Fig. S5A). This indicates that the CCA-adding enzyme monitors the acceptor stem when selecting between CCA and CCACCA addition. Furthermore, the CCA-adding enzyme functions as a CCACCA repair enzyme as it can, for example, add an additional A to a substrate ending in CCACC (Fig. S5B).
Fig. 2. An unstable acceptor stem is isomerized to allow for CCACCA addition.
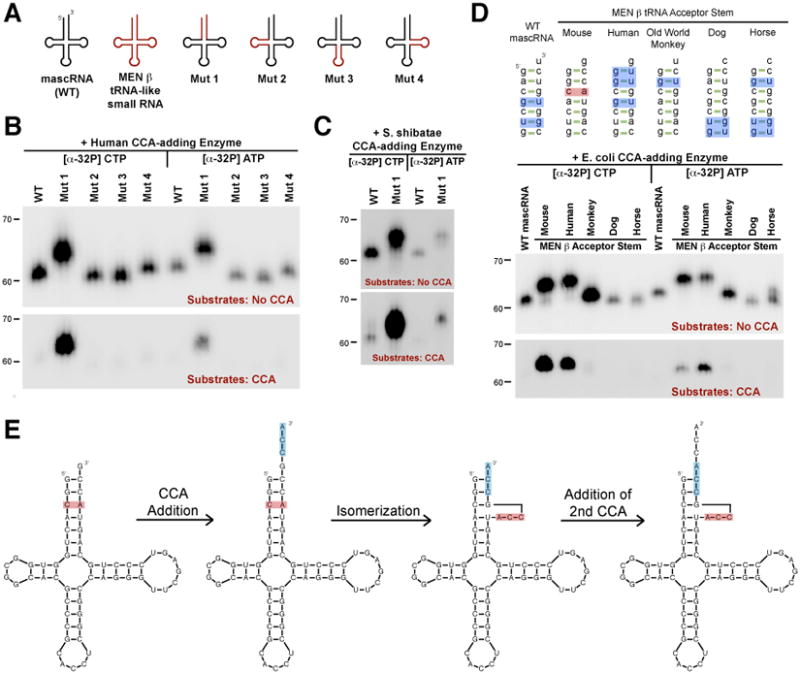
(A) Chimeric transcripts generated. (B and C) Wild-type mascRNA (denoted WT) and the chimeras were incubated in vitro with human (B) or S. shibatae (C) CCA-adding enzyme. [α-32P] CTP or [α-32P] ATP was added to assay for incorporation of each nucleotide into the unlabeled substrates. In the upper panels, substrates lacked CCA so all substrates should undergo, at minimum, one round of CCA addition, with a second round of CCA addition (yielding CCACCA) generating a 3 nucleotide shift in mobility. In the lower panels, CCA was already present so only RNAs that are CCACCA targets yield visible bands. (D) WT mascRNA or chimeras in which the acceptor stem was swapped with that of a MEN β homolog were used as substrates. Mismatches and G-U wobble base pairs are in red and blue, respectively. (E) Model for how CCACCA is added to the mouse MEN β tRNA-like small RNA.
To investigate the features in the mouse MEN β acceptor stem that signal CCACCA addition, we took advantage of the fact that this stem is poorly conserved across species (Fig. 2D and fig. S6). When the mascRNA acceptor stem was swapped with that of the human MEN β tRNA-like transcript, CCACCA addition was also observed in vitro (Fig. 2D). The end of the acceptor stems of the mouse and human MEN β transcripts are significantly unstable due to a C-A mismatch or multiple G-U wobbles, respectively. In contrast, other MEN β homologs have significantly more stable acceptor stems and were CCA targets in vitro. Using COS7 cells, we confirmed that the Old World monkey MEN β homolog is a CCA target and stable transcript in vivo that is easily detected using Northern blot analysis (Fig. S7).
By extensively mutagenizing mascRNA and canonical tRNAs, we determined that CCACCA addition requires a transcript to have an unstable acceptor stem and guanosines at the first and second positions (See SOM, Fig. S8 to S11). Instability facilitates isomerization of the acceptor stem after the first CCA is added, allowing the Cs of the CCA sequence to base pair with G1G2 (Fig. 2E). The A of the CCA sequence becomes the new discriminator base and is positioned in the enzyme's catalytic pocket, ready to undergo a second round of CCA addition using the well-established polymerization mechanisms (8, 17-19). As to why polymerization stops once CCACCA has been added, we propose that additional unpaired nucleotides in the bulge interfere with enzyme recognition.
Could addition of CCACCA be a common reaction for many tRNAs? Between 30 and 45 percent of tRNA genes in a given species begin with G1G2, indicating a strong enrichment for GG at 5′ ends (Fig. 3A and fig. S12). However, it is unlikely that most of these tRNAs are subjected to CCACCA addition under normal conditions as they have stable acceptor stems. Single point mutations that cause a mismatch or an additional G-U wobble in the acceptor stems of two unrelated arginine tRNAs (Fig. 3B and fig. S13) or a cysteine tRNA (Fig. S14) were sufficient to completely convert these transcripts from CCA to CCACCA targets in vitro. Mutant tRNAs ending in CCACCA were rapidly degraded in HeLa nuclear extracts (Fig. 3C and fig. S15), indicative of an efficient decay mechanism that is distinct from cleavage in the anticodon loop (21).
Fig. 3. Destabilizing tRNAs causes CCACCA addition and transcript degradation in vitro.
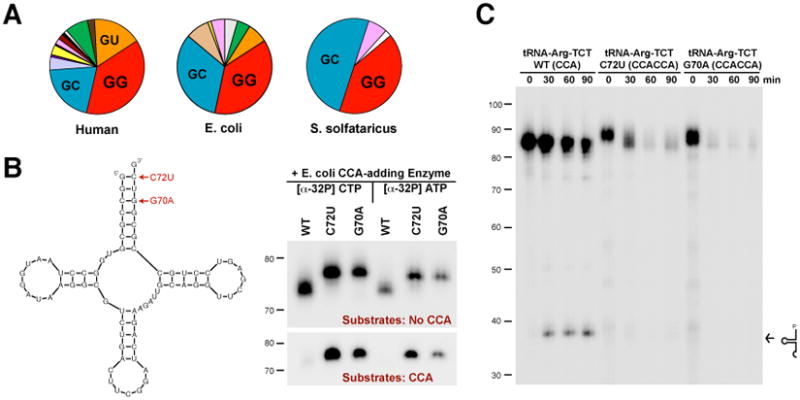
(A) Pie charts showing the first two nucleotides of canonical tRNAs from human (representing 631 tRNAs), E. coli (88 tRNAs), and S. solfataricus (46 tRNAs). (B) tRNAArg(TCG) transcripts containing point mutations were used as in vitro substrates for the E. coli CCA-adding enzyme. (C) Radiolabeled wild-type or mutant arginine tRNAs ending in CCA or CCACCA, respectively, were incubated in HeLa nuclear extracts for the indicated times. Arrow denotes the accumulation of wild-type tRNAs cleaved in the anticodon loop.
The rapid tRNA decay (RTD) pathway functions in yeast as a surveillance mechanism to degrade a subfraction of mature tRNAs (22-24). Originally identified as a mechanism to degrade specific hypomodified tRNAs, the RTD pathway also degrades tRNAs containing mutations that destabilize their structure (25). We noticed that tRNAs sensitive to RTD often conform to the rules for CCACCA addition as they tend to start with GG and have slightly destabilized acceptor stems due to a single G-U wobble near their end (Fig. 4A). We, therefore, hypothesized that CCACCA may be added to tRNAs being degraded by RTD in vivo.
Fig. 4. CCACCA is added to tRNAs being actively degraded in vivo.
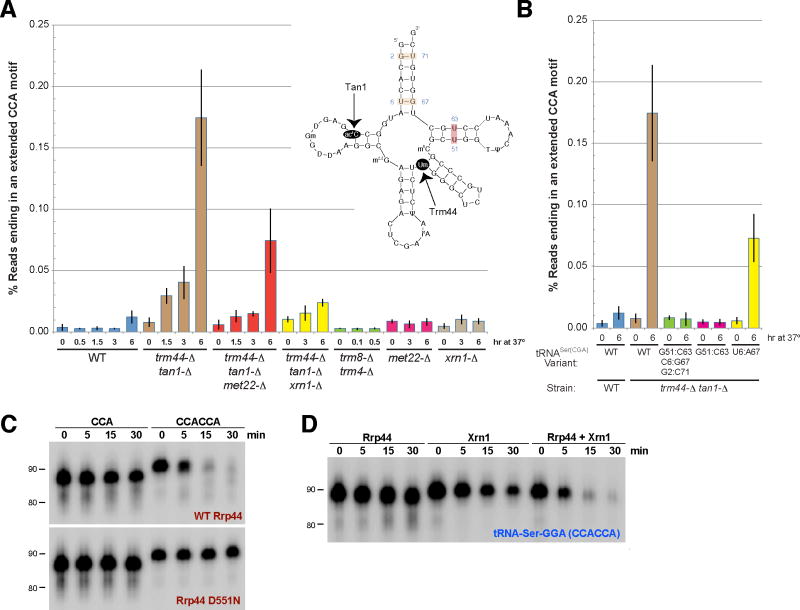
(A) The percentage of tRNASer(CGA) and tRNASer(UGA) transcripts ending in an extended CCA motif in yeast strains grown at 28°C (designated 0 hr) or 37°C for the indicated times was determined. The structure of tRNASer(CGA) is shown. (B) The endogenous tRNASer(CGA) gene was replaced in the trm44-Δ tan1-Δ strain with the variants shown and the strains were subjected to 3′ RACE PCR. Error bars represent standard deviation. (C) A serine tRNA ending in CCACCA was degraded in vitro by yeast Rrp44. The Rrp44 D551N mutant lacks exonuclease catalytic activity. (D) Yeast Rrp44 and Xrn1 cooperate to degrade tRNAs ending in CCACCA in vitro. The enzymes were titrated to identify conditions in which minimal degradation was observed when only a single exonuclease was present.
Here, we focused on tRNASer(CGA) and tRNASer(UGA), two highly similar species that are rapidly degraded in a trm44-Δ tan1-Δ strain grown at high temperatures due to the lack of 2′-O-methylation of U44 and N4 acetylation of C12 (24). By combining 3′ RACE PCR and deep sequencing (Fig. S16), we detected a low, but reproducible, level of extended CCA motifs (defined as CCAC, CCACC, or CCACCA) on tRNASer(CGA) and tRNASer(UGA) in a wild type strain grown at 28°C or 37°C (Fig. 4A and tables S1-2). Consistent with a role for CCACCA addition in this proofreading pathway, switching the trm44-Δ tan1-Δ strain to growth at 37°C triggers RTD of tRNASer(CGA) and tRNASer(UGA) and a progressive increase in transcripts ending in extended CCA motifs, which mirrors the time course of degradation (24). Interestingly, we also detected a significant increase in tRNAs with short poly(A) tails added to their CCA ends when RTD was occurring (Fig. S17 and tables S3-4). Deleting the 5′-3′ exonuclease Xrn1 or Met22, a phosphatase whose substrate is an Xrn1 inhibitor, prevents tRNA degradation by RTD (23) and, likewise, significantly decreased the level of extended CCA motifs and poly(A) tails observed (Fig. 4A).
As increasing the stability of the acceptor or T-stems of tRNASer(CGA) was shown to decrease RTD sensitivity (25), we asked if strengthening the tRNA tertiary structure would also affect CCACCA addition levels in vivo. Changing the U51:U63 mismatch to a G51:C63 base pair near completely abolished CCACCA addition, whereas substitution of U6:G67 by U6:A67 decreased CCACCA levels by ∼60%, consistent with its more modest increase in predicted stability (Fig. 4B and table S2).
We, therefore, suggest that CCACCA or poly(A) addition generates a single-stranded tail that is sufficiently long to be recognized by 3′-5′ exonucleases. Indeed, tRNAs ending in CCACCA, but not CCA, were degraded in vitro by RNase R from E. coli (26) (Fig. S18A) and by yeast Rrp44, the active 3′-5′ exonuclease component of the exosome (27) (Fig. 4C and fig. S18B). Consistent with the genetics and deep sequencing, we found that yeast Xrn1 cooperates with Rrp44 to degrade tRNAs ending in CCACCA in vitro (Fig. 4D and fig. S18C). Rrp44 often pauses once it nears the double-stranded acceptor stem (Fig. S18C) and we propose that Xrn1 recruitment allows efficient degradation of the rest of the transcript. In the absence of Xrn1, it is likely that the CCA-adding enzyme repairs the 3′ end of the tRNA.
Our results suggest a model in which the CCA-adding enzyme adds CCACCA to tRNAs with unstable structures and a 5′ terminal GG sequence, preventing aminoacylation and marking them for degradation (Fig. S19). Many events can destabilize a tRNA, including transcriptional errors, nucleotide modifications that alter base pairing strengths, or conformational changes caused by the binding of proteins or other RNAs in trans. In addition, some tRNAs have genomically encoded mismatches that are corrected by post-transcriptional editing events (28, 29), a process likely proofread by the CCA-adding enzyme as the unedited transcripts are CCACCA targets (Fig. S20). Considering that CCACCA addition is conserved across all three kingdoms of life, surveillance by the CCA-adding enzyme likely represents a universal mechanism for ensuring accurate protein synthesis.
Supplementary Material
Acknowledgments
We thank Alan Weiner for providing CCA-adding enzyme expression plasmids, Alla Leshinsky and Dick Cook for performing the deep sequencing, Elena Conti for providing the Rrp44 protein, as well as Oliver Tam, Assaf Gordon, and Greg Hannon for help with the deep sequencing analysis. We thank Caroline Koehrer, Tom RajBhandary, Paul Schimmel, David Spector, Carol Wilusz, Jeff Wilusz, and members of the Sharp and Phizicky laboratories for discussions and advice. J.E.W. is a fellow of the Leukemia and Lymphoma Society. J.M.W. was supported by NIH Training Grant in Cellular, Biochemical and Molecular Sciences 5T32-GM068411. This work was supported by NIH grants R01-GM34277 (P.A.S.), R01-CA133404 (P.A.S.), and R01-GM52347 (E.M.P.) as well as partially by Cancer Center Support (core) Grant P30-CA14051 from NCI (P.A.S.).
Footnotes
Supporting Online Material: www.sciencemag.org, Materials and Methods, SOM Text, Figs. S1 to S20, Tables S1 to S5, References (30-34)
References and Notes
- 1.Sprinzl M, Cramer F. Prog Nucleic Acid Res Mol Biol. 1979;22:1. doi: 10.1016/s0079-6603(08)60798-9. [DOI] [PubMed] [Google Scholar]
- 2.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Cell. 2006;126:1065. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 3.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. Science. 2000;289 doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 4.Phizicky EM, Hopper AK. Genes Dev. 2010;24 doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deutscher MP. The Enzymes. 1982;15 [Google Scholar]
- 6.Betat H, Rammelt C, Morl M. Cell Mol Life Sci. 2010;67 doi: 10.1007/s00018-010-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong Y, Steitz TA. Curr Opin Struct Biol. 2006;16 doi: 10.1016/j.sbi.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Shi PY, Maizels N, Weiner AM. EMBO J. 1998;17:3197. doi: 10.1093/emboj/17.11.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilusz JE, Freier SM, Spector DL. Cell. 2008;135:919. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunwoo H, et al. Genome Res. 2009;19:347. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilusz JE, Spector DL. RNA. 2010;16:259. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Materials and methods are available as supporting material on Science Online
- 13.Nagaike T, et al. J Biol Chem. 2001;276:40041. doi: 10.1074/jbc.M106202200. [DOI] [PubMed] [Google Scholar]
- 14.Shi PY, Weiner AM, Maizels N. RNA. 1998;4:276. [PMC free article] [PubMed] [Google Scholar]
- 15.Yue D, Weiner AM, Maizels N. J Biol Chem. 1998;273:29693. doi: 10.1074/jbc.273.45.29693. [DOI] [PubMed] [Google Scholar]
- 16.Yue D, Maizels N, Weiner AM. RNA. 1996;2:895. [PMC free article] [PubMed] [Google Scholar]
- 17.Tomita K, et al. Nature. 2004;430:700. doi: 10.1038/nature02712. [DOI] [PubMed] [Google Scholar]
- 18.Xiong Y, Steitz TA. Nature. 2004;430:640. doi: 10.1038/nature02711. [DOI] [PubMed] [Google Scholar]
- 19.Pan B, Xiong Y, Steitz TA. Science. 2010;330:937. doi: 10.1126/science.1194985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Augustin MA, et al. J Mol Biol. 2003;328:985. doi: 10.1016/s0022-2836(03)00381-4. [DOI] [PubMed] [Google Scholar]
- 21.Thompson DM, Parker R. Cell. 2009;138:215. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Alexandrov A, et al. Mol Cell. 2006;21:87. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 23.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Genes Dev. 2008;22:1369. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotelawala L, Grayhack EJ, Phizicky EM. RNA. 2008;14:158. doi: 10.1261/rna.811008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whipple JM, Lane EA, Chernyakov I, D'Silva S, Phizicky EM. Genes Dev. 2011;25:1173. doi: 10.1101/gad.2050711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincent HA, Deutscher MP. J Biol Chem. 2006;281:29769. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 27.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Mol Cell. 2008;29:717. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 28.Leigh J, Lang BF. RNA. 2004;10:615. doi: 10.1261/rna.5195504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lonergan KM, Gray MW. Science. 1993;259:812. doi: 10.1126/science.8430334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.