Abstract
The organization and dynamics of receptors and other molecules in the plasma membrane are not well understood. Here we analyzed the spatio-temporal dynamics of T cell antigen receptor (TCR) complexes and linker for activation of T cells (Lat), a key adaptor molecule in the TCR signaling pathway, in T cell membranes using high-speed photoactivated localization microscopy, dual-color fluorescence cross-correlation spectroscopy and transmission electron microscopy. In quiescent T cells, both molecules existed in separate membrane domains (protein islands), and these domains concatenated after T cell activation. These concatemers were identical to signaling microclusters, a prominent hallmark of T cell activation. This separation versus physical juxtapositioning of receptor domains and domains containing downstream signaling molecules in quiescent versus activated T cells may be a general feature of plasma membrane–associated signal transduction.
A principal function of most T cells is to recognize foreign antigens on other cell surfaces, with the specificity determined by the T cell antigen receptor (TCR). Once the TCR has engaged its ligand and the signaling cascade has been initiated through the associated cluster of differentiation 3 (CD3) subunits, the kinase Zap70 transmits the signal to the adaptor Lat (linker for activation of T cells; A001392). Within seconds, actin-dependent signaling microclusters are formed in the periphery of the contact site between the T cell and antigen-presenting cell, and these microclusters then migrate to the center of the interface and form the immunological synapse1–6. Many signaling molecules, including the TCR and Lat, colocalize in the microclusters during early T cell activation3–6.
On the basis of electron-microscopy studies, varying degrees of preclustering of plasma membrane molecules, including the TCR and Lat, have been proposed7–9. However, there are concerns about potential artifacts with this approach (for example, from fixation, gold particle staining and/or drying). Additionally, fluorescence microscopy and biochemical technologies suggest the coexistence of monomeric and multivalent TCR-CD3 complexes9,10. It is not clear to what extent these multivalent complexes exist or if they form through protein-protein interactions or localization in membrane domains. In the case of biochemical approaches, cell lysis itself could introduce additional artifacts. To overcome such limitations and to gain insight into the organization of plasma membrane proteins in situ, we analyzed the organization of the TCR and Lat in quiescent and activated native T cell plasma membrane sheets and live T cells. We achieved this with a combination of three techniques: a modified high-speed version of photoactivated localization microscopy (hsPALM)11–13, dual-color fluorescence cross-correlation spectroscopy (dcFCCS)14–16 and transmission electron microscopy (TEM)7,8,17,18.
We show that both the TCR and Lat were preclustered into separate and spatially stable membrane domains (protein islands) on quiescent T cells. After antigen recognition, these domains transiently concatenated into microclusters without any substantial change in the size and number of the component domains. These data provide important information about the spatial organization and reorganization of the TCR and Lat in the plasma membrane. Together with published work8, our findings suggest that partitioning receptors and their downstream signaling components into separate membrane domains, and then bringing these domains together, may be an important and general mechanism in the control of cell activation.
RESULTS
TCR and Lat islands concatenate after activation
To obtain high-resolution fluorescence data on TCR and Lat in T cell membranes, we developed a high-speed version of PALM (hsPALM)11. In PALM, the molecules of interest are labeled with photoswitchable fluorophores; here we used the cyan fluorescent protein PSCFP2, whose cyan fluorescence is diminished to approximately one fifth and green fluorescence is increased 400-fold after irradiation with 405-nm light19. In each time interval or frame, a limited subset of fluorophores is photoactivated, excited and photobleached, which allows compilation of a succession of single-molecule images into an image with a resolution about tenfold higher than is possible with diffraction-limited fluorescent imaging (~250 nm). At a rate of 100–250 frames per second in stream-acquisition mode, the photon counts of single fluorophores were sufficient to localize them with an average estimated accuracy of ~25 nm (Supplementary Fig. 1a). PSCFP2 photobleaches irreversibly, so each tagged molecule is observed only once19. We combined up to 1,000 frames to obtain probability density plots for the tagged molecules. Similar techniques have been used to study hemagglutinin and paxillin in live cells12,13.
Our hsPALM produced statistically evaluable results for the TCR (specifically CD3ζ (A000553)) and Lat fused to PSCFP2, with acquisition times of 4–10 s. We chose to characterize the TCR and Lat, as they are not known to physically interact with each other and do not associate with immobile structures, yet are both essential components of the TCR signaling pathway. We derived T cells from short-term cultures of lymph node cells from 5C.C7 TCR–transgenic mice, which yield physiologically normal T cells specific for the major histocompatibility complex class II molecule I-Ek presenting the moth cytochrome c peptide (I-Ek–MCC)20. We analyzed plasma membrane sheets derived from T cells expressing CD3ζ-PSCFP2 or Lat-PSCFP2 generated on a nonactivating surface (poly-L-lysine) or an activating surface (I-Ek–MCC plus the CD28 ligand B7-1 (CD80))18. The removal of the cell bodies eliminates intracellular background fluorescence and ensures that the fluorophores acquired are localized to the plasma membrane. The surfaces contained 18,500–21,500 ligands per μm2, and we found their heterogeneity to be negligible (Supplementary Fig. 1b–d). The activating surfaces initiated Ca2+ flux and tyrosine phosphorylation similar to T cells activated by antigen-presenting cells (data not shown). T cells formed contacts 6–12 μm in width on nonactivating surfaces and 10–20 μm in width on activating surfaces (Supplementary Fig. 2a). The TCR surface concentrations in membrane sheets were ~140 per μm2 on nonactivating surfaces and ~190 per μm2 on activating surfaces after extended contact (15–20 min) and were the same as for live cells on these surfaces.
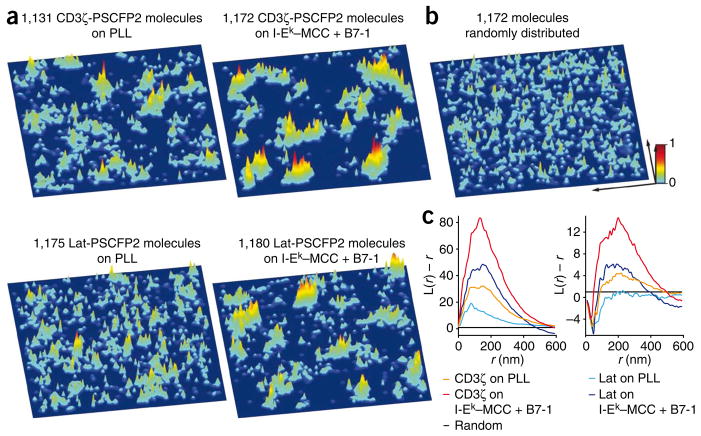
In the hsPALM studies, we visualized the distributions of CD3ζ-PSCFP2 and Lat-PSCFP2 in three-dimensional probability density plots, in which every fluorophore is represented by a normalized Gaussian probability density distribution with a width equal to its positional accuracy of localization. In membrane sheets from quiescent T cells, CD3ζ-PSCFP2 and Lat-PSCFP2 molecules were preclustered into probability density peaks, which are the maxima in probability density plots (Fig. 1a and Supplementary Table 1). Depending on the number of single molecule images combined, 2–7% of fluorophores in a probability density plot were detected outside probability density peaks. These background signals were most probably due to autofluorescence and/or systematic noise. We observed more clustering of CD3ζ-PSCFP2 and Lat-PSCFP2 and microcluster formation in T cell sheets on activating surfaces than on nonactivating surfaces (Fig. 1a and Supplementary Fig. 2). The formation of microclusters was due to T cell activation, as a major histocompatibility complex class I molecule (H-2Kb) tagged with enhanced green fluorescent protein (eGFP) did not form microclusters on activating surfaces or bilayers (Supplementary Fig. 2b). Comparison of experimental data with randomly simulated signals emphasized the pronounced clustering in the former (Fig. 1b). We did cluster analyses of fusion protein distributions with Ripley’s K function21,22, which confirmed preclustering and enhanced clustering after activation (Fig. 1c). We controlled for any movement of these membrane structures during data acquisition and found that at any time, the contribution of each newly photoswitched fluorophore to the structures detected was equal to the contribution of every other fluorophore (Supplementary Fig. 1e). These data show that CD3ζ and Lat molecules are localized in structures that are stationary (within the resolution of PALM) during acquisition times of up to 10 s in quiescent and activated sheets.
Figure 1.
Analysis of the distribution of CD3ζ and Lat in native T cell plasma membrane sheets by hsPALM. (a) Three-dimensional probability density plots for CD3ζ-PSCFP2 (top) and Lat-PSCFP2 (bottom) in native plasma membrane sheets from quiescent T cells (on poly-L-lysine (PLL); left) and activated T cells (on I-Ek–MCC plus B7-1 at a ratio of 1:3; right) on immobilized surfaces. Molecules are presented as a normalized Gaussian probability density distribution with a width equal to their positional accuracy. Height and color (key) represent the probability density at that point (x,y), with the highest probability density of all images set as 1. (b) Simulated three-dimensional probability density plot containing the same number of molecules as the plot in a for CD3ζ-PSCFP2 on an activating surface with identical positional accuracies but randomized positions. Key: x and y arrows, 1 μm. (c) Ripley’s K-function analysis of the clustering of CD3ζ-PSCFP2 and Lat-PSCFP2 molecules (left); values for L(r) – r above 1 (where L is (K/π)0.5 and r is the radius) indicate clustering at distance r. Right, Ripley’s K-function analysis of the clustering of probability density maxima of CD3ζ-PSCFP2 and Lat-PSCFP2; negative values of L(r) – r for small r values in the data curves are due to the dimensions of the peaks themselves. Data are representative of 25–30 experiments (with one sheet per experiment).
Microclusters showed a heterogeneous probability density distribution (Fig. 1a), which suggested they were concatemers of multiple probability density peaks, similar to those observed in plasma membrane sheets from quiescent T cells. Therefore, we determined the positions of probability density peak maxima in the plots and analyzed their clustering by Ripley’s K function as a measurement of concatenation. Not all probability density peaks necessarily originate from independent membrane domains, but they could originate from irregular shaped structures or concentration differences in domains. Clustering of CD3ζ-PSCFP2 probability density peaks in quiescent T cells was either absent or observed only to a small degree, most probably through heterogeneity in the activation history of the T cells. Clustering of CD3ζ-PSCFP2 probability density peaks increased substantially after activation (Fig. 1c). We detected clustering of Lat-PSCFP2 probability density peaks only in activated T cell sheets (Fig. 1c). The total number of probability density peaks for CD3ζ-PSCFP2 or Lat-PSCFP2 was similar in quiescent and activated sheets, with 10–20 per μm2. The median distance of both CD3ζ-PSCFP2 or Lat-PSCFP2 molecules to their nearest probability density peak maximum, which is a measurement of the median ‘radii’ of probability density peaks, was 35–70 nm in quiescent and activated sheets. The median distance between neighboring CD3ζ-PSCFP2 or Lat-PSCFP2 probability density peak maxima was 100–180 nm in quiescent T cell membrane sheets and 60–130 nm in activated T cell membrane sheets. Using the known TCR concentration, we calculated that the CD3ζ-PSCFP2 probability density peaks contained 7–20 TCR-CD3 complexes in quiescent and activated T cell membrane sheets, with 5–25 probability density peaks per microcluster in activated T cell membrane sheets (Table 1). These data suggest that in quiescent T cells, CD3ζ and Lat molecules are preclustered in membrane domains (protein islands) and that these membrane domains are conserved in microclusters after activation.
Table 1.
Comparison of hsPALM and TEM data
| PALM | TEM | ||
|---|---|---|---|
| Median radius of probability density peaks | 35–70 nm | Observed width of domains | 40–300 nm |
| Concentration of probability density peaks on: | Concentration of domains on: | ||
| all immobilized surfaces and bilayers | 10–20 TCR/μm2 | nonactivating immobilized surface | 4–8 TCR/μm2 |
| 10–20 Lat/μm2 | 12–17 Lat/μm2 | ||
| activating immobilized surface | 6–12 TCR/μm2 | ||
| 8–13 Lat/μm2 | |||
| TCRs per probability density peak | 7–20 | TCRs per domain | 7–30 |
| Median distance between neighboring probability density peaks: | Median distance between neighboring domains: | ||
| Quiescent T cells | 100–180 nm (TCR) | Quiescent T cells | 130–260 nm (TCR) |
| 100–180 nm (Lat) | 100–220 nm (Lat) | ||
| Activated T cells (periphery) | 60–130 nm (TCR) | Activated T cells | 80–150 nm (TCR) |
| 60–130 nm (Lat) | 80–150 nm (Lat) | ||
| Activated T cells (cSMAC) | 60–100 nm (TCR) | ||
| TCR or Lat probability density peaks in microcluster on: | TCR or Lat domains in microcluster on: | ||
| Immobilized surface | 5–25 | Immobilized surface | 5–20 |
| Bilayer | 5–15 | ||
Comparison of measured dimensions, distances and numbers for TCR and Lat molecules, probability density peaks and domains obtained by hsPALM or TEM.
Distribution of the TCR and Lat in live T cells
It is possible that the distributions of TCR and Lat in the plasma membrane differ substantially for intact T cells and plasma membrane sheets. Therefore, we analyzed the distributions of CD3ζ-PSCFP2 and Lat-PSCFP2 in live T cells on immobilized surfaces. These hsPALM data confirmed preclustering of CD3ζ-PSCFP2 and Lat-PSCFP2 in plasma membrane sheets (Supplementary Fig. 3) and yielded the same spatial distribution parameters in quiescent and activated T cells.
To address artifacts that could arise from the use of immobile surfaces, we analyzed CD3ζ-PSCFP2 and Lat-PSCFP2 in live T cells on nonactivating fluid lipid bilayers (~16 molecules of the αLβ2 integrin (LFA-1) ligand ICAM-1 per μm2) and within 5 min of adhesion to activating fluid lipid bilayers (~30 I-Ek–MCC, ~8 B7-1 and ~16 ICAM-1 molecules per μm2). Consistent with the results we obtained on immobile surfaces, CD3ζ-PSCFP2 and Lat-PSCFP2 localized in protein islands that concatenated into microclusters after T cell activation (Supplementary Fig. 4a). We confirmed by statistical analysis the clustering of TCR and Lat molecules, as well as their probability density peaks after activation (Supplementary Fig. 4b). The determined spatial distribution parameters were in the same range as those seen with immobilized surfaces, except the number of probability density peaks per microcluster was diminished to 5–15. This was probably due to the shorter lifetime of microclusters on bilayers caused by their translocation to the central supramolecular activation cluster (cSMAC) and the consequently shorter time window during which they could associate with additional protein islands. Notably, these experiments confirmed the idea that CD3ζ and Lat domains are relatively immobile for at least 10 s, even without the restrictions of immobilized surfaces.
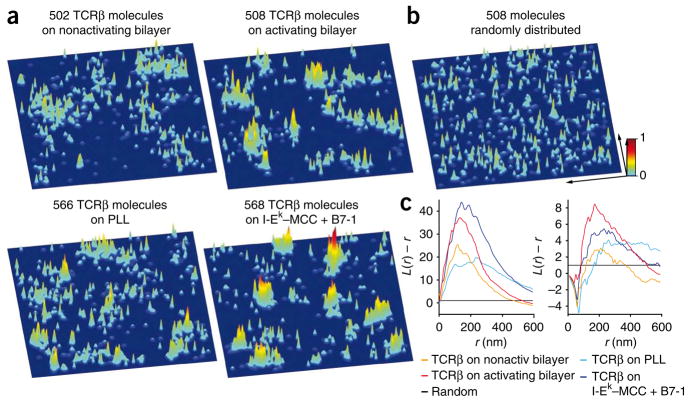
In our CD3ζ-PSCFP2 expression system, ~30% of the fluorophores detected derived from doubly-labeled TCRs because of CD3ζ dimerization. Therefore, we directly labeled endogenous TCR with a single- chain variable fragment (scFv) derived from the H57 antibody23 specific to the TCR β-chain (J.B.H., unpublished observations), crosslinked in a site-specific way to PSCFP2 (anti-TCRβ-scFv–PSCFP2). This probe has the advantage of being an extracellular label and thus it is subject to less background than is ectopically expressed PSCFP2. Notably, the anti-TCRβ-scFv–PSCFP2 probe did not interfere with T cell signaling (data not shown). As in the studies with CD3ζ-PSCFP2, TCRβ localized in probability density peaks in quiescent T cells, which concatenated into microclusters after activation on immobile surfaces and bilayers. Statistical analysis of single-fluorophore and probability density peak positions yielded the same results as the CD3ζ-PSCFP2 data (Fig. 2).
Figure 2.
Analysis of the distribution of endogenous TCRβ in T cells by hsPALM. (a) Three-dimensional probability density plots for T cells labeled with the monovalent anti-TCRβ-scFv–PSCFP2 probe, on nonactivating bilayers (~16 ICAM-1 molecules per μm2; top left) and activating bilayers (~30 I-Ek –MCC plus ~8 B7-1 plus ~16 ICAM-1 molecules per μm2; top right), as well as nonactivating immobilized surfaces (poly-L-lysine; bottom left) and activating immobilized surfaces (I-Ek–MCC plus B7-1 at a ratio of 1:3; bottom right). Results are presented as described in Figure 1. (b) Simulated three-dimensional probability density plot containing the same number of molecules as the plot in a for TCRβ on an activating surface with identical positional accuracies but randomized positions. Key: x and y arrows, 1 μm. (c) Ripley’s K-function analysis of the clustering of TCRβ (left); values for L(r) – r above 1 indicate clustering of molecules at distance r. Right, Ripley’s K-function analysis of the clustering of probability density maxima of TCRβ; negative values of L(r) – r for small r in the data curves are due to the dimensions of the peaks themselves. Nonact, nonactivating. Data representative of 15–20 experiments (with one T cell per experiment).
Avidity studies have suggested that TCR-CD3 complexes on naive T cells undergo long-lived reorganizations after their first encounter with antigen24. The T cells used here had been activated previously, and this might have affected the TCR distribution in quiescent T cells. Therefore, we analyzed the TCR distribution with the anti-TCRβ-scFv–PSCFP2 probe in naive T cells from 5C.C7 mice on nonactivating bilayers. TCRβ was preclustered into probability density peaks, and Ripley’s K function detected substantial clustering (Supplementary Fig. 5). The median radius of probability density peaks was 45–70 nm, and the distance between neighboring probability density peaks was 120–180 nm. We detected no clustering of probability density peaks. These results support the conclusion that TCR preclustering is constitutive and independent of the T cell activation history. Together our in situ data show the existence of TCR and Lat islands in quiescent and naive T cells and their concatenation into microclusters after activation.
Additionally, we examined the effects of actin depolymerization and cholesterol depletion on the distribution of CD3ζ-PSCFP2 and Lat-PSCFP2 in T cells on nonactivating lipid bilayers. In both cases, this led to the formation of large clusters (Supplementary Fig. 6), which suggests that the actin cytoskeleton and cholesterol stabilize TCR and Lat islands and prevent their aggregation. We also analyzed the distribution of the TCR in the cSMAC using the CD3ζ-PSCFP2 fusion protein and the anti-TCRβ-scFv–PSCFP probe (Supplementary Fig. 7). The probability density peaks were clearly visible and densely clustered, as shown by statistical analyses. The radii of probability density peaks were 40–65 nm and the median distance between neighboring probability density peak maxima was 60–100 nm (Table 1). This correlates with the higher TCR concentration in the cSMAC.
Islands concatenate but maintain discrete boundaries
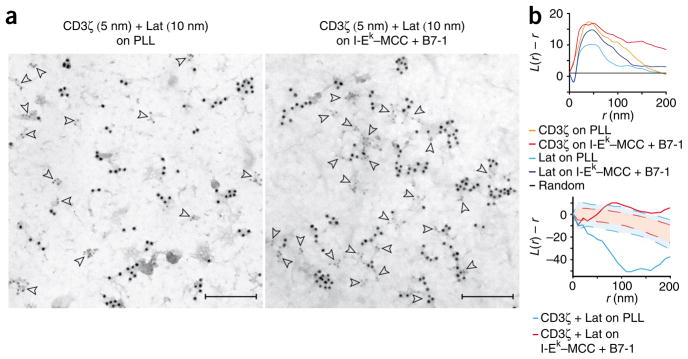
To determine the relationship between TCR and Lat molecules and their respective protein islands, we analyzed fixed T cell membrane sheets with TEM18. This technique gives the highest possible resolution, and ‘ripped’ and fixed plasma membrane sheets accurately reflected live cell membranes, as indicated by hsPALM (Supplementary Fig. 8). TEM analysis confirmed that CD3ζ and Lat molecules were localized in protein islands in quiescent T cell membrane sheets with little contact between CD3ζ and Lat islands (Fig. 3a and Supplementary Fig. 9). In both labeling combinations, the lower staining efficiency of secondary antibodies conjugated to gold particles 10 nm in diameter resulted in apparently unclustered gold particles. However, when we labeled either TCR or Lat with gold particles 5 nm in diameter, we observed single particles only at background amounts. Together these data confirm that TCR and Lat are independently preclustered in quiescent T cells. After activation, CD3ζ and Lat islands formed concatemers with 5–20 CD3ζ or Lat islands per microcluster and more frequent contacts between them (Fig. 3a). We observed little or no mixing of CD3ζ and Lat islands. Analysis of the gold particle distribution by Ripley’s K function showed clustering in sheets from quiescent T cells and enhanced clustering in sheets from activated T cells for CD3ζ and Lat (Fig. 3b). Colocalization of CD3ζ and Lat molecules, as analyzed with the bivariate Ripley’s K function, showed their explicit separation in sheets from quiescent T cells (Fig. 3b). After activation, CD3ζ and Lat molecules showed substantial association at distances above 40–100 nm, which was due to association of the protein islands without apparent overlap and/or mixing of their contents. This number closely reflected the average protein island radii and fit well with the radii of the probability density peaks determined in the hsPALM studies.
Figure 3.
TEM analysis of the concatenation of CD3ζ and Lat islands. (a) Localization of CD3ζ and Lat in plasma membrane sheets generated from quiescent T cells (on poly-L-lysine; left) and activated T cells (on I-Ek–MCC plus B7-1 at a ratio of 1:3; right), detected (arrowheads) with 5-nm gold particles (CD3ζ) or 10-nm gold particles (Lat). (b) Ripley’s K-function analysis of the clustering of CD3ζ and Lat molecules (top); values for L(r) – r above 1 indicate clustering of molecules at distance r. Below, bivariate Ripley’s K-function analysis of the coclustering of CD3ζ and Lat molecules (solid blue lines, quiescent T cells; solid red lines, activated T cells). Values above the 99% confidence envelope (blue shaded areas (quiescent) and red shaded areas (activated)) indicate statistically significant association or attraction at distance r; values below the 99% confidence envelope indicate statistically significant repulsion or explicit separation at distance r. Scale bars, 200 nm. Data are representative of 25–30 experiments (with one sheet per experiment).
We detected four to eight and six to twelve CD3ζ islands per μm2 in quiescent and activated T cell membrane sheets, respectively. These data, together with the lower number of gold particles detected in quiescent T cell membrane sheets, show that antibody-based detection of CD3ζ is less efficient in quiescent sheets than in activated sheets. This might be due to steric inhibition, as the inactive form of CD3ζ may be buried in the plasma membrane25–27. We detected 12–17 and 8–13 Lat islands per μm2 in quiescent and activated sheets, respectively, with the same detection efficiency for Lat in both cases. Thus, the number of Lat islands decreased after activation, whereas their average size increased (Fig. 3a). In the hsPALM studies, we found the number of probability density peaks to be relatively constant and higher than the number of protein islands detected in the TEM images. The electron-microscopy data support the assumption that not all maxima in the probability density plots derive from independent domains but that because of stable borders, shape and/or concentration differences, juxtaposed protein islands can still be detected as separate probability density maxima by hsPALM. Comparison of hsPALM and TEM statistics suggested that there were 1.2–1.5 probability density peaks per protein island on average. Therefore, we estimate that TCR islands contain 7–30 TCRs. The width of protein islands ranges from 40 nm to 300 nm, which is in agreement with the median distances of fluorophores to their nearest peak measured by hsPALM. After activation, the distance between islands decreased from 130–260 nm to 80–150 nm for CD3ζ islands and from 100–220 nm to 80–150 nm for Lat islands (Table 1). Experiments with gold-conjugated antibodies of reversed particle sizes yielded similar results (Supplementary Fig. 9).
Movement of the TCR and Lat in live T cells
To understand the spatio-temporal relationship between TCR and Lat molecules and their respective protein islands, we used dcFCCS to analyze the movement of these molecules together in live T cells on lipid bilayers. In dcFCCS, there is simultaneous measurement of fluctuations in fluorescence intensity for two different fluorophores in a small sample area ~0.5 μm in diameter. Fluctuations in fluorescence intensity are caused by the exchange of fluorophores between the observation area and its surroundings. Thus, a measured cross-correlation function G with nonzero values for G(τ = 0) (where τ is the delay time) indicates synchronized or correlated movement of the two different fluorophores. In contrast to dcFCCS studies of the movement of molecules in solution, the correlated movement of molecules in a matrix such as the plasma membrane does not require physical interactions; all that is required is that the molecules move in concert. We used fusion proteins of CD3ζ and Lat with either eGFP or the red fluorescent protein mCherry. Correlated movement can be quantified as the fraction F′ or F″, and the magnitude or frequency of fluctuations can be quantified as the parameter N (Supplementary Methods). We controlled for bleed-through fluorescence, photo-bleaching effects, fluorophore blinking, similar expression amounts and maximum cross-correlation (Supplementary Methods and Supplementary Fig. 10).
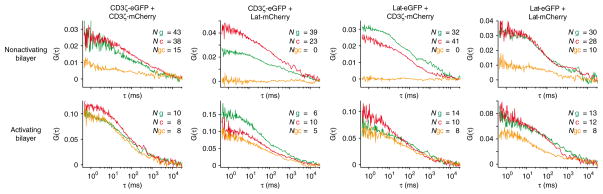
We analyzed by dcFCCS T cells expressing all four possible combinations of CD3ζ and Lat tagged with eGFP or mCherry. The fact that we detected a high fraction of mobile CD3ζ and Lat molecules by dcFCCS, but found in the hsPALM studies that CD3ζ and Lat islands were stable in their position during acquisition times, indicates that protein islands are indeed membrane domains and not protein complexes. Long-range diffusion of molecules is probably sustained by their frequent exchange between protein islands of similar content, which could be due to ‘hop diffusion’28,29. Autocorrelation curves for CD3ζ and Lat did not fit well to single diffusion rates, which showed diffusion patterns characteristic of molecules constrained in substructures. We used the decay of correlation curves and the sample area to determine an approximate diffusion coefficient for CD3ζ and Lat of ~0.5 μm2/s in quiescent and activated T cells. For T cells on nonactivating bilayers, the value for N was 50–70. We detected cross-correlation between CD3ζ itself with an F′ of 0.25 ± 0.1 and an F″ of 0.13 ± 0.1, and between Lat itself with an F′ of 0.23 ± 0.1 and an F″ of 0.15 ± 0.1 (Fig. 4 and Supplementary Table 2). Although dimerization of CD3ζ might have influenced these numbers, the low occurrence of CD3ζ-eGFP or CD3ζ-mCherry made this unlikely. Furthermore, published studies with Fab fragments specific to the TCR β-chain have detected slightly more correlation by two-color coincidence detection microscopy in quiescent T cells30. We observed only background cross-correlation between CD3ζ and Lat in quiescent T cells, with an F′ of 0.08 ± 0.1 and an F″ of 0.04 ± 0.1 (Fig. 4 and Supplementary Table 2). These data further confirm that CD3ζ and Lat localize in different protein islands. After binding of T cells to activating bilayers for 1–3 min, N decreased to 10–20. The correlation between CD3ζ itself increased to an F′ of 0.89 ± 0.1 and an F″ of 0.78 ± 0.1, and the correlation between Lat itself increased to an F′ of 0.56 ± 0.1 and an F″ of 0.39 ± 0.1. CD3ζ and Lat correlated after activation with an F′ of 0.64 ± 0.1 and an F″ of 0.42 ± 0.1 (Fig. 4 and Supplementary Table 2). The data from activating bilayers show that CD3ζ and Lat molecules move in concert with themselves and each other after T cell activation and reflect the concatenation of CD3ζ and Lat islands observed in the hsPALM and TEM studies.
Figure 4.
Analysis of the correlation of CD3ζ and Lat in live T cells on bilayers by dcFCCS. Cross-correlation of CD3ζ and Lat in T cells expressing all possible combinations of CD3ζ and Lat proteins fused with eGFP or mCherry; T cells were allowed to settle on nonactivating bilayers (~16 ICAM-1 molecules per μm2; top) and activating bilayers (~30 I-Ek–MCC plus ~8 B7-1 plus ~16 ICAM-1 molecules per μm2; bottom), and dcFCCS measurements were obtained within 3 min of adhesion where the plasma membrane contacts the bilayer. Green, eGFP autocorrelation curves; red, mCherry autocorrelation curves; yellow, cross-correlation curves for eGFP and mCherry. Cross-correlation curves with nonzero values for G(τ) indicate correlated movement. Numbers in plots indicate entities containing eGFP alone (Ng), mCherry alone (Nc), or eGFP and mCherry (Ngc). Data are representative of five to seven experiments (with one T cell per experiment).
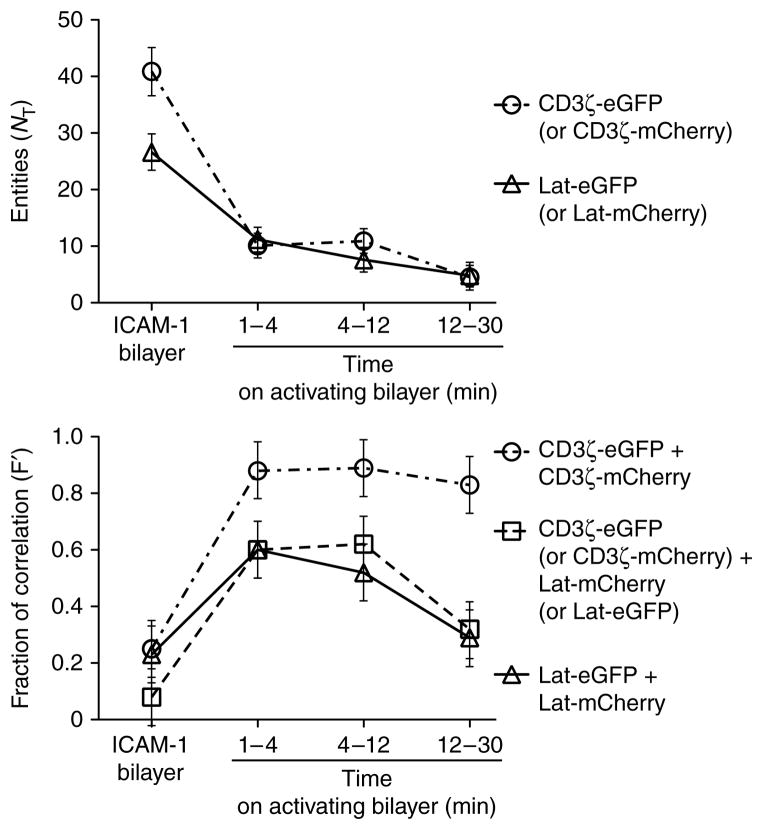
To understand the dynamics of concatenation, we analyzed T cells for up to 30 min after activation in the periphery and measured a progressive decrease in N for CD3ζ and Lat. The correlation between CD3ζ and Lat, and between Lat itself, increased within 1 min of activation, remained high between 1 min and 12 min and then decreased to approximately 50% of the maximum correlation (Fig. 5). The short time frame in which this correlation occurred suggested that little or no mixing of CD3ζ and Lat took place on a molecular level. The correlation of CD3ζ with itself increased within a minute of activation and remained high for all later times (Fig. 5). The separation of TCR and Lat into the cSMAC and periphery, respectively, coincides with the changes in correlation at later time points and is the most likely cause of these changes.
Figure 5.
Kinetics of the correlation between CD3ζ and Lat themselves and each other in T cells on nonactivating and activating bilayers. Above, total entities (NT) containing CD3ζ, Lat or both (time windows, horizontal axis) on a nonactivating bilayer (~16 ICAM-1 molecules per μm2) or activating bilayer (~30 I-Ek–MCC, ~8 B7-1 molecules and ~16 ICAM-1 molecules per μm2). Below, fraction of correlated movement (F′) between CD3ζ and Lat themselves and each other (same bilayers and time windows as above). Data are representative of two to three experiments with about five measurements each.
Our data have shown that the TCR and Lat are localized in distinct plasma membrane domains (protein islands) that concatenate transiently after T cell activation and that these concatemers constitute the microclusters associated with TCR signaling3–6. We have provided an example in which protein islands containing T cell signaling molecules, in this case TCR and Lat, concatenate rapidly after antigen recognition to form microclusters (Supplementary Fig. 11). These move toward the center of the T cell–antigen-presenting cell interaction site. Just before they reach the center of the synapse, the concatemers fall apart and some of the protein islands move on to form the cSMAC, whereas others remain in the periphery4,5.
DISCUSSION
Although it has been shown before that the TCR and Lat are both present in the same microclusters after T cell activation, their organization before activation and their spatial relationship to each other in microclusters has remained unclear. Here we have shown that these molecules are organized in distinct and positionally stable membrane domains (protein islands) on quiescent T cell membranes with little or no colocalization. As measured by hsPALM and TEM, these domains have an average radius of 35–70 nm and are up to 300 nm in their longest dimension (as observed by TEM). These dimensions are in accordance with the confinement zones described before28,29,31, as well as the protein islands that described before18. We found that TCR probability density peaks contained 7–20 TCRs and therefore protein islands contained 7–30 TCRs. We were not able to make similar estimates regarding Lat because of uncertainties about the total number of Lat molecules per T cell. This clustered arrangement of TCRs would provide multiple nearby receptors to engage a ligand once a given complex of TCR–peptide–major histocompatibility complex has dissociated. Thus, given the short half-life of most TCR-ligand interactions and the relatively few ligands required for T cell activation32, protein islands could be important in facilitating the maximum amount of signaling. The idea of localization in plasma membrane domains is further supported by the finding that the recruitment of TCRs into microclusters reaches a plateau at ligand densities much less than the TCR density6 and the correlated downregulation of engaged and unengaged TCRs33,34.
These data indicate that these domains represent the smallest activation unit and are recruited to microclusters independently of the number of ligand-engaged TCRs in them. Microclusters are composed of 5–15 concatenated protein islands on lipid bilayers or 5–25 protein islands on immobilized surfaces, which translates into 70–200 or 70–300 TCRs per microcluster, respectively. This is similar to the reported number of 40–150 TCRs per microcluster on bilayers5. The formation of microclusters on immobilized surfaces suggests that the recruitment of protein islands to microclusters is entirely ligand independent. Our TEM and dcFCCS data have shown that TCR and Lat islands concatenated to form microclusters while retaining their separate character. Using the dimensions of the TCR domains and the number of molecules per domain, we calculate that the concentration of TCRs in the protein islands is on the order of 1,200 TCRs per μm2, versus 140–190 TCRs per μm2 for the entire T cell. This suggests that TCR and Lat concentrations in domains are constant regardless of changing T cell morphology, expression of TCR or Lat, or activation status. This stability in local concentrations of these molecules could ensure that activation thresholds stay within predetermined boundaries and that signal transduction remains reproducible despite changing cellular circumstances.
The TCR and Lat domains were relatively stable in size and position in the time frame of our hsPALM measurements (10 s). Long-range diffusion, as measured by dcFCCS, and as seen by single-molecule tracking of CD2, Lat and the kinase Lck35, can be explained by continuous exchange of molecules between these membrane domains. This exchange probably takes place when domains of compatible properties contact and is probably similar to hop diffusion28,29. Hop diffusion between concatenated TCR or Lat islands is the most likely cause of the detected correlation between TCR and Lat molecules despite their localization in different types of protein islands. The greater density of membrane domains in microclusters and hop diffusion could contribute to the temporary trapping of CD2, Lat and Lck in them35.
The finding that TCR and Lat islands concatenated after T cell activation raises the question of how this process is regulated. Published work has shown that a key intermediary between the TCR-CD3 complex and Lat is Zap70, which binds phosphorylated immunoreceptor tyrosine-based activation motifs on CD3 molecules when the TCR has engaged a ligand, becomes activated by Lck and then phosphorylates Lat36,37. The formation of concatemers could be a primary signaling event that recruits protein islands containing downstream signaling molecules (such as Lat) to the TCR-CD3 islands. Alternatively, activation of Lat islands (for example, through translocation of Zap70) could activate the cytoskeletal machinery, which might then bring the various domains into contact to form microclusters.
Reports showing the coexistence of monomeric and multivalent TCRs indicate that ~45% of TCRs are monomers, ~30% are detected in groups of two or three and only ~10% are found in clusters with sizes comparable to the clusters shown here9. Because of the low antibody staining efficiencies typical of previous approaches, the extent of TCR preclustering measured was probably an underestimate. The interaction we saw here with TCR and Lat may be a general property of receptors that use Lat. TEM studies with fixed plasma membrane sheets from mast cells have found that the receptor FcεRI and Lat are clustered separately in quiescent membrane sheets7,8. After receptor crosslinking with soluble antibodies and multivalent antigen, the FcεRI and Lat domains are joined together. Although those studies did not show the same extent of preclustering and concatenation as we have shown here, this could be due to the different cell type used or the different mode of cell activation.
In this study we have shown that the architecture of the plasma membrane is not only important for the exclusion or accumulation of T cell signaling molecules (for example, through rafts) but also controls the signaling process by the segregation and controlled association of particular membrane domains. Although the molecules and signaling events involved in the regulation of these membrane reorganizations are unknown at present, the division of signaling cascades into building blocks through localization into distinct membrane domains and their spatio-temporal regulation may be a general feature of plasma membrane organization and signaling.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/natureimmunology/.
Supplementary Material
Acknowledgments
We thank B. Wilson for advice on TEM and plasma membrane sheet preparation; J. Perrino from the Cell Sciences Imaging Facility for expertise and service; and F. Tynan for comments on the manuscript. Supported by the US National Institutes of Health (AI 55277 to M.M.D.), the US National Science Foundation (J.T.G.), the Howard Hughes Medical Institute (M.M.D. and J.T.G.) and the Human Frontier Science Program (B.F.L.).
Footnotes
Accession codes. UCSD-Nature Signaling Gateway (http://www.signaling-gateway.org): A001392 and A000553.
Note: Supplementary information is available on the Nature Immunology website.
AUTHOR CONTRIBUTIONS
B.F.L. and M.M.D., conceptualization and manuscript preparation; B.F.L., PALM experiments and analysis, TEM studies, molecular biology, cell biology and protein chemistry; M.A.M., PALM conceptualization and analysis software; M.B.F. and J.T.G., dcFCCS conceptualization; M.B.F., dcFCCS software and analysis; M.B.F. and B.F.L., dcFCCS experiments; and J.B.H., anti-TCRβ-scFv and microscope setup.
References
- 1.Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 2.Grakoui A, et al. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 3.Bunnell SC, et al. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campi G, Varma R, Dustin ML. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J Exp Med. 2005;202:1031–1036. doi: 10.1084/jem.20051182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokosuka T, et al. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat Immunol. 2005;6:1253–1262. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- 6.Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 2006;25:117–127. doi: 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson BS, Pfeiffer JR, Oliver JM. Observing FcεRI signaling from the inside of the mast cell membrane. J Cell Biol. 2000;149:1131–1142. doi: 10.1083/jcb.149.5.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson BS, Pfeiffer JR, Surviladze Z, Gaudet EA, Oliver JM. High resolution mapping of mast cell membranes reveals primary and secondary domains of FcεRI and LAT. J Cell Biol. 2001;154:645–658. doi: 10.1083/jcb.200104049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schamel WW, et al. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J Exp Med. 2005;202:493–503. doi: 10.1084/jem.20042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Miguel G, et al. Multivalent structure of an αβT cell receptor. Proc Natl Acad Sci USA. 1999;96:1547–1552. doi: 10.1073/pnas.96.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 12.Hess ST, et al. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc Natl Acad Sci USA. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shroff H, Galbraith CG, Galbraith JA, Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magde D, Elson EL, Webb WW. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 15.Bacia K, Kim SA, Schwille P. Fluorescence cross-correlation spectroscopy in living cells. Nat Methods. 2006;3:83–89. doi: 10.1038/nmeth822. [DOI] [PubMed] [Google Scholar]
- 16.Groves JT, Parthasarathy R, Forstner MB. Fluorescence imaging of membrane dynamics. Annu Rev Biomed Eng. 2008;10:311–338. doi: 10.1146/annurev.bioeng.10.061807.160431. [DOI] [PubMed] [Google Scholar]
- 17.Sanan DA, Anderson RG. Simultaneous visualization of LDL receptor distribution and clathrin lattices on membranes torn from the upper surface of cultured cells. J Histochem Cytochem. 1991;39:1017–1024. doi: 10.1177/39.8.1906908. [DOI] [PubMed] [Google Scholar]
- 18.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci USA. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chudakov DM, et al. Photoswitchable cyan fluorescent protein for protein tracking. Nat Biotechnol. 2004;22:1435–1439. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- 20.Seder RA, Paul WE, Davis MM, Fazekas de St Groth B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J Exp Med. 1992;176:1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ripley BD. Modeling spatial patterns. J R Stat Soc [Ser A] 1977;B39:172–212. [Google Scholar]
- 22.Ripley BD. Tests of randomness for spatial point patterns. J R Stat Soc [Ser A] 1979;B41:368–374. [Google Scholar]
- 23.Liu J, et al. Crystallization of a deglycosylated T cell receptor (TCR) complexed with an anti-TCR Fab fragment. J Biol Chem. 1996;271:33639–33646. doi: 10.1074/jbc.271.52.33639. [DOI] [PubMed] [Google Scholar]
- 24.Fahmy TM, Bieler JG, Edidin M, Schneck JP. Increased TCR avidity after T cell activation: a mechanism for sensing low-density antigen. Immunity. 2001;14:135–143. [PubMed] [Google Scholar]
- 25.Aivazian D, Stern LJ. Phosphorylation of T cell receptor zeta is regulated by a lipid dependent folding transition. Nat Struct Biol. 2000;7:1023–1026. doi: 10.1038/80930. [DOI] [PubMed] [Google Scholar]
- 26.Sigalov AB, Aivazian DA, Uversky VN, Stern LJ. Lipid-binding activity of intrinsically unstructured cytoplasmic domains of multichain immune recognition receptor signaling subunits. Biochemistry. 2006;45:15731–15739. doi: 10.1021/bi061108f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu C, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3ε cytoplasmic tyrosine-based motif. Cell. 2008;135:702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sako Y, Kusumi A. Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. J Cell Biol. 1994;125:1251–1264. doi: 10.1083/jcb.125.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusumi A, Ike H, Nakada C, Murase K, Fujiwara T. Single-molecule tracking of membrane molecules: plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules. Semin Immunol. 2005;17:3–21. doi: 10.1016/j.smim.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 30.James JR, et al. Single-molecule level analysis of the subunit composition of the T cell receptor on live T cells. Proc Natl Acad Sci USA. 2007;104:17662–17667. doi: 10.1073/pnas.0700411104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simson R, Sheets ED, Jacobson K. Detection of temporary lateral confinement of membrane proteins using single-particle tracking analysis. Biophys J. 1995;69:989–993. doi: 10.1016/S0006-3495(95)79972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 33.Exley M, Wileman T, Mueller B, Terhorst C. Evidence for multivalent structure of T cell antigen receptor complex. Mol Immunol. 1995;32:829–839. doi: 10.1016/0161-5890(95)00046-h. [DOI] [PubMed] [Google Scholar]
- 34.Bonefeld CM, et al. TCR comodulation of nonengaged TCR takes place by a protein kinase C and CD3γ di-leucine-based motif-dependent mechanism. J Immunol. 2003;171:3003–3009. doi: 10.4049/jimmunol.171.6.3003. [DOI] [PubMed] [Google Scholar]
- 35.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu DH, Morita CT, Weiss A. The Syk family of protein tyrosine kinases in T cell activation and development. Immunol Rev. 1998;165:167–180. doi: 10.1111/j.1600-065x.1998.tb01238.x. [DOI] [PubMed] [Google Scholar]
- 37.Samelson LE. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu Rev Immunol. 2002;20:371–394. doi: 10.1146/annurev.immunol.20.092601.111357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.