Abstract
Objective
To investigate the relationship between hMLH1 promoter methylation and changes in chromatin composition. To study how the occupancy of methyl CpG binding domain proteins (MBDs) and histone acetylation/methylation in hMLH1 promoter may participate in hMLH1 silencing.
Methods
64 endometrial cancer samples were screened for hMLH1 mRNA expression. hMLH1 promoter methylation status was confirmed by methylation-specific PCR in cancers with high and low levels of hMLH1 expression. Chromatin immunoprecipitation was performed to compare the MBD occupancy and histone modifications between the methylated/silenced and unmethylated/active hMLH1 genes in multiple primary endometrial cancers.
Results
We demonstrated that MeCP2, MBD1 and MBD2, but not MBD3 and MBD4, specifically bind to methylated hMLH1 promoters. Hyperacetylated histones H3 and H4 were found to be associated with the unmethylated and transcriptionally active hMLH1 promoters. While H3 lysine-4 methylation was present in unmethylated hMLH1 promoters, H3 lysine-9 methylation was found exclusively in methylated promoters. Western blot analysis showed that similar global levels of MBDs and histones were present in the two cancer groups with high and low hMLH1 expression.
Conclusions
A distinct combination of MBDs and histone modification is associated with the silencing of the hMLH1 gene. The changes in hMLH1 chromatin composition are closely related to methylation status of hMLH1 promoters. These changes are not accounted by the global expression levels of MBDs and histones in endometrial cancers.
Keywords: Endometrial cancer, DNA methylation, hMLH1, MBD, Epigenetic
Introduction
Endometrial cancers occur with increased rates in women with hereditary nonpolyposis colorectal cancer (HNPCC), an autosomal dominant disorder that is linked to germ line mutations in one or more of the mismatch repair (MMR) genes including MLH1, MSH2 and MSH6 [1]. A direct consequence of DNA repair deficiency is the accumulation of DNA replication errors, or mutator phenotype, represented by high frequency of microsatellite instability (MSI-H). In one study, 61% of patients with MSI-H colorectal–endometrial double primary cancers were found carrying a mutation in at least one of the MMR genes [1]. In sporadic endometrial cancers, MSI-H was found in 20–30% of cases [2,3]. However, somatic mutation of MMR genes occurs in less than 10% of HNPCC-unrelated, MSI-H endometrial cancers [4,5]. An accumulating body of evidence supports epigenetic silencing, in particular hMLH1 silencing, as an alternative mechanism leading to the loss of the mismatch repair functions in sporadic endometrial cancers [6]. Human MLH1 (hMLH1) gene expression is controlled by a GC-rich promoter containing a classical CpG island [7,8]. DNA hypermethylation of this CpG island has been found to be closely associated with the loss of hMLH1 expression and the development of the MSI-H phenotype [3,9,10]. Methylation of hMLH1 was observed in 92% of endometrioid adenocarcinomas with MSI-H [2]. Consistent with this observation from clinical samples, in vitro studies have indicated that reintroduction of hMLH1 into hMLH1-negative cells was able to rescue the DNA repair function [11,12].
The hMLH1 hypermethylation appears to be an early event in the development of human malignancies. An age-related hypermethylation of the 5′ region of hMLH1 was detected in normal colonic mucosa of MSI-H colorectal cancers [13]. Esteller et al. reported an abnormal methylation of hMLH1 in some cases of atypical hyperplasia that coexist with endometrial carcinomas [14]. In an independent investigation, Horowitz et al. observed hMLH1 promoter methylation in areas of atypical endometrial hyperplasia lacking detectable MSI-H [15]. They concluded that hMLH1 hypermethylation represents an early event of endometrial cancer preceding the occurrence of the apparent MSI-H phenotype [15]. Recently, we demonstrated significant overexpression of DNMT3B and DNMT1, the enzymes catalyzing cytosine methylation, in primary Type I endometrial cancers as well as their corresponding cell lines [16]. Interestingly, the less well-differentiated grade III cancers express higher DNMT levels than the well-differentiated grade I cancers. These studies suggest that hMLH1 methylation/silencing may represent one of the critical events leading to malignant transformation. Consequently, knowledge of this epigenetic mechanism may hold the key for a better understanding of the pathogenesis of endometrial cancers.
DNA methylation-mediated gene silencing is a complicated process that relies on a coordinated action by multiple factors. Studies have shown that methyl CpG binding domain proteins (MBDs) are able to recruit histone deacetylase (HDAC) to local chromatin domains [17–19]. HDAC will convert the surrounding histones to their deacetylated form. Chromatins with deacetylated histones adopt a “closed” conformation that is associated with inactivated gene transcription [20,21]. Fahrner et al. investigated the dependency of hMLH1 expression on histone modification and DNA methylation in RKO colon cancer cells and found that deacetylation and methylation of lysine-9 in H3 were present in the methylated hMLH1 promoter [22]. Inhibition of DNA methyltransferase, but not histone deacetylase, led to an order of events that was initiated with DNA demethylation followed by gene re-expression and histone code reversal [22]. This temporal order of changes suggested a dominant role of DNA methylation in the control of hMLH1 transcription. Kondo et al. studied multiple methylated/silenced genes including hMLH1 in three colon cancer cell lines and found that reduced H3 lysine-4 methylation and increased lysine-9 acetylation and methylation are critical for the maintenance of methylation-associated gene silencing [23]. Since these studies concentrated on alterations in histone modification, the involvement of MBDs remains unclear. More importantly, the previous studies were performed on a limited number of cell lines, leaving uncertainties concerning the situation in primary cancers. Therefore, in this study, we investigated the relationship between DNA methylation and chromatin composition in primary endometrial cancers. Using a modified chromatin immunoprecipitation protocol, we performed a comprehensive analysis on MBDs occupancy as well as histone acetylation/methylation in two groups of endometrial cancers. One group represents cases with high hMLH1 expression from unmethylated promoters and the other with silenced hMLH1 expression from hypermethylated promoters. The comparative studies have identified a specific combination of MBDs binding and histone modification code associated with hMLH1 inactivation.
Materials and methods
Reagents
Antibodies against MeCP2, histone H3 and H4, acetyl-histone H3, acetyl-histone H4, dimethyl-histone H3 (Lys4) and dimethyl-histone H3 (Lys9) were obtained from Upstate Biotechnology Inc. (Lake Placid, NY). Rabbit antibody for β-actin and rabbit and goat antibodies for MBD1, MBD2, MBD3 and MBD4 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Tissue preparation
The use of human tissues in this study was approved by the Institutional Review Board of Mayo Foundation. In accordance with the Minnesota Statute for Use of Medical Information in Research, only those patients who consented to the use of their medical records were included in this analysis. Snap-frozen endometrial cancer specimens were obtained from 64 patients following hysterectomy and kept at −80°C. The histological grade of these cancers is shown in Fig. 1. All specimens were reviewed by a single pathologist and confirmed to be of endometrioid cancer histology. The endometrial cancer tissues were dissected to remove normal tissues and cut into 10 µm sections for RNA isolation, protein extraction and ChIP experiments.
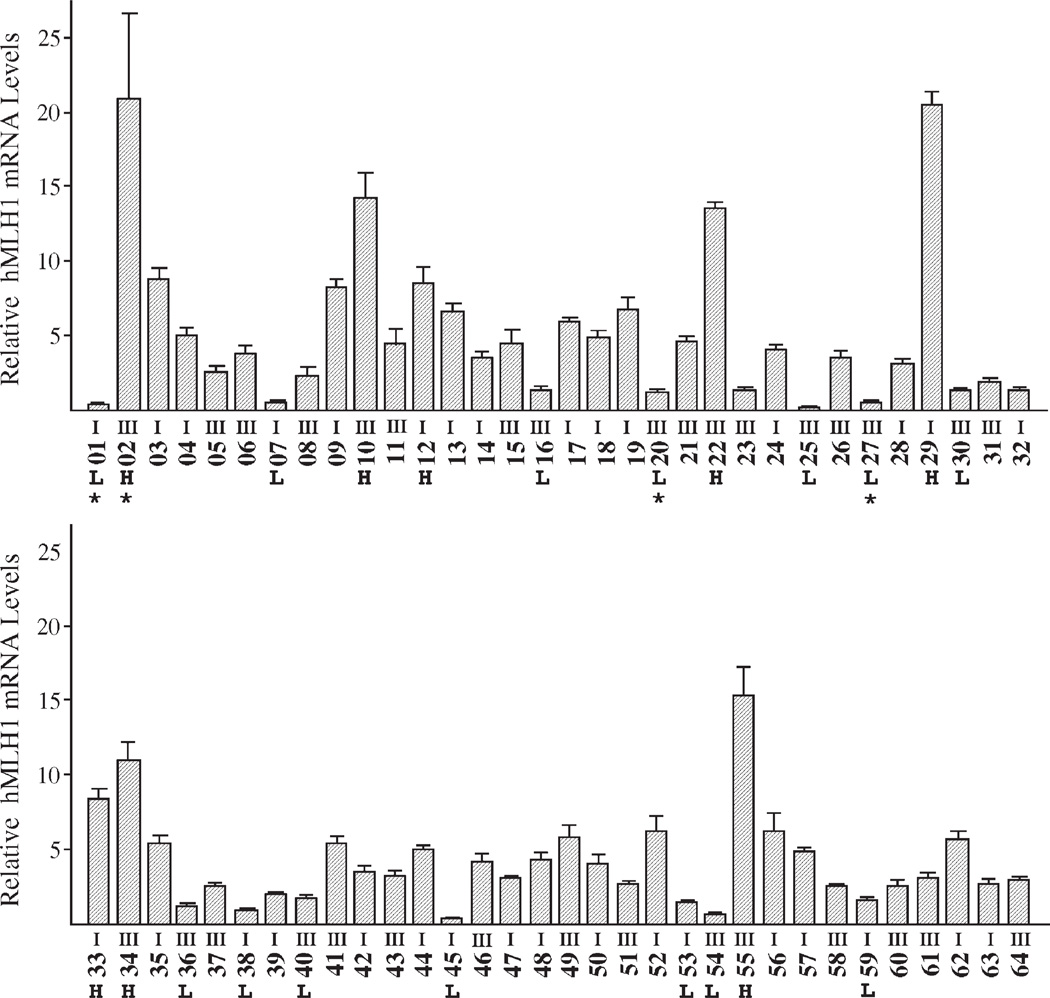
Fig. 1.
hMLH1 levels in endometrial cancers. Sixty four endometrial cancer samples were screened for hMLH1 mRNA expression. The histological grade of the cancers was indicated for each sample. Real-time PCR was performed on hMLH1 and GAPDH as described in Materials and methods. Cancer samples with the highest (H) and lowest (L) hMLH1 mRNA levels were selected for further analysis. Note that samples marked by “*” failed to provide sufficient tissues and were excluded for further analysis.
Real-time PCR
RNA isolation, quantification and cDNA synthesis were performed as previously described [16]. The hMLH1 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA levels were measured by real-time PCR using primers: hMLH1-forward, 5′-GAAAACTGAAAGCCCCTCCT; hMLH1-reverse, 5′-ACGGTTGAGGCATTGGGTAGT. GAPDH-forward, 5′-GAAGGTGAAGGTCGGAGTC; GAPDH-reverse, 5′-GAAGATGGTGATGGGATTTC. Real-time PCR was performed on the ABI Sequence Detector-770 (Applied Biosystems, Foster City, CA) using CYBR Green PCR Master Mix (Stratagene, Cedar Creek, TX) under the following conditions: initial denaturing: 95°C for 10 min followed by 40 cycles of denaturing at 95°C for 30 s, annealing at 56°C for 40 s and extension at 72°C for 30 s. The threshold cycle number (CT) for hMLH1 was normalized against GAPDH internal reference gene by the formula: ΔCT=CTMLH1−CTGAPDH. The difference between hMLH1 and GAPDH was further converted to relative fold (F=2ΔCT). Standardized hMLH1 mRNA levels were arbitrarily amplified by a factor of 10,000 for the convenience of data presentation. Average hMLH1 levels and standard errors were determined from three independent experiments.
Methylation-specific PCR
Genomic DNA was subjected to sodium bisulfite conversion using the EZ DNA methylation kit (Zymo Research, Orange, CA). The converted DNA was eluted with 10 µl of 1× TE from DNA affinity columns and 2 µl used for methylation-specific PCR using published primers [12]. The same PCR conditions as those for real-time PCR (see above) were applied. PCR products were documented by agarose gel electrophoresis and ethidium bromide staining.
Western blot analysis
Tissue extract preparations and SDS polyacrylamide gel electrophoresis were carried out as previously described [16]. MeCP2, MBD1, MBD3, MBD4, total H3 and H4 and acetylated H3 and H4 were detected using specific antibodies following the manufacturer’s instructions. Chemiluminescence detection was performed with the ECLplus™ Western Blotting Detection System (Amersham Corp, Arlington Heights, IL). The blots were re-probed with β-actin antibody and the results provided controls for protein loading.
Chromatin immunoprecipitation (ChIP) assays
Critical technical parameters of ChIP assay for endometrial tissues, including the amount of tissue, time of formaldehyde cross-linking and sonication conditions, were extensively optimized in pilot studies. Tissue samples (300 mg) were cross-linked by addition of 1% formaldehyde and incubated for 10 min at room temperature. After centrifugation (13,000 × g for 10 min), the supernatant was removed and pellets were washed twice with ice-cold PBS supplemented by 1× protease inhibitor cocktail (1 mM PMSF, 1 µg/ml aprotinin and 1 µg/ml pepstatin A). Pellets were resuspended in 300 µl of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1) and subjected to sonication. Ten-second pulses at 10-s intervals for four times (Sonic Dismembrator, Model 500, Fisher Scientific) were used to achieve chromatin fragmentation of 200 and 1000 bp.
Sonicated samples were centrifuged at 13,000 × g at 4°C for 10 min and supernatants transferred to 15 ml tubes. The samples are diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1% Triton X-10, 2 mM EDTA, 16.7 mM Tris–Cl, pH 8.1, 150 mM NaCl), and 20 µl of aliquots of each sample was removed to serve as the input controls. To reduce nonspecific background, the DNA–protein complexes were pre-cleared by incubation with 75 µl of Protein A agarose beads (50% slurry containing salmon sperm DNA). The pre-absorption was carried out at 4°C with constant rotation for 2 h. Anti-MBDs or anti-histone antibodies (20 µl) were added to the samples, and primary antibody binding performed at 4°C for overnight with constant mixing. In negative controls, antiserum from non-immunized mouse was used instead of specific antibodies. To collect immune complexes, 60 µl of Protein A agarose-salmon sperm DNA (50% slurry) is added to each tube, and incubation continued for 2 h at 4°C. Agarose beads were recovered by gentle centrifugation at 2000 rpm for 2 min. The beads are washed sequentially with 1 ml buffer for 5 min in the following order: two times with low salt buffer (0.1% SDS, 1% Triton X-10, 2 mM EDTA, 20 mM Tris–Cl, pH 8.1, 150 mM NaCl), two times with high salt buffer (0.1% SDS, 1% Triton X-10, 2 mM EDTA, 20 mM Tris–Cl, pH 8.1, 500 mM NaCl), once with LiCl buffer (0.25 mM LiCl, 1% deoxycholic acid, 1 mM EDTA, 10 mM Tris, pH 8.10) and once with 1× TE buffer. After washing, 500 µl fresh 1% SDS and 0.1 M NaHCO3 were used to elute immune complexes. Formaldehyde cross-links were reversed by adding 20 µl 5 M NaCl to 500 µl eluates and heating at 65°C for 4 h. DNA fragments were recovered by ethanol precipitation following proteinase K digestion and phenol/chloroform extraction.
PCR was performed with hMLH1 promoter-specific primers: MLH1-forward, 5′-AACGCCTTGCAGGACGCTTA, and MLH1-reverse, 5′-TGAAGAGAGAGCTGCTGCTCG. PCR conditions were: 94°C for 5 min for initial denature followed by 35 cycles of denature at 94°C for 45 s, annealing at 56°C for 45 s and extension at 72°C for 1 min. PCR products were visualized by 2% agarose gels electrophoresis and ethidium bromide staining. ChIP experiments were repeated three times with positive and negative controls.
Data analysis
The results of Western blot and ChIP experiments were documented with an HP Q3190A scanner and analyzed by densitometry measurement using the NIH Image program. The signals were standardized against the input controls, and their relative levels were compared using Student's t tests with the assumption of two-tail distribution and two samples with equal variance. The statistical significance (P<0.05) is marked by asterisk in the figures. Average values and standard errors were calculated from at least three repeated ChIP experiments.
Results
Measurement of hMLH1 expression in endometrial cancers
In order to investigate the relationship between hMLH1 hypermethylation and chromatin composition, we needed to know the hMLH1 expression levels in individual cancers. Using the quantitative real-time PCR technique, we examined 64 endometrial cancer tissues. As shown in Fig. 1, endometrial cancers contain varied hMLH1 mRNA levels. No statistically significant difference of hMLH1 levels was found between grade I and grade III cancers. Based on the hMLH1 expression pattern, these primary cancers were considered to be associated with relatively high, low, and intermediate levels of hMLH1 mRNA. The samples with high hMLH1 expression were considered to likely contain unmethylated hMLH1 promoters, whereas those with low level expression may possess methylated promoters. The samples with intermediate hMLH1 levels were not subsequently examined as they may indicate heterogeneity of the tissue containing significant amounts of normal endometrial or stromal cells. Potential contamination by normal cells in the “intermediate” group may complicate downstream analysis. Therefore, we elected to focus on comparison of the two extreme groups with the highest and lowest hMLH1 levels, and the tissues with intermediate hMLH1 levels were excluded for further analysis.
Confirmation of hMLH1 DNA methylation
We examined the hMLH1 promoter methylation status by methylation-specific PCR (MSP). Of the 14 cancers with relatively low hMLH1 expression, 11 samples provided sufficient tissue for DNA isolation, chemical conversion, and methylation-specific PCR analysis. Among these samples, 7 were found to contain completely methylated hMLH1 promoter by MSP criteria (Fig. 2). The rest of the samples contained partially methylated or unmethylated DNA. Of the 8 samples containing relatively high hMLH1 mRNA levels, 7 samples provided sufficient DNA for methylation studies. We found that all of these samples contain unmethylated hMLH1 promoters. These results were consistent with previous observations [2,3,9,10] and provided further support for a close association between hMLH1 silencing and promoter hypermethylation.
Fig. 2.
Confirmation of hMLH1 methylation. DNA methylation in the selected samples was examined by methylation-specific PCR. All the 7 cancers (H1–H7) expressing the highest levels of hMLH1 contain an unmethylated hMLH1 gene. Cancers with the lowest hMLH1 mRNA were found to contain completely methylated (L1–L7), partially methylated (marked by “*”) or unmethylated (marked by “**”) promoters.
ChIP analysis of chromatin composition
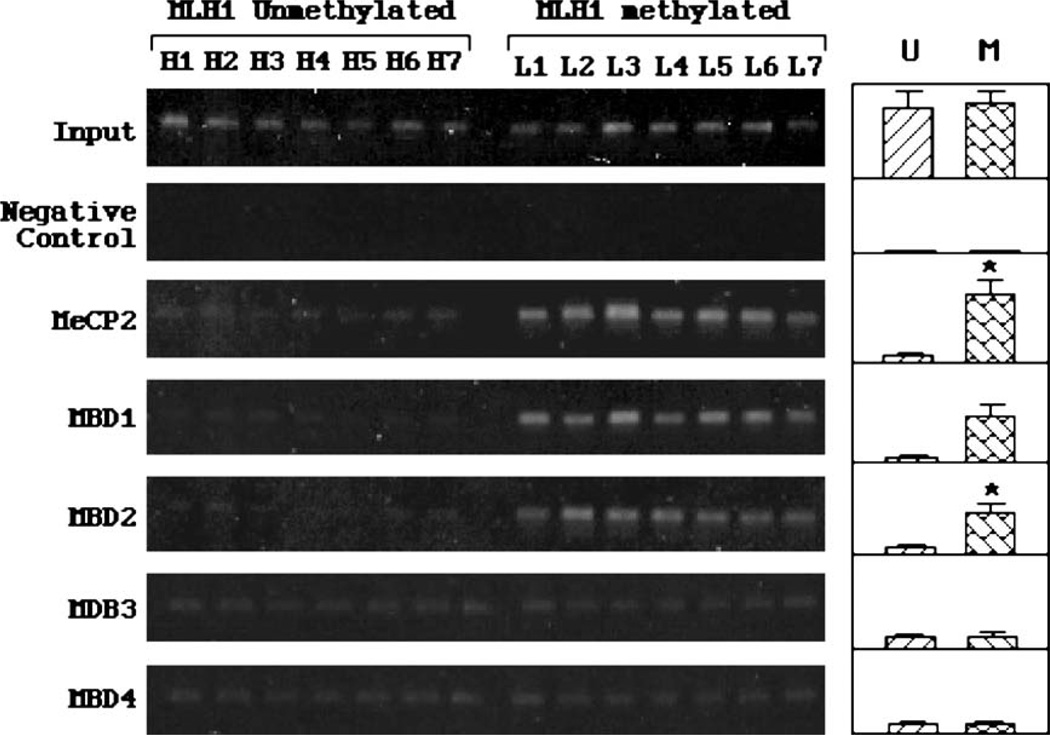
The MBDs occupancy and histone acetylation/methylation at hMLH1 promoter region were compared between the seven hMLH1-unmethylated and seven hMLH1-methylated samples. Fig. 3 shows the results of ChIP experiments using MBDs isoform-specific antibodies. Results from input positive control and non-antibody negative controls indicated the specificity of the ChIP experiments. High levels of promoter occupancy by MECP2, MBD1 and MBD2 were present in cancer samples containing methylated hMLH1 promoters, but absent in the unmethylated promoters. A diminished binding by MBD3 and MBD4 in both methylated and unmethylated promoters was observed. These results indicate that MECP2, MBD1 and MBD2, but not MBD3 and MBD4, are directly involved in the inactivation of methylated hMLH1 promoters in primary endometrial cancers. Potential implications of the differential MBD occupancy observed in the two groups of cancers were described in Discussion.
Fig. 3.
MBDs occupancy of the hMLH1 promoters. Chromatin immunoprecipitation experiments were performed on the cancer samples with unmethylated (H1–H7) or methylated (L1–L7) hMLH1 promoters. The left panels present typical ChIP results of three or more repeated experiments. The right panels show the results from densitometry analysis on average levels of MDB binding signals from multiple cancer samples. Significantly higher (P ≤ 0.05 as indicated by asterisk) average levels of occupancy by MeCP2, MBD1 and MBD2 were found in hMLH1-methylated cancers. Minimal binding of MDB3 and MBD4 was observed in both groups of cancer samples.
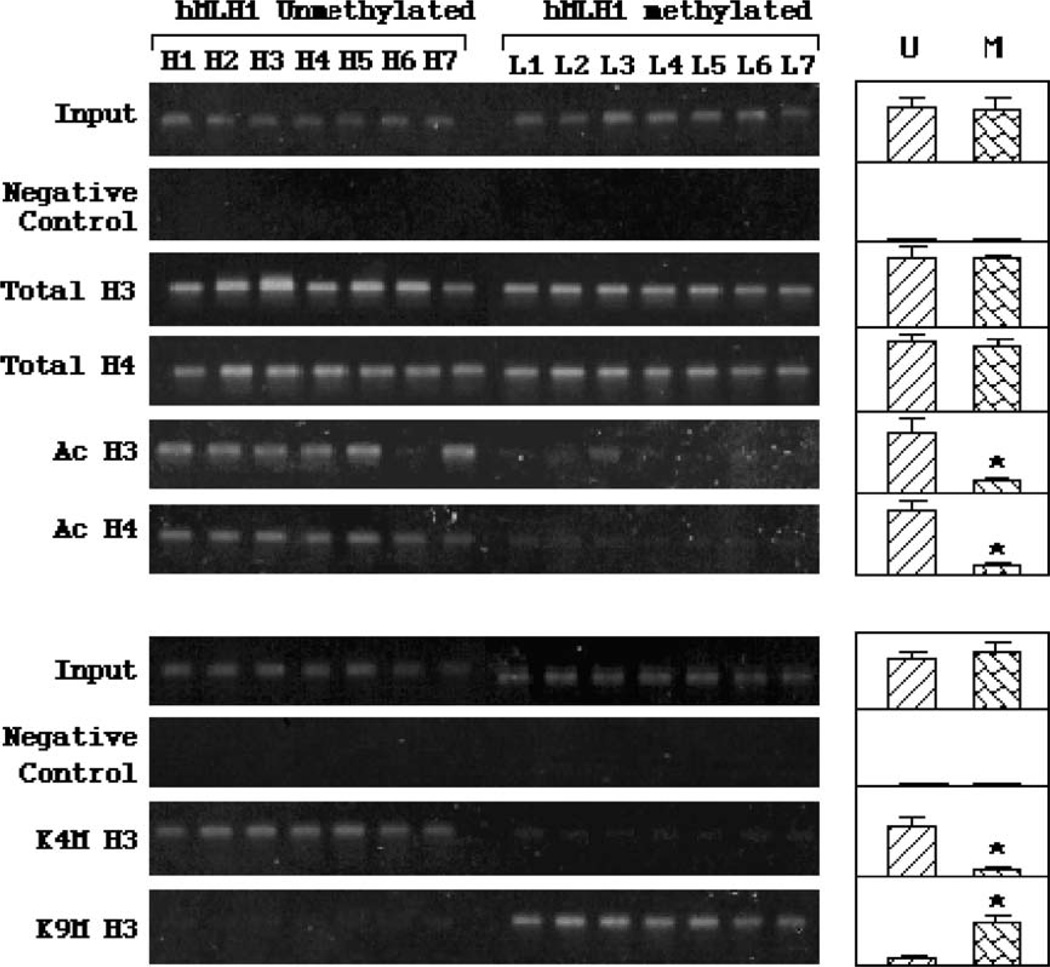
In addition to MBDs occupancy, we examined the histone H3 and H4 acetylation and methylation code of hMLH1 promoters in the two groups (Fig. 4). While immunoprecipitation with total H3 and H4 antibodies produced equally strong PCR signals in the two groups, antibodies against acetylated H3 and H4 detected much stronger signals in the hMLH1-unmethylated than -methylated cancers. Densitometry analysis indicated a highly significant difference in H3 and H4 acetylation levels between the two groups. H3 methylation at lysine-4 (K4M H3) was mostly observed in the unmethylated group. Very low levels of H3 K4M were detected in the hMLH1-methylated group. In a sharp contrast, the lysine-9 methylation (K9M H3) was only found in hMLH1-methylated, but not unmethylated, cancer samples. These results demonstrated an intimate correlation between DNA methylation and histone covalent modification in endometrial cancers.
Fig. 4.
ChIP studies on covalent histone modification in hMLH1 promoters. The left images are example results of three or more repeated experiments. The results of densitometry analysis on multiple patient samples are shown in the right panels. Statistical analysis indicates that similar levels of hMLH1 promoter DNA were recovered by antibodies against total histone H3 or H4 from the two cancer groups containing either unmethylated or methylated hMLH1 promoters. However, significantly higher (P ≤ 0.05 as indicted by asterisks) levels of acetylated histone H3 (Ac H3) and H4 (Ac H4), and K4 methylation in H3 (K4M H3), were found in unmethylated hMLH1 promoters. In contrast, decreased levels of K9 methylation (K9M H3) were found to be associated with the methylated hMLH1 promoters.
Cellular levels of MBDs and histones
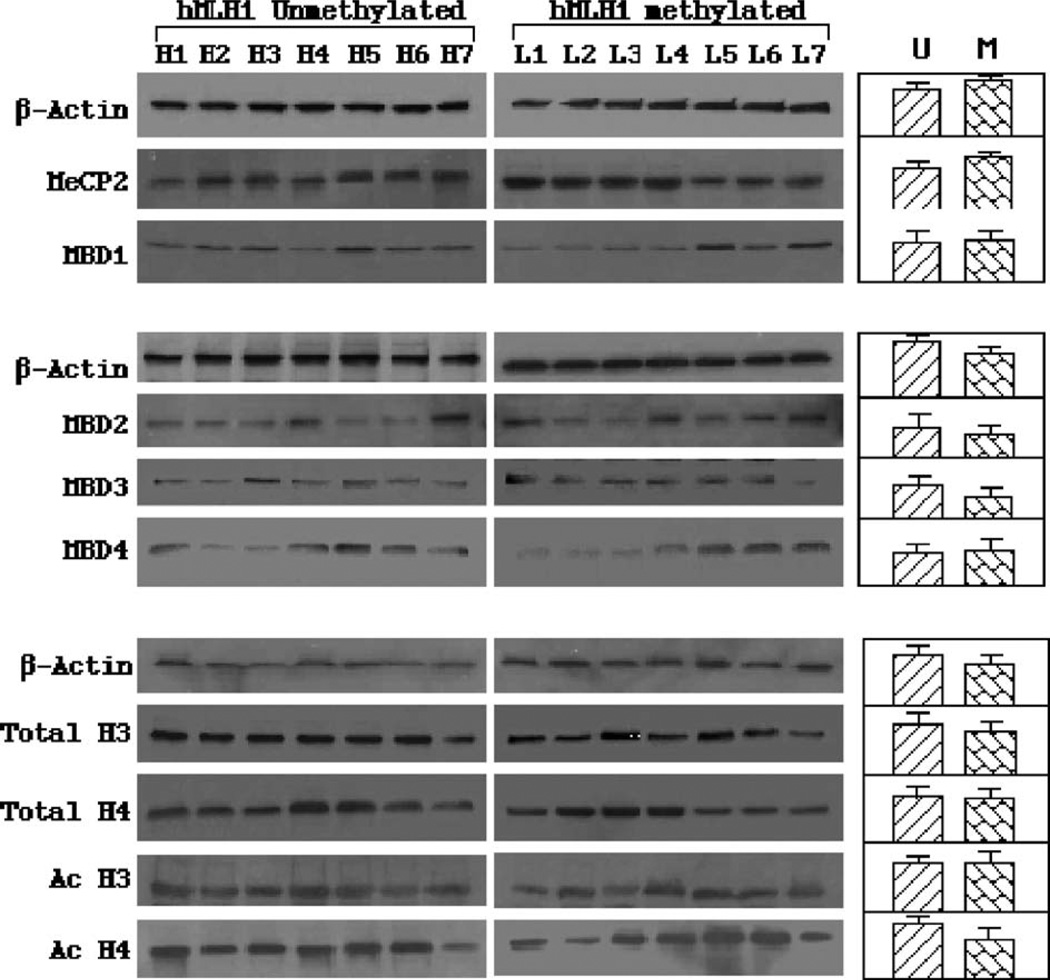
We wanted to know if the differential MBD binding and histone modification of the two groups were caused by alterations on the cellular levels of the corresponding proteins. Western blot analyses were performed on cancer samples used for ChIP studies (Fig. 5). The hMLH1-methylated and -unmethylated groups were found to contain similar concentrations of MBDs and histones. Thus, the changes in hMLH1 chromatin composition are not likely caused by global alterations in the protein expression. These results suggested that hMLH1 silencing is mostly controlled at the local chromatin level by gene-specific mechanisms.
Fig. 5.
Total cellular levels of MBD and histone proteins. Western blot analysis was performed on MBDs, total histone H3 and H4 and acetylated H3 and H4. The left panels are some example results representing at least three repeated Western blot experiments. The right panels show the result of densitometry analysis on multiple cancer samples. No statistically significant difference in average concentrations of MBDs and histones was found between cancer groups containing unmethylated and methylated MLH1 genes. For each blot, the β-actin protein levels were also measured for loading controls.
Discussion
The most important finding of this study is a clear correlation between MBD binding and hMLH1 expression. Previous studies have demonstrated both direct and indirect inhibitory effects of DNA methylation on gene transcription. Methyl groups can directly affect DNA structure and interfere with DNA-binding activities of transcription factors [24,25]. Using gel shift and ChIP assays, Chang et al. have shown that methylation modification of the IGFBP promoter resulted in reduced Sp-1/Sp-3 binding to their cognate sites and decreased IGFBP transcription [26]. Overexpression of MeCP2 further suppressed the IGFBP promoter activity by competition between MeCP2 and Sp-1/Sp-3 [26]. It has been shown that unmethylated hMLH1 promoter is controlled by coordinated actions of an enhancer and consensus CCAAT sites located at nucleotides −282 to −151 relative to the transcription start site [8]. The CCAAT box is a well-characterized positive cis element recognized by a ubiquitously expressed transcription factor NF-Y (CBF) [27,28]. Interestingly, methylation of CpG in this region was found to be invariably correlated with the absence of hMLH1 expression in colon cancer cells [29]. Although the core sequences of the CCAAT sites do not contain CpG dinucleotides, interference of NF-Y with CCAAT element by MBD-binding could not be excluded. MeCP2, MBD1 and MBD2 may also induce hMLH1 silencing by actively recruiting HDAC to the promoter region [17–19]. This mechanism is supported by the fact that all the three MBDs that we identified on methylated hMLH1 promoters are capable of forming complexes with HDAC [17,29]. Furthermore, a simultaneous increase in MBDs occupancy (Fig. 3) and histone deacetylation (Fig. 4) were observed in methylated hMLH1 promoters. It is important to point out that mechanisms unrelated to DNA methylation may participate in hMLH1 transcription regulation. Indeed, in this study, we found that one cancer (sample 59, Figs. 1 and 2) expresses relatively low level of hMLH1 but contains completely unmethylated promoter. In addition, monoallelic hMLH1 DNA methylation has been reported in some cases of colon cancers [30]. These situations, however, are not covered by the current study that focused on cancers with biallelically methylated hMLH1 genes.
MeCP2, MBD1, MBD2, MBD3 and MBD4 all contain the 70 amino acid methyl CpG-binding domain capable of interacting with methylated CpG sites. Limited sequence divergences in the methyl CpG-binding domain, however, result in rather striking changes in DNA-binding functions [31]. Variations in the DNA ligand such as spacing between methyl CpG as well as flanking sequences may also influence MBD–DNA interactions [31]. Indeed, previous studies have suggested gene-specific actions of MBDs. For example, MBD2, but not MBD1 or MeCP2, was found to interact with methylated P16/Ink4A, P14/ARF [32] and GSTP1 [33]. MBD3 is part of the NuRD complex involved in control of gene expression in early development [34]. The current study demonstrated that MeCP2, MBD1 and MBD2, but not MBD3 and MBD4, bind to methylated hMLH1 promoters. Therefore, we provided additional support for different function of MBDs in gene-specific silencing. At this time, it is not clear if MeCP2, MBD1 and MBD2 coexist in a single hMLH1 promoter or whether these MBDs bind to the hMLH1 promoter in a cell-cycle-specific manner.
A strong association of methyl-H3 K9 with methylated hMLH1 has been observed in colon cancer cell lines [21,22]. Interestingly, this change was accompanied by decreased H3 K4 methylation in methylated hMLH1 promoters. But unlike histone acetylation, the relationship between histone methylation alterations and MBD binding to methylated DNA is less characterized, and many aspects of the mechanism remain unclear. Different models have been proposed for the cross-talk between histone methylation and DNA methylation. Tamara et al. demonstrated that replacement of H3 lysine-9 with leucine or arginine led to marked reduction in DNA methylation levels in Neurospora crassa, suggesting that histone methylation may affect DNA methylation [35]. Recent studies indicating recruitment of DNMT by HP1 (heterochromatin protein) and SUV39h (histone methyl transferase) in mouse embryonic cells provided a mechanism for histone methylation-mediated DNA methylation in mammalian cells [36]. In contrast, studies in human colon cancer cell lines showed that treatment by DNMT inhibitors rapidly reduced the H3 K9 methylation at multiple methylated loci including that of hMLH1, suggesting a dominant role of DNA methylation [21]. Further studies will be required to determine the primary epigenetic events leading to the silencing of hMLH1 in endometrial cancers.
Altered MBDs expression has been observed in non-small-cell lung cancers [37]. We tested the possibility of whether global alterations in MBD expression and/or histone modification in cancer cells may contribute to the different chromatin composition observed in the two groups. Their similar levels of MBDs and histone modifications suggested that hMLH1 chromatin composition is not caused by differences in the expression or availability of the various proteins. It is interesting to observe that, while MeCP2 and histones showed relatively small inter-group differences, MBD1, MBD2, MBD3 and MBD4 exhibit divergent protein levels among individual cancers. Nevertheless, no direct connection between the cellular concentration and hMLH1 promoter binding could be found in either group. For example, L4 and L7 showed similarly low levels of MBD1 occupancy, but the two cancers contained quite different levels of MBD1 proteins. In fact, the MBD1 level in L7 is among the highest. Overall, our data showed no evidence that global levels of MBDs and histones were limiting parameters that could decide the chromatin composition of the hMLH1 promoter.
In summary, we have characterized alterations of chromatin composition in multiple primary endometrial cancers and identified characteristic changes of MBDs occupancy and histone modifications in methylated hMLH1 genes. These data provided important information on epigenetic mechanisms leading to the MSI-H phenotype in endometrial cancers. Given the potentially reversible nature of epigenetic changes [38–40], these findings may be useful for designing novel therapeutic strategies targeting the restoration of hMLH1 expression and associated DNA repair function in endometrial cancers.
Acknowledgments
The authors thank Ying Zhao for her technical support. This work was supported by NIH grant R01 HD41577 (S.-W. Jiang), University of Texas M.D. Anderson Cancer Center Uterine Cancer SPORE Development Award (S.-W. Jiang) and Mayo Clinic and Foundation Eagles Funds for Cancer Research (S.-W. Jiang and K.C. Podratz).
References
- 1.Cederquist K, Emanuelsson M, Goransson I, Holinski-Feder E, Muller-Koch Y, Golovleva I, et al. Mutation analysis of the MLH1, MSH2 and MSH6 genes in patients with double primary cancers of the colorectum and the endometrium: a population-based study in northern Sweden. Int J Cancer. 2004;109:370–376. doi: 10.1002/ijc.11718. [DOI] [PubMed] [Google Scholar]
- 2.Hirasawa A, Aoki D, Inoue J, Imoto I, Susumu N, Sugano K, et al. Unfavorable prognostic factors associated with high frequency of microsatellite instability and comparative genomic hybridization analysis in endometrial cancer. Clin Cancer Res. 2003;9:5675–5682. [PubMed] [Google Scholar]
- 3.Esteller M, Levine R, Baylin SB, Ellenson LH, Herman JG. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–2417. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]
- 4.Gurin CC, Federici MG, Kang L, Boyd J. Causes and consequences of microsatellite instability in endometrial carcinoma. Cancer Res. 1999;59:462–466. [PubMed] [Google Scholar]
- 5.Banno K, Susumu N, Hirao T, Yanokura M, Hirasawa A, Aoki D, et al. Identification of germline MSH2 gene mutations in endometrial cancer not fulfilling the new clinical criteria for hereditary nonpolyposis colorectal cancer. Cancer Genet Cytogenet. 2003;146:58–65. doi: 10.1016/s0165-4608(03)00157-2. [DOI] [PubMed] [Google Scholar]
- 6.Atkin NB. Microsatellite instability. Cytogenet Cell Genet. 2001;92:177–181. doi: 10.1159/000056898. [DOI] [PubMed] [Google Scholar]
- 7.Quaresima B, Faniello MC, Baudi F, Cuda G, Grandinetti C, Tassone P, et al. Transcriptional regulation of the mismatch repair gene hMLH1. Gene. 2001;275:261–265. doi: 10.1016/s0378-1119(01)00656-4. [DOI] [PubMed] [Google Scholar]
- 8.Warnick CT, Dabbas B, Ilstrup SJ, Ford CD, Strait KA. Cell type-dependent regulation of hMLH1 promoter activity is influenced by the presence of multiple redundant elements. Mol Cancer Res. 2003;1:610–618. [PubMed] [Google Scholar]
- 9.Strazzullo M, Cossu A, Baldinu P, Colombino M, Satta MP, Tanda F, et al. High-resolution methylation analysis of the hMLH1 promoter in sporadic endometrial and colorectal carcinomas. Cancer. 2003;98:1540–1546. doi: 10.1002/cncr.11651. [DOI] [PubMed] [Google Scholar]
- 10.Simpkins SB, Bocker T, Swisher EM, Mutch DG, Gersell DJ, Kovatich AJ, et al. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum Mol Genet. 1999;8:661–666. doi: 10.1093/hmg/8.4.661. [DOI] [PubMed] [Google Scholar]
- 11.Russo MT, Blasi MF, Chiera F, Fortini P, Degan P, Macpherson P, et al. The oxidized deoxynucleoside triphosphate pool is a significant contributor to genetic instability in mismatch repair-deficient cells. Mol Cell Biol. 2004;24:465–474. doi: 10.1128/MCB.24.1.465-474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakagawa H, Nuovo GJ, Zervos EE, Martin EW, Jr, Salovaara R, Aaltonen LA, et al. Age-related hypermethylation of the 5′ region of MLH1 in normal colonic mucosa is associated with microsatellite-unstable colorectal cancer development. Cancer Res. 2001;61:6991–6995. [PubMed] [Google Scholar]
- 14.Esteller M, Catasus L, Matias-Guiu X, Mutter GL, Prat J, Baylin SB, et al. hMLH1 promoter hypermethylation is an early event in human endometrial tumorigenesis. Am J Pathol. 1999;155:1767–1772. doi: 10.1016/S0002-9440(10)65492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horowitz N, Pinto K, Mutch DG, Herzog TJ, Rader JS, Gibb R, et al. Microsatellite instability, MLH1 promoter methylation, and loss of mismatch repair in endometrial cancer and concomitant atypical hyperplasia. Gynecol Oncol. 2002;86:62–68. doi: 10.1006/gyno.2002.6724. [DOI] [PubMed] [Google Scholar]
- 16.Jin F, Dowdy SC, Xiong Y, Eberhardt NL, Podratz KC, Jiang SW. Upregulation of DNA methyltransferase 3B expression in endometrial cancers. Gynecol Oncol. 2005;96:531–538. doi: 10.1016/j.ygyno.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 17.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 18.Sekimata M, Takahashi A, Murakami-Sekimata A, Homma Y. Involvement of a novel zinc finger protein, MIZF, in transcriptional repression by interacting with a methyl-CpG-binding protein, MBD2. J Biol Chem. 2001;276:42632–42638. doi: 10.1074/jbc.M107048200. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki M, Yamada T, Kihara-Negishi F, Sakurai T, Oikawa T. Direct association between PU.1 and MeCP2 that recruits mSin3A-HDAC complex for PU.1-mediated transcriptional repression. Oncogene. 2003;22:8688–8698. doi: 10.1038/sj.onc.1207182. [DOI] [PubMed] [Google Scholar]
- 20.Somech R, Izraeli S, A JS. Histone deacetylase inhibitors—A new tool to treat cancer. Cancer Treat Rev. 2004;30:461–472. doi: 10.1016/j.ctrv.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–7218. [PubMed] [Google Scholar]
- 23.Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003;23:206–215. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka Y, Kurumizaka H, Yokoyama S. CpG methylation of the CENP-B box reduces human CENP-B binding. FEBS J. 2005;272:282–289. doi: 10.1111/j.1432-1033.2004.04406.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhu WG, Srinivasan K, Dai Z, Duan W, Druhan LJ, Ding H, et al. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Mol Cell Biol. 2003;23:4056–4065. doi: 10.1128/MCB.23.12.4056-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang YS, Wang L, Suh YA, Mao L, Karpen SJ, Khuri FR, et al. Mechanisms underlying lack of insulin-like growth factor-binding protein-3 expression in non-small-cell lung cancer. Oncogene. 2004;23:6569–6580. doi: 10.1038/sj.onc.1207882. [DOI] [PubMed] [Google Scholar]
- 27.Hung HL, High KA. Liver-enriched transcription factor HNF-4 and ubiquitous factor NF-Y are critical for expression of blood coagulation factor X. J Biol Chem. 1996;271:2323–2331. doi: 10.1074/jbc.271.4.2323. [DOI] [PubMed] [Google Scholar]
- 28.Dugast C, Weber MJ. NF-Y binding is required for transactivation of neuronal aromatic l-amino acid decarboxylase gene promoter by the POU-domain protein Brn-2. Brain Res Mol Brain Res. 2001;89:58–70. doi: 10.1016/s0169-328x(01)00063-8. [DOI] [PubMed] [Google Scholar]
- 29.Deng G, Chen A, Hong J, Chae HS, Kim YS. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999;59:2029–2033. [PubMed] [Google Scholar]
- 30.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 31.Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magdinier F, Wolffe AP. Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc Natl Acad Sci U S A. 2001;98:4990–4995. doi: 10.1073/pnas.101617298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin X, Nelson WG. Methyl-CpG-binding domain protein-2 mediates transcriptional repression associated with hypermethylated GSTP1 CpG islands in MCF-7 breast cancer cells. Cancer Res. 2003;63:498–504. [PubMed] [Google Scholar]
- 34.Sakai H, Urano T, Ookata K, Kim MH, Hirai Y, Saito M, et al. MBD3 and HDAC1, two components of the NuRD complex, are localized at Aurora-A-positive centrosomes in M phase. J Biol Chem. 2002;277:48714–48723. doi: 10.1074/jbc.M208461200. [DOI] [PubMed] [Google Scholar]
- 35.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 36.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 37.Muller-Tidow C, Kugler K, Diederichs S, Klumpen S, Moller M, Vogt U, et al. Loss of expression of HDAC-recruiting methyl-CpG-binding domain proteins in human cancer. Br J Cancer. 2001;85:1168–1174. doi: 10.1054/bjoc.2001.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalgo ML, Hayashida T, Bender CM, Pao MM, Tsai YC, Gonzales FA, et al. The role of DNA methylation in expression of the p19/p16 locus in human bladder cancer cell lines. Cancer Res. 1998;58:1245–1252. [PubMed] [Google Scholar]
- 39.Graziano F, Arduini F, Ruzzo A, Mandolesi A, Bearzi I, Silva R, et al. Combined analysis of E-cadherin gene (CDH1) promoter hypermethylation and E-cadherin protein expression in patients with gastric cancer: implications for treatment with demethylating drugs. Ann Oncol. 2004;15:489–492. doi: 10.1093/annonc/mdh108. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert J, Gore SD, Herman JG, Carducci MA. The clinical application of targeting cancer through histone acetylation and hypomethylation. Clin Cancer Res. 2004;10:4589–4596. doi: 10.1158/1078-0432.CCR-03-0297. [DOI] [PubMed] [Google Scholar]