Abstract
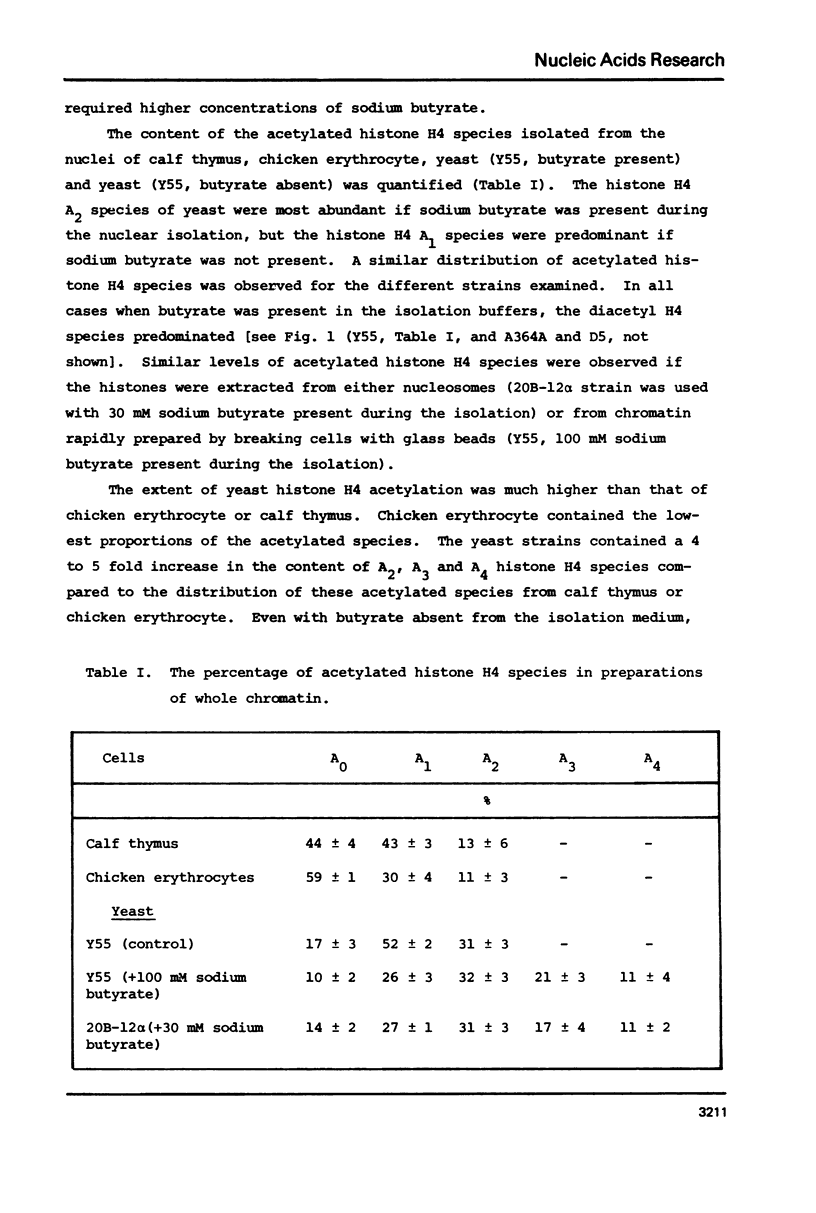
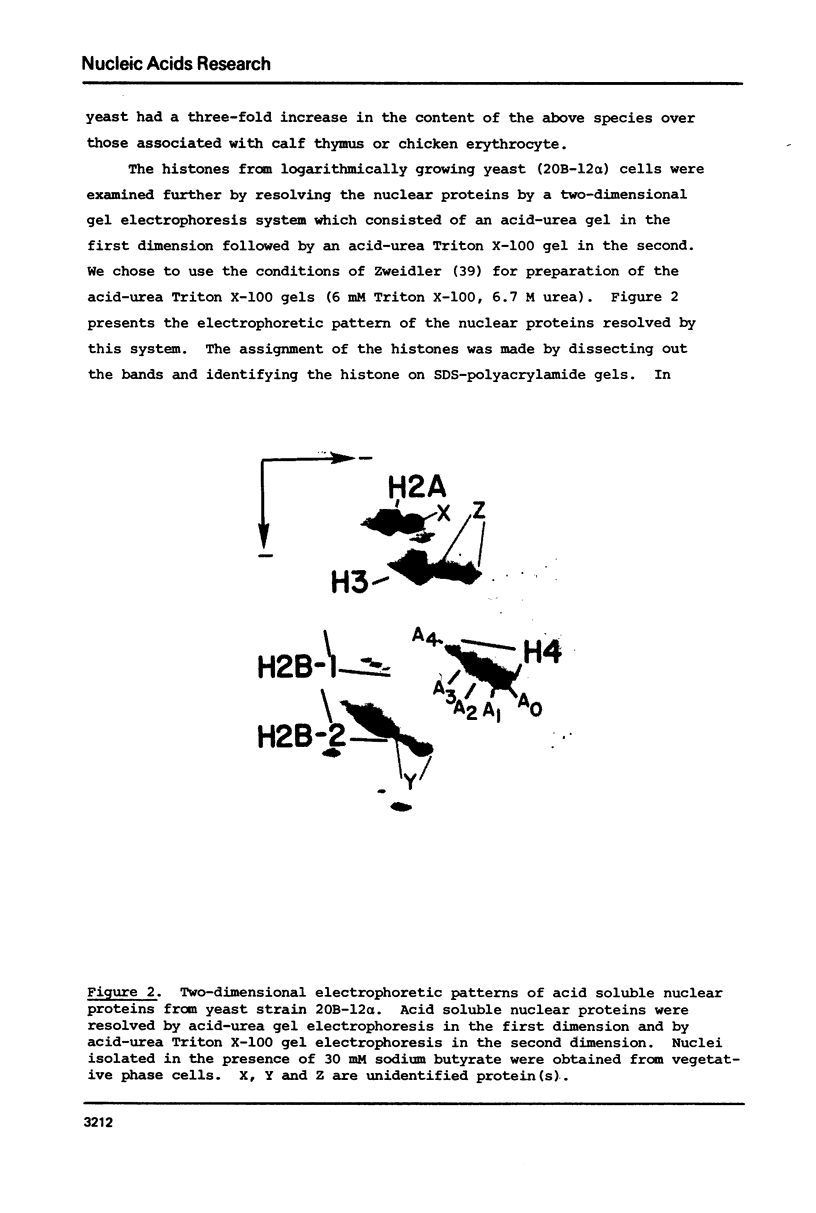
The content of the acetylated histone species associated with the highly transcriptionally active chromatin of yeast was examined. We found yeast chromatin to contain very high levels of the acetylated species for histones H3, H4 and possibly the H2B variants, H2B-1 and H2B-2. Sixty-three percent of the histone H4 species was represented by the di-, tri- and tetra-acetylated forms. These results make yeast chromatin among the most highly acetylated of any chromatins reported thus far. In addition, the results are consistent with the idea that hyperacetylation of histones allows chromatin to be transcribed at an increased rate.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bode J., Henco K., Wingender E. Modulation of the nucleosome structure by histone acetylation. Eur J Biochem. 1980 Sep;110(1):143–152. doi: 10.1111/j.1432-1033.1980.tb04849.x. [DOI] [PubMed] [Google Scholar]
- Boffa L. C., Vidali G., Mann R. S., Allfrey V. G. Suppression of histone deacetylation in vivo and in vitro by sodium butyrate. J Biol Chem. 1978 May 25;253(10):3364–3366. [PubMed] [Google Scholar]
- Brandt W. F., Patterson K., von Holt C. The histones of yeast. The isolation and partial structure of the core histones. Eur J Biochem. 1980 Sep;110(1):67–76. doi: 10.1111/j.1432-1033.1980.tb04841.x. [DOI] [PubMed] [Google Scholar]
- Candido E. P., Reeves R., Davie J. R. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978 May;14(1):105–113. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- Chahal S. S., Matthews H. R., Bradbury E. M. Acetylation of histone H4 and its role in chromatin structure and function. Nature. 1980 Sep 4;287(5777):76–79. doi: 10.1038/287076a0. [DOI] [PubMed] [Google Scholar]
- Chestier A., Yaniv M. Rapid turnover of acetyl groups in the four core histones of simian virus 40 minichromosomes. Proc Natl Acad Sci U S A. 1979 Jan;76(1):46–50. doi: 10.1073/pnas.76.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousens L. S., Gallwitz D., Alberts B. M. Different accessibilities in chromatin to histone acetylase. J Biol Chem. 1979 Mar 10;254(5):1716–1723. [PubMed] [Google Scholar]
- Covault J., Chalkley R. The identification of distinct populations of acetylated histone. J Biol Chem. 1980 Oct 10;255(19):9110–9116. [PubMed] [Google Scholar]
- Davie J. R., Candido E. P. Acetylated histone H4 is preferentially associated with template-active chromatin. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3574–3577. doi: 10.1073/pnas.75.8.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie J. R., Candido E. P. DNase I sensitive chromatin is enriched in the acetylated species of histone H4. FEBS Lett. 1980 Feb 11;110(2):164–168. doi: 10.1016/0014-5793(80)80063-9. [DOI] [PubMed] [Google Scholar]
- Dobson M. E., Ingram V. M. In vitro transcription of chromatin containing histones hyperacetylated in vivo. Nucleic Acids Res. 1980 Sep 25;8(18):4201–4219. doi: 10.1093/nar/8.18.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner C., Traut R. R., Mason D. T., Wikman-Coffelt J. Quantification of Coomassie Blue stained proteins in polyacrylamide gels based on analyses of eluted dye. Anal Biochem. 1975 Feb;63(2):595–602. doi: 10.1016/0003-2697(75)90386-3. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky M. A., Glover C., Johmann C. A., Keevert J. B., Mathis D. J., Samuelson M. Histones and chromatin structure in Tetrahymena macro- and micronuclei. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):493–503. doi: 10.1101/sqb.1978.042.01.052. [DOI] [PubMed] [Google Scholar]
- Jackson V., Shires A., Chalkley R., Granner D. K. Studies on highly metabolically active acetylation and phosphorylation of histones. J Biol Chem. 1975 Jul 10;250(13):4856–4863. [PubMed] [Google Scholar]
- Jones E. W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977 Jan;85(1):23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl L., Lyness T., Dixon G. H., Levy-Wilson B. Distribution of high mobility group proteins among domains of trout testis chromatin differing in their susceptibility to micrococcal nuclease. J Biol Chem. 1980 Feb 10;255(3):1090–1095. [PubMed] [Google Scholar]
- Lohr D., Hereford L. Yeast chromatin is uniformly digested by DNase-I. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4285–4288. doi: 10.1073/pnas.76.9.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie A. J., Dixon G. H. Synthesis, acetylation, and phosphorylation of histone IV and its binding to DNA during spermatogenesis in trout. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1975–1979. doi: 10.1073/pnas.69.7.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardian J. K., Isenberg I. Yeast inner histones and the evolutionary conservation of histone-histone interactions. Biochemistry. 1978 Sep 5;17(18):3825–3833. doi: 10.1021/bi00611a023. [DOI] [PubMed] [Google Scholar]
- Mathis D. J., Oudet P., Wasylyk B., Chambon P. Effect of histone acetylation on structure and in vitro transcription of chromatin. Nucleic Acids Res. 1978 Oct;5(10):3523–3547. doi: 10.1093/nar/5.10.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. T., Burgess D. R. SDS microslab linear gradient polyacrylamide gel electrophoresis. Anal Biochem. 1978 Jul 1;87(2):386–396. doi: 10.1016/0003-2697(78)90688-7. [DOI] [PubMed] [Google Scholar]
- Moore M., Jackson V., Sealy L., Chalkley R. Comparative studies on highly metabolically active histone acetylation. Biochim Biophys Acta. 1979 Jan 26;561(1):248–260. doi: 10.1016/0005-2787(79)90508-2. [DOI] [PubMed] [Google Scholar]
- Nelson D. A., Beltz W. R., Rill R. L. Chromatin subunits from baker's yeast: isolation and partial characterization. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1343–1347. doi: 10.1073/pnas.74.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. A., Perry W. M., Chalkley R. Sensitivity of regions of chromatin containing hyperacetylated histones to DNAse I. Biochem Biophys Res Commun. 1978 May 15;82(1):365–363. doi: 10.1016/0006-291x(78)90617-4. [DOI] [PubMed] [Google Scholar]
- Nelson D., Covault J., Chalkley R. Segregation of rapidly acetylated histones into a chromatin fraction released from intact nuclei by the action of micrococcal nuclease. Nucleic Acids Res. 1980 Apr 25;8(8):1745–1763. doi: 10.1093/nar/8.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Pospelov V. A., Svetlikova S. B., Vorob'ev V. I. Nucleosome-nucleosome interaction in chromatin. FEBS Lett. 1979 Mar 1;99(1):123–128. doi: 10.1016/0014-5793(79)80263-x. [DOI] [PubMed] [Google Scholar]
- Riggs M. G., Whittaker R. G., Neumann J. R., Ingram V. M. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature. 1977 Aug 4;268(5619):462–464. doi: 10.1038/268462a0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Wangh L. J., Allfrey V. G. Processing of newly synthesized histone molecules. Science. 1975 Oct 10;190(4210):117–128. doi: 10.1126/science.1166303. [DOI] [PubMed] [Google Scholar]
- Sealy L., Chalkley R. DNA associated with hyperacetylated histone is preferentially digested by DNase I. Nucleic Acids Res. 1978 Jun;5(6):1863–1876. doi: 10.1093/nar/5.6.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealy L., Chalkley R. The effect of sodium butyrate on histone modification. Cell. 1978 May;14(1):115–121. doi: 10.1016/0092-8674(78)90306-9. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978 Apr;13(4):691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- Sommer Yeast chromatin: search for histone H1. Mol Gen Genet. 1978 May 31;161(3):323–331. doi: 10.1007/BF00331008. [DOI] [PubMed] [Google Scholar]
- Spiker S. A modification of the acetic acid-urea system for use in microslab polyacrylamide gel electrophoresis. Anal Biochem. 1980 Nov 1;108(2):263–265. doi: 10.1016/0003-2697(80)90579-5. [DOI] [PubMed] [Google Scholar]
- Vidali G., Boffa L. C., Bradbury E. M., Allfrey V. G. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc Natl Acad Sci U S A. 1978 May;75(5):2239–2243. doi: 10.1073/pnas.75.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S., Isenberg I. High mobility group proteins of Saccharomyces cerevisiae. Biochemistry. 1980 May 13;19(10):2236–2240. doi: 10.1021/bi00551a037. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Allen J. R., Riedel G., Van Holde K. E. The effects of salt concentration and H-1 depletion on the digestion of calf thymus chromatin by micrococcal nuclease. Nucleic Acids Res. 1979;6(5):1843–1862. doi: 10.1093/nar/6.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Stein A. Folding of DNA by histones which lack their NH2-terminal regions. J Biol Chem. 1978 Jun 10;253(11):3857–3861. [PubMed] [Google Scholar]
- Wray W., Stubblefield E. A new method for the rapid isolation of chromosomes, mitotic apparatus, or nuclei from mammalian fibroblasts at near neutral pH. Exp Cell Res. 1970 Mar;59(3):469–478. doi: 10.1016/0014-4827(70)90656-7. [DOI] [PubMed] [Google Scholar]
- Zweidler A. Resolution of histones by polyacrylamide gel electrophoresis in presence of nonionic detergents. Methods Cell Biol. 1978;17:223–233. [PubMed] [Google Scholar]