Abstract
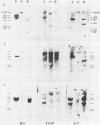
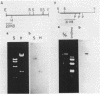
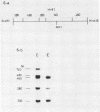
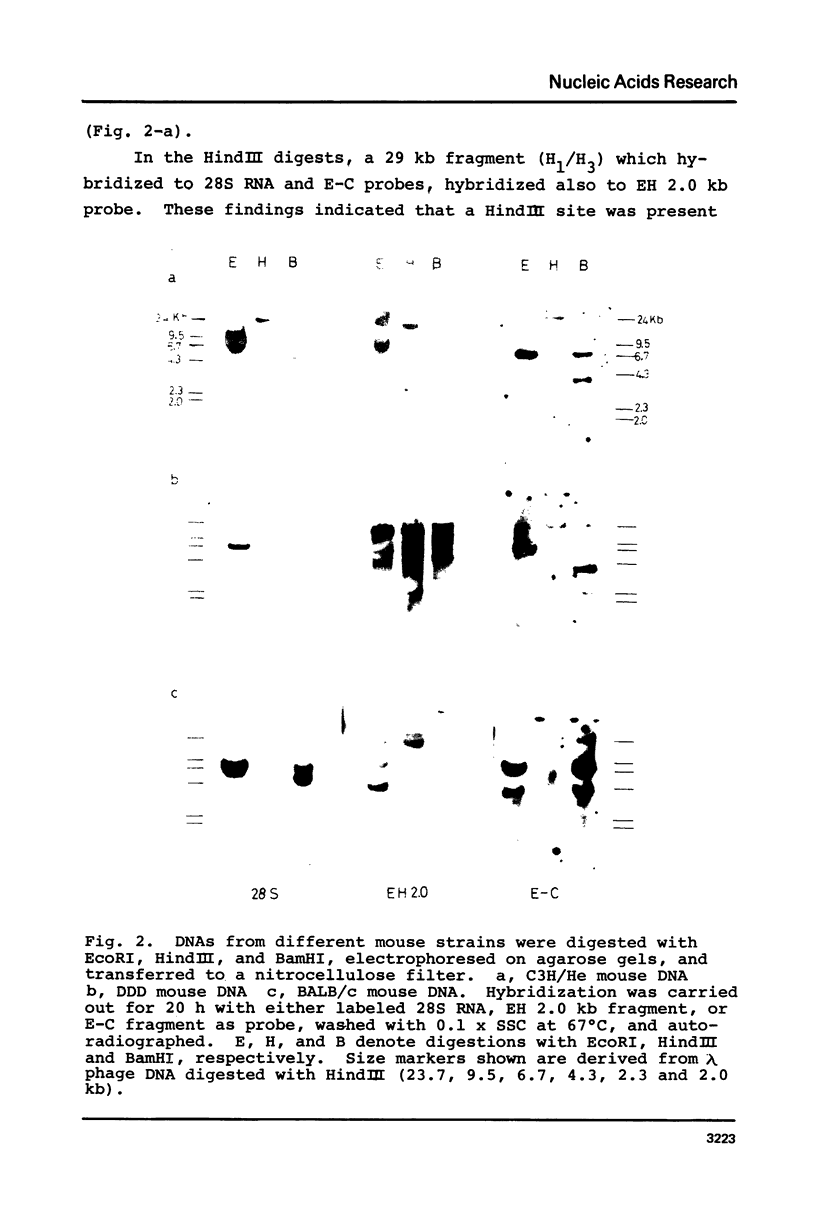
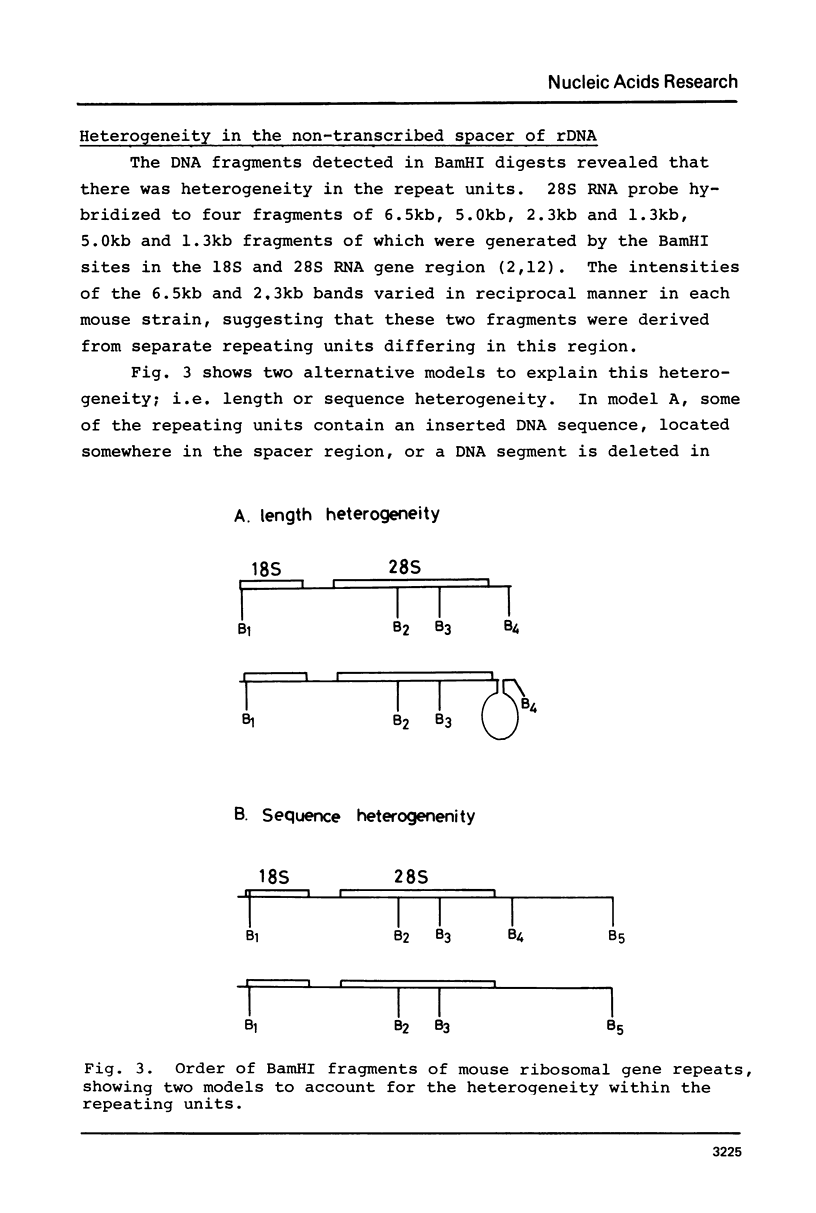
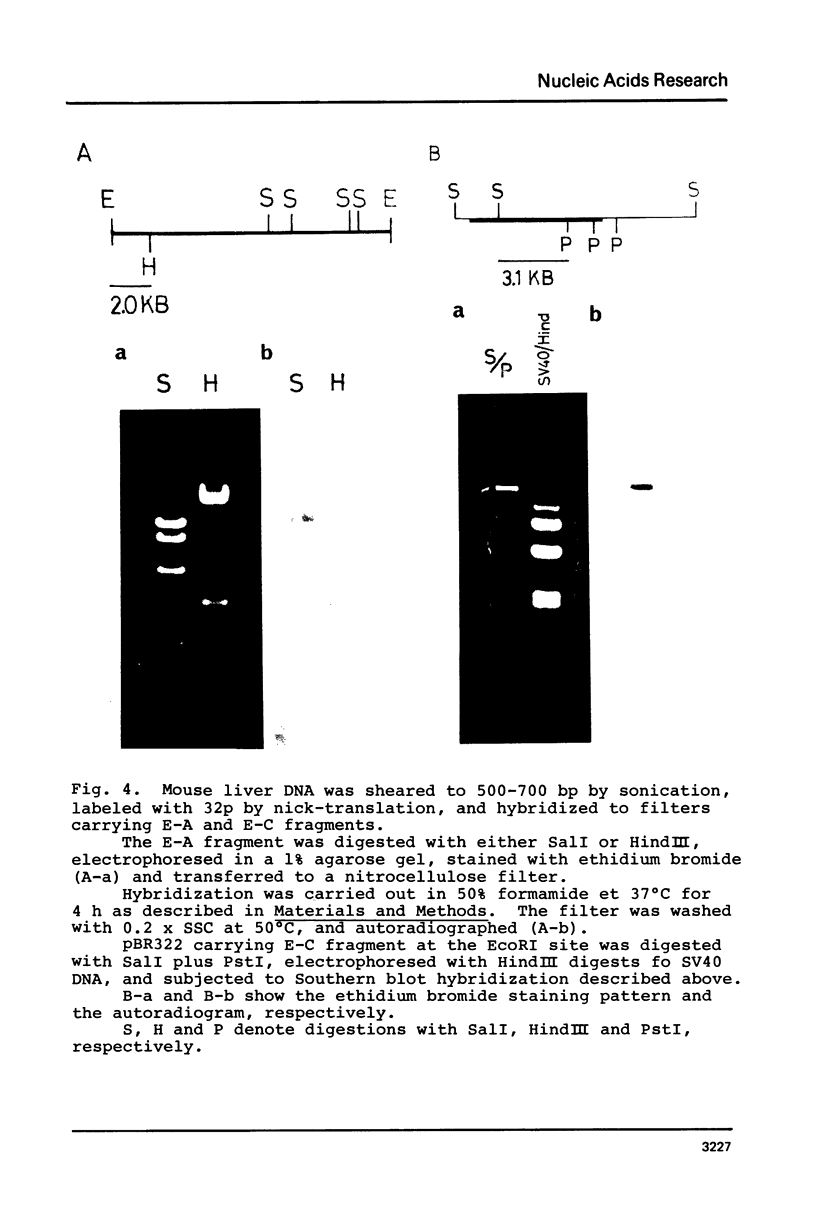
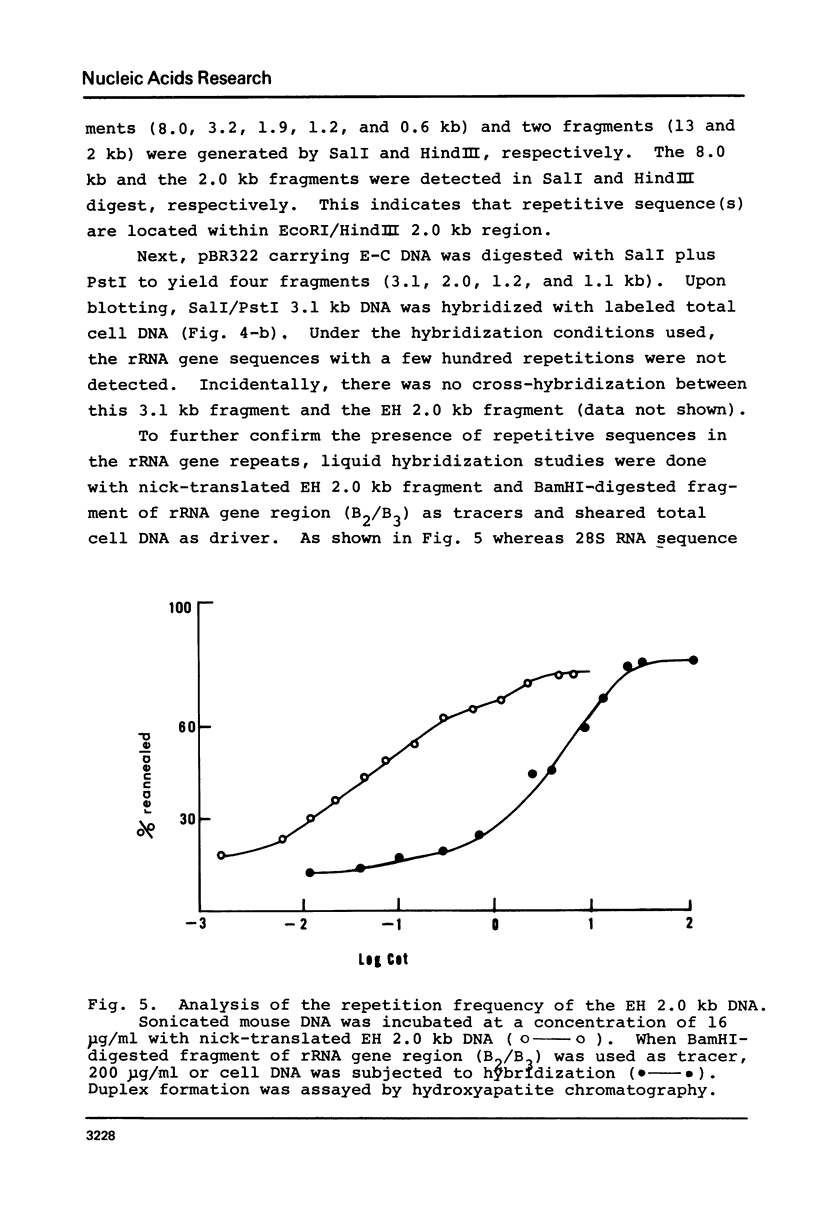
The organization of the ribosomal RNA (rRNA) genes of the mouse was determined by Southern blot hybridization using cloned rDNA fragments as probes, which could encompass the entire spacer region between two rRNA gene regions. The rRNA genes are organized into tandem repeats of nearly uniform length of about 44 kb. The heterogeneity detected in the nontranscribed spacer appears to be caused by its sequence rather than its length difference. At least three kinds of repetitive sequences are present in the non-transcribed spacer region; two of them are located 13 kb upstream from the 5'-end of 18S RNA gene and the other located 1 to 4 kb downstream from the 3'-end of 28S RNA gene.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnheim N., Kuehn M. The genetic behaviour of a cloned mouse ribosomal DNA segment mimics mouse ribosomal gene evolution. J Mol Biol. 1979 Nov 15;134(4):743–763. doi: 10.1016/0022-2836(79)90483-2. [DOI] [PubMed] [Google Scholar]
- Arnheim N., Seperack P., Banerji J., Lang R. B., Miesfeld R., Marcu K. B. Mouse rDNA nontranscribed spacer sequences are found flanking immunoglobulin CH genes and elsewhere throughout the genome. Cell. 1980 Nov;22(1 Pt 1):179–185. doi: 10.1016/0092-8674(80)90166-x. [DOI] [PubMed] [Google Scholar]
- Arnheim N., Southern E. M. Heterogeneity of the ribosomal genes in mice and men. Cell. 1977 Jun;11(2):363–370. doi: 10.1016/0092-8674(77)90053-8. [DOI] [PubMed] [Google Scholar]
- Botchan P., Reeder R. H., Dawid I. B. Restriction analysis of the nontranscribed spacers of Xenopus laevis ribosomal DNA. Cell. 1977 Jul;11(3):599–607. doi: 10.1016/0092-8674(77)90077-0. [DOI] [PubMed] [Google Scholar]
- Cory S., Adams J. M. A very large repeating unit of mouse DNA containing the 18S, 28S and 5.8S rRNA genes. Cell. 1977 Aug;11(4):795–805. doi: 10.1016/0092-8674(77)90292-6. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Fritsch E. F., Lawn R. M., Maniatis T. Molecular cloning and characterization of the human beta-like globin gene cluster. Cell. 1980 Apr;19(4):959–972. doi: 10.1016/0092-8674(80)90087-2. [DOI] [PubMed] [Google Scholar]
- Glover D. M., Hogness D. S. A novel arrangement of the 18S and 28S sequences in a repeating unit of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):167–176. doi: 10.1016/0092-8674(77)90212-4. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Manning R. F., Samols D. R., Gage L. P. The genes for 18S, 5.8S and 28S ribosomal RNA of Bombyx mori are organized into tandem repeats of uniform length. Gene. 1978 Oct;4(2):153–166. doi: 10.1016/0378-1119(78)90027-6. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Kominami R., Honjo T., Muramatsu M. Cloning and determination of a putative promoter region of a mouse ribosomal deoxyribonucleic acid fragment. Biochemistry. 1980 Aug 5;19(16):3780–3786. doi: 10.1021/bi00557a020. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Thomas M., Kramer R. A., Davis R. W. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978 Aug 15;123(3):387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Brown D. D., Wellauer P. K., Dawid I. B. Patterns of ribosomal DNA spacer lengths are inherited. J Mol Biol. 1976 Aug 25;105(4):507–516. doi: 10.1016/0022-2836(76)90231-x. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tartof K. D., Dawid I. G. Similarities and differences in the structure of X and Y chromosome rRNA genes of Drosophila. Nature. 1976 Sep 2;263(5572):27–30. doi: 10.1038/263027a0. [DOI] [PubMed] [Google Scholar]
- Tiemeier D. C., Tilghman S. M., Leder P. Purification and cloning of a mouse ribosomal gene fragment in coliphage lambda. Gene. 1977;2(3-4):173–191. doi: 10.1016/0378-1119(77)90016-6. [DOI] [PubMed] [Google Scholar]
- Trendelenburg M. F., Scheer U., Zentgraf H., Franke W. W. Heterogeneity of spacer lengths in circles of amplified ribosomal DNA of two insect species, Dytiscus marginalis and Acheta domesticus. J Mol Biol. 1976 Dec;108(2):453–470. doi: 10.1016/s0022-2836(76)80130-1. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Brown D. D., Reeder R. H. The molecular basis for length heterogeneity in ribosomal DNA from Xenopus laevis. J Mol Biol. 1976 Aug 25;105(4):461–486. doi: 10.1016/0022-2836(76)90229-1. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Isolation and sequence organization of human ribosomal DNA. J Mol Biol. 1979 Mar 5;128(3):289–303. doi: 10.1016/0022-2836(79)90089-5. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977 Feb;10(2):193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]