Summary
Induced pluripotent stem (iPS) cells can be obtained from fibroblasts upon expression of Oct4, Sox2, Klf4 and c-Myc. To understand how these factors induce pluripotency, we carried out genome-wide analyses of their promoter binding and expression in iPS and partially reprogrammed cells. We find that target genes of the four factors strongly overlap in iPS and embryonic stem (ES) cells. In partially reprogrammed cells, many genes co-occupied by c-Myc and any of the other three factors already show an ES-like binding and expression pattern. In contrast, genes that are specifically co-bound by Oct4, Sox2 and Klf4 in ES cells and encode pluripotency regulators severely lack binding and transcriptional activation. Among the four factors, c-Myc promotes the most ES cell-like transcription pattern when expressed individually in fibroblasts. These data uncover temporal and separable contributions of the four factors during the reprogramming process and indicate that ectopic c-Myc predominantly acts before pluripotency regulators are activated.
Introduction
Reprogramming of human and mouse fibroblasts to induced pluripotent stem (iPS) cells has been achieved by the expression of only four transcription factors, Oct4, Sox2, Klf4 and c-Myc, subsequently referred to as the “four factors” (Maherali et al., 2007; Okita et al., 2007; Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007; Yu et al., 2007). iPS cells hold great promise for the study and therapy of human diseases (Dimos et al., 2008; Park et al., 2008) because they are highly similar to embryonic stem (ES) cells in their ability to self-renew and give rise to all the three germ layers. Molecularly, reprogramming results in the remodeling of the somatic cell transcriptional and chromatin programs to the ES-like state, including the reactivation of the somatically silenced X chromosome, demethylation of the Oct4 and Nanog promoter regions and genome-wide resetting of histone H3 lysine 4 and 27 trimethylation (Maherali et al., 2007; Takahashi et al., 2007; Wernig et al., 2007). A key question raised by transcription factor-induced reprogramming is how the four factors act to bring about this change.
Two studies demonstrated that reprogramming of murine fibroblasts is a gradual process that follows a defined series of molecular events (Brambrink et al., 2008; Stadtfeld et al., 2008). Upon retroviral transduction of the factors fibroblast-specific genes become repressed, exemplified by the downregulation of the somatic marker Thy1, followed first by the activation of embryonic markers such as alkaline phosphatase (AP) and SSEA1 and later by the induction of pluripotency genes such as Nanog and Oct4. While Thy1 down-regulation occurs in the majority of cells, gain of SSEA1 and activation of pluripotency regulators happens with very low efficiency suggesting that as yet unknown epigenetic barriers need to be overcome to induce pluripotency (Huangfu et al., 2008; Meissner et al., 2007; Mikkelsen et al., 2008).
In ES cells, chromatin immunoprecipitation (ChIP) experiments have demonstrated that Klf4, Sox2 and Oct4 together with other ES-specific transcription factors such as Nanog often co-occupy target genes, including their own promoters, which suggests that they cooperate in regulatory feedback loops to maintain self-renewal and pluripotency (Boyer et al., 2005; Chen et al., 2008; Jiang et al., 2008; Kim et al., 2008; Loh et al., 2006). These transcription factors can function both as activators of self-renewal and pluripotency genes and as repressors of lineage commitment genes suggesting that they could activate the ES-specific transcription program and repress fibroblast expression during reprogramming. Based on limited target overlap in ES cells, it was proposed that the function of c-Myc differs from that of Oct4, Sox2 and Klf4 (Chen et al., 2008; Kim et al., 2008). However, to date no studies have been performed to analyze how the four factors bind and function in iPS cells. It is conceivable that they have a different binding pattern in these cells, given their diverse origin. It is also possible that they fulfill transient roles during reprogramming that cannot be inferred from the analysis of the pluripotent end state and necessitate the study of intermediate stages of the process. Understanding the contribution of each factor to the different steps of reprogramming and an in-depth characterization of the iPS state should shed light on the molecular nature of reprogramming.
Here, we studied the function of the four factors at three stages of the reprogramming process: the initial phase, an intermediate step represented by partially reprogrammed cell lines and at the final iPS cell stage. Partially reprogrammed cells express markers of the intermediate reprogramming stage and have failed to transcriptionally activate pluripotency regulators, but can be converted to a pluripotent state with small molecules that affect chromatin modifications or modulate signal transduction (Silva et al., 2008; Mikkelsen et al., 2008). By comparing binding of the factors and expression changes at the three stages, our analysis provides insight into the specific contribution of each factor to the reprogramming process.
Results
The four factors bind similar sets of target genes in iPS and ES cells
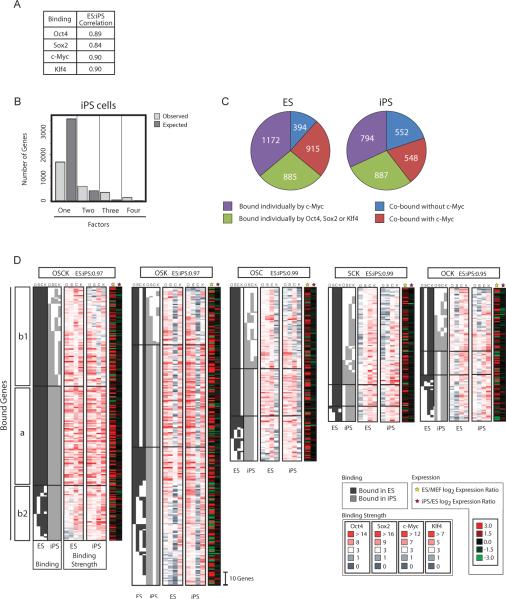
To characterize the completely reprogrammed state, we determined the promoter regions that are bound by the four factors in two ES and two iPS cell lines using ChIP and promoter microarrays and compared binding patterns between these cell types. The iPS cell lines analyzed here (clones 1D4 and 2D4) have completely suppressed retroviral transgene expression and entered a self-sustaining pluripotent state that can give rise to adult chimeras with germline contribution (Maherali et al., 2007). For each factor, binding between the two iPS cell lines is as highly correlated as between the two ES cell lines (Supp Table 1). Hence, the replicate data sets were averaged before determining bound genes for each factor and cell type (all averaged binding data are provided in Supp Table 2 and raw data files are summarized in Supp Table 3). A gene was called bound using an algorithm that considers information of neighboring probes (Materials and Methods; Boyer et al., 2005). It should be noted that genes that fall below the threshold for being called bound might still be targeted by the various factors, perhaps transiently, reflecting weaker binding events. However, we use the phrase “bound” gene to refer to only those genes that passed our binding criteria. The reliability of our binding calls was validated by a comparison of our ES cell targets to previously published observations (Boyer et al., 2005; Chen et al., 2008; Jiang et al., 2008; Kim et al., 2008; Loh et al., 2006). For example, a gene ontology (GO) analysis indicated that c-Myc targets in ES cells include many metabolic regulators while Oct4, Sox2 and Klf4 targets are skewed toward transcriptional regulators of development and differentiation; and the known DNA binding motifs for Oct4, Sox2, Klf4 and c-Myc are highly enriched in the promoter regions that are targeted by these factors (data not shown). Furthermore, real-time PCR analysis confirmed strong enrichments at loci that were called bound (Supp Fig 1) and bound targets identified in merged data sets were reliably found in the individual replicates indicating high reproducibility of the data (Supp Fig 2A).
The four factors exhibit overall similarity in their binding between ES and iPS cells at the genome-wide level (Fig 1A). The number of target genes co-occupied by three or all four factors in iPS cells is much higher than expected by random chance (476 genes) and the number of targets bound by just one factor is much lower (1681 genes, χ2 test p = 0.0057, Fig 1B), like in ES cells (Chen et al., 2008; Kim et al., 2008). Notably, while we found 794 genes bound by c-Myc alone, approximately half of the genes that are occupied by multiple transcription factors (548 genes) are also targets of this factor (Fig 1C), suggesting that, surprisingly, c-Myc co-regulates many genes together with the other three factors in iPS cells (χ2 test p = 1.9 e−291).
Figure 1. Target overlap between ES and iPS cells.
(A) Pearson correlation of binding strength at the probe level for each factor between iPS and ES cells across all genes.
(B) Bar chart indicates the number of genes observed to be bound by only one, any combination of two or three, and all four factors in iPS cells (light grey) and the expected number of target genes calculated assuming that each factor binds independently of the others (dark grey).
(C) Pie charts indicate the number and proportion of target genes bound by single or multiple factors in iPS or ES cells, divided according to the presence of c-Myc.
(D) Comparison of binding patterns in ES and iPS cells for genes bound by the indicated combination of three or four factors (denoted by single letter nomenclature with O=Oct4, S=Sox2, C=c-Myc, and K=Klf4), The three segments of each cluster are based on the overlap of binding events between ES and iPS cells: section (a) depicts genes bound by the same set of factors in ES and iPS cells, (b1) and (b2) contain genes that have binding by additional and/or fewer factors in either cell type. Binding of a gene is marked by a dark (ES) or light grey block (iPS). Heat maps plot the corresponding binding strength. For each factor, the lowest maximum binding strength among all genes and cell types analyzed in this study was set to the darkest red color. For each binding cluster, expression ratios ES cells/fibroblasts (MEFs) (yellow star) and iPS/ES cells (red star) and the Pearson correlation coefficient comparing expression of the genes within a cluster between ES and iPS cells are given (top).
To analyze co-occupancy by the four factors in more detail, we classified target genes according to the combination of factors bound. This categorization yielded a total of 15 groups, with one group containing genes bound by all four factors (single letter code OSCK for Oct4, Sox2, c-Myc and Klf4), four groups with genes bound by different combinations of three factors (OSC, OSK, OCK and SCK), six groups of genes that are bound by two factors (OS, OC, OK, SC, SK and CK) and four gene groups that are bound by only one (Supp Fig 3). Next, we asked whether genes that are bound by a particular combination of factors in iPS cells are associated with the same set of factors in ES cells and vice versa (Fig 1D and Supp Fig 4). Across all 15 groups, the binding pattern is similar between the two cell types. Focusing on the groups of genes co-bound by three or four factors (Fig 1D), up to 64% of ES cell targets are bound by the same sets of factors in iPS cells (Fig 1D, section a). However, the binding pattern is not identical because there are genes that are bound by fewer factors in iPS cells than ES cells (Fig 1D, section b1), as there are genes bound by fewer factors in ES cells than iPS cells (Fig 1D, section b2). Nevertheless, only very few genes that are associated with three or four factors in ES cells are not bound by any factor in iPS cells and vice versa. Indeed, when allowing differential binding by one factor between ES and iPS cells, up to 87% of the genes are bound by comparable sets of factors. These findings also apply to genes bound by two factors (Supp Fig 4A). Interestingly, genes that are bound by a specific factor in one cell type but not in the other have a lower binding strength than those genes that are bound in both cell types (Fig 1D, Supp Fig 2, and 4A) supporting the notion that binding patterns in iPS and ES cells are highly similar. Furthermore, target genes are similarly expressed in both cell types suggesting that differences in factor binding are too small to dramatically affect transcription (Fig 1D and Supp Fig 4A).
The similarity of binding events between both cell types is further reflected by analogous functional classifications for genes in the 15 binding groups in iPS and ES cells (Supp Fig 3). For both cell types, GO analysis demonstrates that genes associated with c-Myc, by itself or in combination with the other factors, are significantly enriched for regulators of metabolic processes including the control of translation, RNA splicing, cell cycle and energy production, while genes bound by combinations of Oct4, Sox2 and Klf4, in the absence of c-Myc, are mainly implicated in transcriptional control of development. Together, these data indicate that Oct4, Sox2, Klf4 and c-Myc binding patterns in iPS cells resemble those of ES cells, suggesting that pluripotency relies on binding of the four factors to a conserved set of target genes. The extensive co-binding among all four factors indicates that c-Myc participates in the transcriptional network formed by Oct4, Sox2 and Klf4 much more than expected.
Extensive failure to activate ES-like expression in partially reprogrammed cells
To begin to understand events that occur during the reprogramming process, we decided to analyze partially reprogrammed cell lines. These cells emerge at intermediate stages of reprogramming, when reprogrammed cells are selected based on morphology or reporter expression, and have been informative for understanding the barriers of the reprogramming process (Meissner et al., 2007; Mikkelsen et al., 2008; Takahashi and Yamanaka, 2006). We selected two stable, clonal, partially reprogrammed cell lines for our analysis (1A2 and 1B3) that were previously derived from female fibroblasts carrying a GFP/puromycin resistance reporter cassette under the control of the endogenous Nanog promoter (Maherali et al., 2007; Supp Fig 5). These partially reprogrammed cells have an ES like morphology surrounded by a few round cells, are positive for SSEA1 and, similar to ES cells, have a doubling time of 13 hours (Supp Fig 5). The four factors are expressed from the exogenous retroviral constructs and endogenous pluripotency regulators such as Nanog and Oct4 and the inactive X chromosome are not reactivated (Supp Fig 5). These cell lines can be maintained in culture without a change in morphology and the expression profile of a pool of 10 colonies is highly similar to that of the population demonstrating homogeneity (Supp Fig 6). A small fraction of GFP-positive, i.e. highly Nanog-expressing, cells (~1%) that support chimerism stochastically arises in these cell lines (Supp Fig 6; Maherali et al., 2007), and upon addition of a specific MEK inhibitor used in recent reprogramming experiments (Silva et al., 2008) an efficient transition to GFP-positive colonies is observed (Supp Fig 6). These data demonstrate that our partially reprogrammed cells reflect an intermediate stage of the reprogramming process that can transition to the completely reprogrammed iPS state.
We first characterized the gene expression pattern of the two partially reprogrammed cell lines. They are highly similar in expression to those analyzed recently by Mikkelsen et al. (Mikkelsen et al., 2008), even when derived from somatic cells other than fibroblasts, supporting the idea that partially reprogrammed cells are trapped at a common intermediate state (Supp Fig 6). Fibroblast-specific genes are more efficiently downregulated in partially reprogrammed cells (correlation ES/MEF : partial/MEF =0.628) than ES-specific genes are activated (correlation ES/MEF : partial/MEF =0.324; Supp Fig 7, Supp Table 4). A GO analysis revealed that ES-specific metabolic regulators are more completely activated in partial reprogrammed cells than transcriptional regulators (Supp Fig 7), in agreement with the observation that key transcriptional regulators of pluripotency are turned on only during the final steps of the reprogramming process (Brambrink et al., 2008; Stadtfeld et al., 2008).
Four factor binding in partially reprogrammed cells
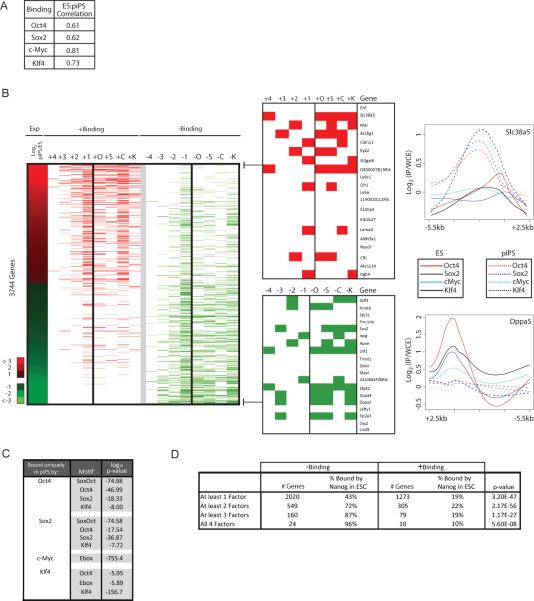
To address whether differential gene expression between partially reprogrammed cells and ES cells is due to binding of the four factors to different sets of genes, we determined their target genes in both partially reprogrammed cell lines. Analogous to expression, binding is highly correlated between these cell lines supporting the notion that these cells represent a stable and homogenous intermediate reprogramming state (Supp Table 1). We therefore merged binding data from both cell lines to generate the set of bound genes and found that the genome-wide binding of each factor is much less conserved between ES cells and partially reprogrammed cells than between iPS and ES cells (Fig 2A, compare with 1A).
Figure 2. Differential binding between partially reprogrammed cells and ES cells correlates with expression differences.
(A) Pearson correlation of binding strength at the probe level for each factor between ES cells and partially reprogrammed cells across all genes.
(B) The heat map in the Exp column depicts the log2 expression ratio between partially reprogrammed cells (piPS) and ES cells for genes more than two-fold differentially expressed between these cell types. For these genes, +Binding columns illustrate binding events in partially reprogrammed cells that were not found in ES cells (in red); with +1 to +4 indicating the number of factors that were bound in partially reprogrammed cells but not in ES cells and the letter indicating the factor (Oct4 (+O), Klf4 (+K), Sox2 (+S), or c-Myc (+C)). Similarly, in −Binding columns, genes that were bound in ES cells but not in partially reprogrammed cells are marked in green; with −1 to −4 indicating the number of factors that were bound in ES cells and not in partially reprogrammed cells and the letter indicating the factor. Binding differences for the most differentially expressed genes between ES and partially reprogrammed cells are presented in a zoomed view to the right and graphs to the far right plot log2(IP/input) ratios of probes over the 8kb promoter region for one gene from each subgroup.
(C) For binding events found in partially reprogrammed cells but not ES cells, bound regions were scanned for the presence of the DNA binding motifs of the four factors (with the Ebox motif being bound by c-Myc). Significantly enriched motifs are indicated.
(D) Genes bound by fewer factors in partially reprogrammed cells than ES cells (− Binding) or by more factors (+ Binding) were divided according to the number of factors differentially bound between these cell types. The number of genes per group and the fraction of these genes that is bound by Nanog in ES cells are presented. Significance was computed using the difference of proportions test.
To study the relationship of factor binding and expression in detail, we focused on the 3744 genes that are more than two-fold differentially expressed between partially reprogrammed cells and ES cells (Fig 2B, Supp Table 6). Approximately one third of these genes (1139 genes) are direct binding targets of the factors. Genes that are higher expressed in partially reprogrammed cells than in ES cells are often bound by more factors in the intermediate state than in ES cells. In contrast, genes that are more highly expressed in ES cells often are bound by fewer factors in partially reprogrammed cells than in ES cells (Fig 2B). The higher the amplitude of differential expression the more factors tend to be differentially bound between ES and partially reprogrammed cells (Fig 2B) suggesting that binding by multiple factors leads to stronger transcriptional activation. These data suggest that genes are more strongly expressed in partially reprogrammed cells compared to ES cells due to targeting of the four factors to promoter regions that they do not normally bind in ES cells and conversely the failure to activate ES cell-specific genes results from the lack of binding of the factors in partially reprogrammed cells. Differential binding of the factors in partially reprogrammed cells is also reflected by a different spectrum of functional classifications for target genes in partially reprogrammed cells (Supp Fig 3).
It is critical to understand why the four factors target different genes in partially reprogrammed cells than in ES cells. We found that promoter regions bound uniquely in partially reprogrammed cells contain the known DNA binding motif for the occupying factor suggesting that targeting to these genes in partially reprogrammed cells is not simply due to non-specific or indirect binding but instead may be guided by direct interactions with their respective DNA binding motif (Fig 2C). Given that the expression of the four factors is three to eight-fold higher in partially reprogrammed cells than ES or iPS cells, these sites are perhaps low affinity binding sites. In contrast, promoter regions that are bound in ES cells but not in partially reprogrammed cells may need other proteins that are not expressed in partially reprogrammed cells to allow cooperative binding of the factors. One such candidate is the transcription factor Nanog, which co-localizes with Oct4 and Sox2 at a subset of promoter regions in ES cells (Boyer et al., 2005; Chen et al., 2008; Kim et al., 2008) and is not expressed in our partially reprogrammed cell lines (Supp Fig 5). Indeed, we found that genes that lack ES-like binding by the factors in partially reprogrammed cells are more often targets of Nanog in ES cells than genes where binding uniquely occurs in partially reprogrammed cells (Fig 2D). In particular, genes that are bound by three or four factors in ES cells and completely lack binding in partially reprogrammed cells are often Nanog targets in ES cells (Fig 2D). Thus, the absence of Nanog, and likely that of other transcription factors, could contribute to the lack of ES-specific binding in partially reprogrammed cells.
Oct4, Sox2 and Klf4 targeting is particularly impaired in partially reprogrammed cells
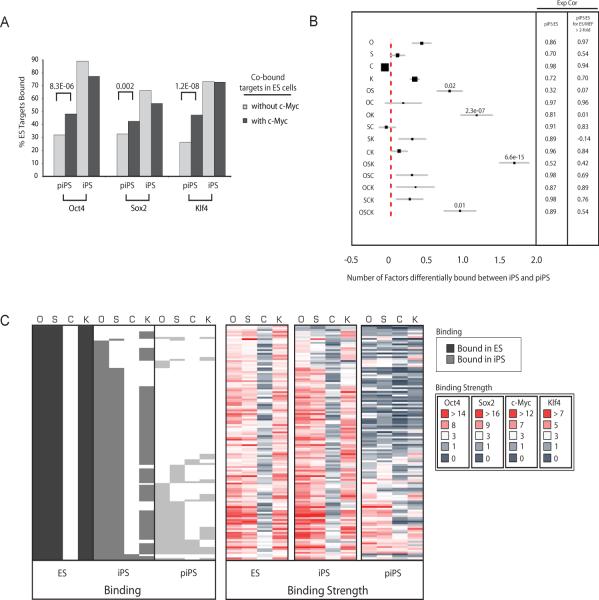
Partially reprogrammed cells have fewer bona-fide ES cell targets than iPS cells for Oct4 (38% of its ES cell targets are bound in partially reprogrammed cells vs 79% in iPS cells), Sox2 (35% vs 54%), and Klf4 (38% vs 67%), while c-Myc binds 67% in partially reprogrammed cells and 59% of its ES targets in iPS cells. Thus there is a widespread difference in binding between ES cells and partially reprogrammed cells that impinges more on Oct4, Sox2, and Klf4 than c-Myc. This observation led us to address whether c-Myc co-occupancy affects binding of the other three factors. We found that Oct4, Sox2 and Klf4 binding in partially reprogrammed cells is more similar to their binding in ES cells when c-Myc is co-bound at the promoter (Fig 3A). For example, 48% of Oct4 ES cell targets are bound in partially reprogrammed cells when the target was co-bound by c-Myc in ES cells, compared to only 32% when co-binding in ES cells occurred with Klf4 and/or Sox2. Thus, c-Myc association divides ES cell target genes of the four factors into two groups and appears to be a measure of their targeting efficiency in partially reprogrammed cells. The difference in Oct4, Sox2 and Klf4 binding based on c-Myc co-occupancy is not observed when ES and iPS cell binding is compared (Fig 3A).
Figure 3. OSK targets are most differentially bound between iPS cells and partially reprogrammed cells.
(A) Genes bound by two, three or four factors in ES cells were divided into those co-occupied by c-Myc and those lacking c-Myc. For both groups of genes, the bar plot presents the fraction of Oct4, Sox2 or Klf4 ES targets that are bound by the respective factor in partially reprogrammed cells (piPS) or iPS cells. Significant differences for binding in piPS, computed using the difference of proportions test, are indicated.
(B) The graph displays the average number of factors differentially bound between iPS and partially reprogrammed cells for target genes grouped according to their factor association in ES cells (given to the left), within a 95% confidence interval. Filled squares reflect the relative number of genes in each target group. Significance was determined using the binomial distribution. Pearson correlation coefficients (Exp Cor) for expression between ES and partially reprogrammed cells (piPS) were computed for genes in each group and for those genes in a given group that are expressed at least two-fold higher in ES cells compared to MEFs.
(C) Binding calls and binding strength heatmaps of Oct4, Sox2, c-Myc and Klf4 in ES cells, iPS cells and partially reprogrammed cells (piPS, lightest grey) for OSK ES targets as described in Fig 1D.
To further analyze these binding differences, we measured the average number of factors differentially bound between iPS cells and partially reprogrammed cells for genes in each ES cell binding group (Fig 3B). The most significant differences in binding between iPS and partially reprogrammed cells exist in those genes that are co-occupied by Oct4, Sox2 and Klf4 (OSK), followed by genes associated with both Oct4 and Klf4 (OK). Genes bound by all four factors (OSCK) or Oct4 and Sox2 (OS) just pass the significance threshold. In general, genes in these groups also have the lowest correlation in expression between pluripotent and partially reprogrammed cells (Fig 3B). Genes bound by OSK are characterized by high binding strength in ES and iPS cells and largely unbound in partially reprogrammed cells (Fig 3C). Indeed, with only 24 OSK target genes in partially reprogrammed cells compared to 193 genes in iPS cells and 129 in ES cells, co-occupancy by OSK hardly exists in partially reprogrammed cells. These data show that there is widespread differential binding between iPS/ES cells and partially reprogrammed cells that most dramatically occurs in genes occupied by OSK. This finding was confirmed when differential binding between ES/iPS and partially reprogrammed cells was analyzed for genes in each iPS-specific binding group (data not shown).
Oct4, Sox2 and Klf4 co-occupancy is associated with strong transcriptional activation
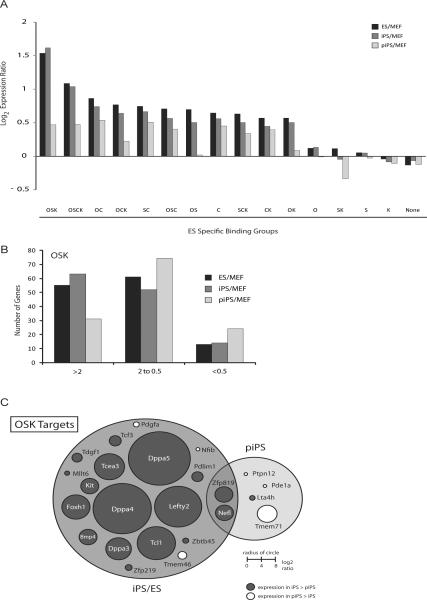
Genes co-occupied by OSK in ES cells, on average, have the highest increase in expression between iPS cells and fibroblasts, i.e. upon reprogramming (Fig 4A). As expected, these genes are not upregulated to the ES/iPS cell level in partially reprogrammed cells. In particular, OSK targets that are most activated in the transition of fibroblasts to iPS cells are the ones that fail to be induced (Fig 4B). These genes are not bound in partially reprogrammed cells and include highly expressed regulators of the pluripotent state (for example Tcl1, Dppa3, Dppa4, Dppa5) and embryonic development (for example Foxh1, Lefty2) (Fig 4C).
Figure 4. OSK targets are most differentially expressed between ES/iPS cells and partially reprogrammed cells.
(A) Bar chart depicting the mean of the log2 expression ratio between the ES (black), iPS (dark grey) or partially reprogrammed cells (piPS, light grey) and fibroblasts (MEF) for genes bound by the indicated combination of factors in ES cells or for all unbound genes (None).
(B) The graph divides OSK targets of ES cells according to their expression ratio between indicated cell types.
(C) Venn diagram depicting genes bound by OSK in iPS and ES cells but not in partially reprogrammed cells (piPS) (left), genes bound in partially reprogrammed cells but not in ES and iPS cells (right), and genes bound by OSK in all three cell types (overlapping area). Circles are scaled and colored according to the expression difference between iPS cells and piPS.
The expression analysis also revealed that the four factors appear to have a larger impact on the activation of gene expression during the reprogramming process because co-bound genes are usually more highly expressed in iPS and ES cells than in fibroblasts (Fig 4A). These findings extend across the three cell types analyzed here (data not shown), indicating that an intrinsic property of reprogramming factor co-binding is to activate genes, irrespective of the cell type. Consistent with published observations (Chen et al., 2008; Kim et al., 2008), genes occupied by c-Myc are activated more strongly than those bound singly by Oct4, Sox2 or Klf4.
Binding is incompatible with low histone H3K4 trimethylation
We reasoned that the chromatin state of promoter regions could influence the binding properties of the factors during reprogramming. In general, transcribed genes are marked by the “activating” histone H3 lysine 4 trimethylation (H3K4me3), while repressed genes, especially those encoding developmental regulators, are associated with histone H3 lysine 27 trimethylation (H3K27me3). In repressed genes, H3K27me3 is often found in combination with H3K4me3 forming bivalent chromatin domains (Bernstein et al., 2006). Our previous analysis demonstrated that these histone methylation marks are reset to the ES cell pattern in iPS cells (Maherali et al., 2007). To test the relationship between promoter histone methylation and binding of the four factors, we analyzed H3K4me3 and H3K27me3 patterns in fibroblasts, partially reprogrammed cells, iPS and ES cells for all promoter regions bound by the four factors.
We found that the differential binding of the four factors between partially reprogrammed cells and ES/iPS cells cannot generally be explained by differences in K4 and K27 methylation patterns between the pluripotent cells, partially reprogrammed cells or fibroblasts. Specifically, the proportion of genes enriched for K27 methylation in ES cells and partially reprogrammed cells is very similar for ES cells targets (Fig 5A) indicating that the presence of H3K27me3 per se does not interfere with binding of the factors. Similarly, the majority of ES cell targets of the four factors is methylated at K4 in ES cells, a pattern that is also observed in fibroblasts (Supp Fig 8) and partially reprogrammed cell (data not shown). We noted that c-Myc, alone or with other factors, mainly associates with genes that have less repressive chromatin given that they are methylated at K4 and lack K27 methylation in all cell types including fibroblasts (Supp Fig 8, Fig 5A). This result potentially explains why c-Myc binds in a more ES like-pattern in partially reprogrammed cells.
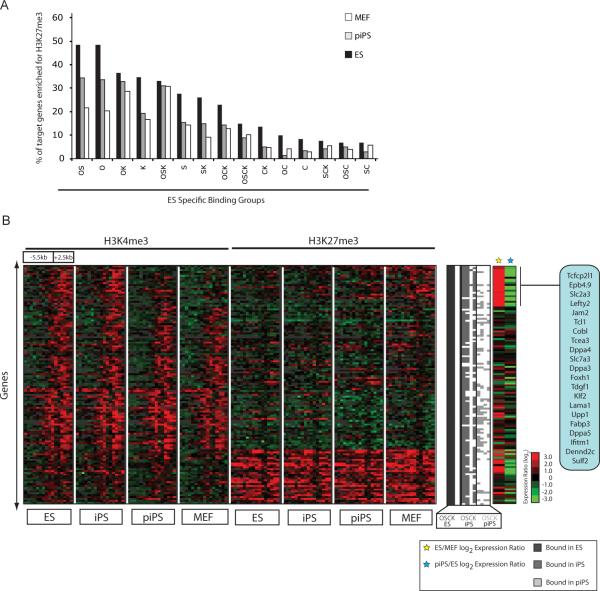
Figure 5. A distinctive histone methylation pattern for OSK target genes that are most dramatically upregulated in ES/iPS cells compared to fibroblasts.
(A) Bar chart depicts the fraction of genes with enrichment of histone H3 K27 trimethylation in their promoter region in different cell types, for genes that are bound by the indicated combination of factors in ES cells.
(B) Hierarchical clustering of histone H3 K4 and K27 trimethylation patterns in ES cell, iPS cells, partially reprogrammed cells (piPS) and fibroblasts for genes bound by OSK in ES cells. Each row represents the methylation pattern along the −5.5kb to +2.5kb promoter region relative to the transcription start site (TSS), reiterated eight times to present the data for each cell type and methylation mark. Each 8 kb promoter region is divided into sixteen 500bp fragments that display the average log ratio of probe signal intensity with green, red, and grey representing lower-than-average, higher-than-average, and missing values for enrichment due to lack of probes in those regions, respectively. Binding of the factors in ES (dark grey), iPS (light grey), and partially reprogrammed cells (lightest grey) is presented to the right of the histone methylation data. Two sets of log2 expression ratios are appended (ES/MEF and piPS/ES) and genes most highly expressed in ES cells relative to MEF are marked and listed.
When specifically analyzing the methylation pattern in OSK cell ES targets (Fig 5B), we found a distinct histone methylation signature for genes that are most dramatically upregulated between ES/iPS cells and fibroblasts. Characterized by high H3K27me3 and low H3K4me3 enrichment in fibroblasts, these genes are strongly positive for H3K4me3 and have lost H3K27me3 in ES/iPS cells in accordance with their high transcriptional activation (Fig 5B, highlighted genes). In partially reprogrammed cells, this set of genes exhibits an intermediate pattern of histone methylation and has a severe lack of binding of the three factors (Fig 5B). A similar pattern of histone methylation change is also evident in a subset of OK, OS and OSCK bound target genes that exhibit a strong induction of expression during reprogramming (data not shown and Supp Fig 8). Together, these data indicate that highly expressed targets of the factors, which encode many important regulators of pluripotency and embryonic development (Fig 5B), are characterized by a specific histone H3K4/K27 methylation pattern that may be related to the lack of binding by the factors in partially reprogrammed cells.
c-Myc is a major contributor to initial reprogramming events
Our analysis of binding and expression in partially reprogrammed cells suggested that c-Myc does not greatly contribute to the activation of pluripotency regulators. Next, we wanted to directly test the contribution of each factor to the early phase of the reprogramming process and followed the levels of the somatic surface marker Thy1 upon retroviral expression of a single factor in fibroblasts. Within two days of infection, the population of Thy1-positive fibroblasts decreased most dramatically in cells expressing only c-Myc and by day four, more than half of those cells had lost the Thy1 marker (Fig 6A). This strong effect was not seen in fibroblasts infected with Klf4, Oct4 or Sox2 (Fig 6A) although the infection efficiency of each virus was approximately the same (data not shown). When the four factors were added to fibroblasts in combinations, Thy1 downregulation was most affected when c-Myc and Klf4 were included in the mixture (Supp Fig 9A). Collectively these results demonstrate that c-Myc mediates downregulation of the fibroblast marker most efficiently among the four factors.
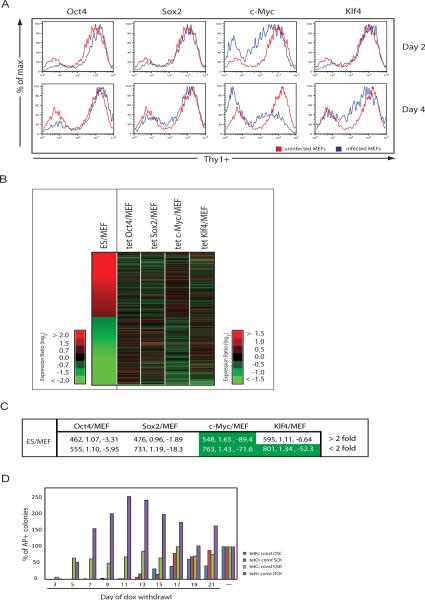
Figure 6. The role of c-Myc in early steps of the reprogramming process.
(A) FACS plots showing Thy1 surface levels of fibroblasts two or four days post infection with a retrovirus carrying the indicated factor (blue) in comparison to uninfected fibroblasts (red).
(B) Heatmap of expression differences between ES cells and fibroblasts (MEFs) and between fibroblast lines induced for c-Myc, Klf4, Oct4 or Sox2 expression for three days with doxycycline relative to an uninduced fibroblast control, for all genes more than 2-fold differentially expressed between ES cells and MEFs, ordered by decreasing expression ratio.
(C) Overlap of genes 2-fold up or down between ES/MEF and 2-fold up or down between MEFs induced to express Oct4, Sox2, Klf4 or c-Myc and uninduced MEFs. The first number presents the genes in the overlap, the second number the enrichment (described in Materials and Methods), and the third number log10 of the p-value. Significant enrichments are highlighted in green.
(D) R26-rtTA/wt fibroblasts were infected with three constitutively expressed retroviruses (const) and a dox-inducible retrovirus for the fourth factor (tet). Doxycycline (dox) was added until the indicated day and AP-positive colonies scored on day 25.
To determine if this observation applies genome-wide, we investigated global expression changes that occur in fibroblasts upon individual expression of c-Myc, Klf4, Sox2 or Oct4. We first established an inducible expression system that allowed us to start with a homogenous population of fibroblasts expressing one factor. The factors, under the control of a doxycycline- inducible promoter, were cloned into a retroviral vector that included a hygromycin resistance gene. Fibroblasts heterozygous for the reverse tetracycline transactivator in the constitutively active Rosa26 locus (R26 rtTA/wt) were infected with individual inducible viruses, selected for hygromycin resistance, and three days after addition of doxycycline profiled for expression, along with an uninfected control. Addition of doxycycline induced expression of the factors in almost 100% of the cells (data not shown). In agreement with the Thy1 result, c-Myc-induced expression changes resembled those found between ES cells and fibroblasts most closely when compared to the effects of the other three factors (Fig. 6B), both for genes that become up and downregulated between ES cells and fibroblasts (Fig. 6C). Furthermore, the expression changes upon ectopic c-Myc expression are most similar to those occurring in fibroblasts overexpressing all four factors, particularly for those genes that become repressed (Supp Fig 9B). The genes downregulated in fibroblasts upon c-Myc expression include collagens and are involved in signaling and organ development. Together with the analysis of factor binding and expression in the partially and completely reprogrammed state, these results indicate that c-Myc enhances early steps of reprogramming by repressing fibroblast-specific expression and upregulating the metabolic program of the embryonic state.
Previously it was shown that reprogramming only occurs when all four factors are expressed for at least 8–10 days (Brambrink et al., 2008; Stadtfeld et al., 2008), but the temporal requirement for each individual factor remained untested. Based on our data, we wanted to test whether ectopical c-Myc expression is required for a shorter time period during reprogramming than that of Oct4, Sox2 and Klf4. R26 rtTA/wt fibroblasts were infected with combinations of retroviruses that allowed the doxycycline-inducible expression of only one factor and constitutive expression of the other three. Doxycycline was added 24 hours post infection to induce expression of the “fourth” reprogramming factor and subsequently withdrawn at different time points, leading to efficient downregulation of induced transcripts within 24 hours as determined by real time PCR (data not shown). AP-positive colonies were scored 25 days post infection as a measure of reprogramming (Fig 6D). Fibroblasts induced to express c-Myc for five days gave rise to AP-positive colonies that only modestly increased in numbers when c-Myc expression was maintained for longer periods. In contrast, induction of c-Myc for only three days resulted in dramatically fewer colonies, similar to reprogramming experiments in which cMyc is completely omitted (Nakagawa et al., 2008; Wernig et al., 2008; data not shown), suggesting that a five day pulse of ectopic c-Myc expression is sufficient to obtain c-Myc- dependent reprogramming. In contrast, exogenous expression of Oct4 and Klf4 was required for at least 12 days. Surprisingly, ectopic Sox2 expression for only five days was enough to give rise to AP-positive colonies, although unlike for c-Myc, these colonies were more heterogeneous in morphology and greatly increased in number with prolonged exogenous expression, a result that deserves future investigation. Taken together, these data indicate that the four factors differ in their contribution to the reprogramming process and suggest that ectopic c-Myc is only required initially to attain a high reprogramming efficiency.
Discussion
This study aimed to uncover the contribution of the reprogramming factors to the induction of pluripotency. During the reprogramming process, fibroblast markers are repressed and early embryonic markers like SSEA1 activated before expression of pluripotency regulators and the self-sustaining pluripotent state are attained (Brambrink et al., 2008; Stadtfeld et al., 2008). In agreement with these observations, fibroblast-specific genes are efficiently silenced in partially reprogrammed cells, while the embryonic program is not fully induced. Clones obtained in fibroblast reprogramming experiments with ectopic expression of only Oct4, Klf4 and c-Myc are also characterized by repression of fibroblast-specific genes although not to the same extent as in our four factor induced partially reprogrammed cell lines (Supp Fig 10). Thus, silencing of the somatic cell expression program appears to be an important initial step required for the induction of the ES-like expression program. Our data indicate a major contribution of c-Myc to this first step. We found that c-Myc promotes the most ES-like expression changes, including the repression of fibroblast-specific genes, of the four factors (Fig 7A;i). Mechanisms by which c-Myc induces transcriptional repression are much less understood than its function as a transcriptional activator (Wanzel et al., 2003). Global repressive effects of c-Myc could be mediated through its interaction with the transcription factor Miz (Wu et al., 2003) or direct binding and activation of a transcriptional repressor. Interestingly, when murine fibroblasts were treated for seven days with valporic acid (VPA), a histone deacetylase inhibitor that can replace c-Myc function during reprogramming, their expression started to resemble an ES-like state, including the repression of highly transcribed fibroblast-specific genes (Huangfu et al., 2008). Thus, c-Myc expression or VPA treatment may lay the framework for the efficient repression of the somatic expression and induction of the ES cell expression program.
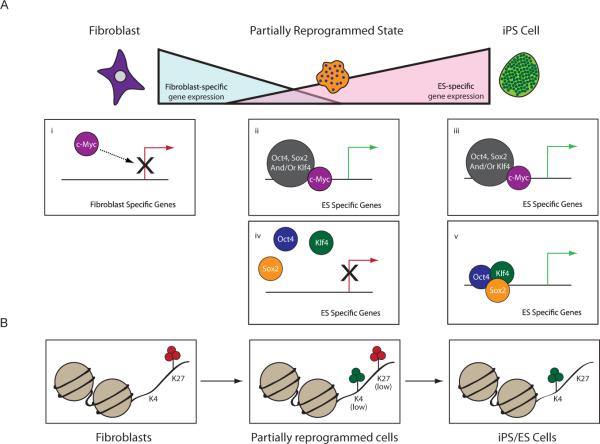
Figure 7. Role of the four factors during reprogramming.
(A) c-Myc is a major contributor to the down-regulation of fibroblast-specific genes at the beginning of the reprogramming process (i). In iPS cells, c-Myc binds and activates many ES-cell specific genes with roles in metabolic regulation, often with the other three factors (iii). For many of these target genes, binding and activation is also found in partially reprogrammed cells (ii). Target genes that are co-bound by Oct4, Sox2 and Klf4 in ES/iPS cells encode some of the most highly expressed regulators of pluripotency (v) and are not bound and activated at the partially reprogrammed state (iv). Our data suggest that the targeting of OSK to these genes represents a barrier to the reprogramming process and that c-Myc-bound genes largely become activated during earlier steps.
(B) State of histone H3K4 and K27 trimethylation within promoter regions of genes that become most highly activated during reprogramming and that are co-bound by Oct4, Sox2 and Klf4 in ES/iPS cells but not partially reprogrammed cells.
The comparison of binding patterns of the factors in iPS/ES cells and partially reprogrammed cell lines further strengthens the conclusion that the contribution of c-Myc to the reprogramming process is separable from that of the other three factors. The metabolism-related embryonic expression program is induced by binding of combinations of all four factors (Fig 7A; ii and iii), while the activation of many regulators of the pluripotent state is dependent on co-binding of only Oct4, Klf4, and Sox2 (Fig 7A;v), which does not occur in partially reprogrammed cells (Fig 7A;iv). Indeed, ectopic expression of c-Myc is only required for the first few days of the reprogramming process. We propose that lack of Oct4, Sox2 and Klf4 co-binding in partially reprogrammed cells contributes to the failure to establish the pluripotent state. During reprogramming, Oct4, Sox2 and Klf4 can likely only stochastically overcome this binding block contributing to the low efficiency of this conversion.
An important question is why Oct4, Sox2 and Klf4 cannot associate with their ES cell target genes in partially reprogrammed cells. One explanation could be the absence of other factors, such as Nanog, that allow targeting of the reprogramming factors. These factors could form large complexes necessary for cooperative binding to target genes or could induce a conformational change of DNA allowing subsequent binding by Oct4, Sox2 and Klf4. Alternatively, the target promoters may have repressive chromatin structures that prevent appropriate binding of the factors. Indeed, our data demonstrate that a gain in histone H3K4 methylation is specifically associated with the activation of many pluripotency regulators (Fig 7B). In partially reprogrammed cells, the methylation status is not in an ES-like pattern and the reprogramming factors do not bind appropriately suggesting that the establishment of the ES-like histone methylation pattern is either a requirement for or coincides with the recruitment of the factors. Elucidating how the methylation pattern transitions to an ES-like state and how Oct4, Sox2 and Klf4 are recruited to genes encoding pluripotency regulators should allow for the development of more efficient reprogramming strategies.
Interestingly, c-Myc is dispensable for fibroblast reprogramming as both human and murine iPS cells can be obtained in the absence of c-Myc, albeit with dramatically reduced efficiency and kinetics (Nakagawa et al., 2008; Wernig et al., 2008). The fact that activation of transcriptional regulators of pluripotency is largely independent of c-Myc binding provides an explanation for why c-Myc is not absolutely required for reprogramming. Our data indicate that reprogramming is delayed and inefficient in the absence of ectopic c-Myc expression because c-Myc greatly enhances the initial steps of reprogramming. Similarly, the fact that Oct4, Sox2 and Klf4 are targeted in a more ES-like pattern in partially reprogrammed cells to genes that are co-bound by c-Myc suggests that its presence facilitates the binding of the other factors. In addition, c-Myc normally fulfills many other functions, for example in regulating DNA replication (Dominguez-Sola et al., 2007) and global histone acetylation (Knoepfler et al., 2006) which may facilitate the reprogramming process more indirectly.
Once reprogramming is complete the binding pattern of the four factors is similar to that of ES cells suggesting that a common binding signature has to be achieved to attain pluripotent gene expression. Differences in binding between these ES and iPS cells occur generally in genes that are bound more weakly. In the future, it will be important to determine whether any of these binding differences reflect an epigenetic memory of the starting cell population that may influence differentiation behavior.
Experimental Procedures
Cell culture and reprogramming experiments
ES (V6.5 and E14), iPS (1D4 and 2D4), partially reprogrammed cells (1A2 and 1B3) and three primary mouse embryonic fibroblast (MEF) lines derived from d14.5 embryos were cultured according to standard methods. Reprogrammed cell lines were grown in the presence of 1ug/ml puromycin. For new reprogramming experiments, the cDNAs of the four factors were cloned into the pMX (constitutive expression) and pRetroX-Tight-Hyg (Clontech, dox-inducible) retroviral vectors, which were individually transfected into PlatE packaging cells using Fugene (Roche). Viral supernatants harvested 48 hours post infection were used to infect MEFs in the presence of 10ug/ml polybrene. Where indicated, MEFs heterozygous for R26 rtTA were used and cultures supplemented with 2ug/ml dox. Dox-containing media was changed every three days until withdrawal. To obtain expression data upon induction of only one single factor, R26 rtTA MEFs were infected with the respective pRetro-virus and selected for 1 week in 100ug/ml hygromycin before dox-induction.
Flow Cytometry
Thy1-positive fibroblasts were sorted on a FACS ARIA (BD Biosciences) and subsequently infected. At the indicated time point, cells were harvested, passed through a 40um cell strainer, incubated with PE-conjugated rat anti-Thy1 antibody (eBiosciences), and analyzed on a LSR cytometer (BD Biosciences) using the FloJo software (TreeStar).
ChIP on Chip
Chromatin fragments associated with the respective transcription factor or histone mark were immunoprecipitated with specific antibodies (Oct4 (sc5279) and c-Myc (sc764) both SantaCruz, Sox2 (Chemicon 5603), Klf4 (R&D Biosystems AF3158), Nanog (Abcam 21603), H3K4 trimethylation (Abcam 8580) and K27 trimethylation (Upstate 07-449) and hybridized on an Agilent promoter microarray (G4490) as described previously (Maherali et al., 2007). Probe signals were extracted and normalized using Agilent's Feature Extraction and ChIP Analytics software and binding calls made following a heuristic described previously (Boyer et al., 2005). Briefly, the algorithm takes the p-values of two signal measures (normalized log ratio of the signal intensity), X and X□, as input. X is the signal measure of an individual probe and X□ the average signal measure of a probe and its closest upstream and downstream neighbors within 1 kb. Replicate samples were combined by taking the geometric mean of the X and X□ p-values for each probe. A probe was called bound if the p-value of X□ is < .001 and one of the following conditions is met: the p-value of X < .001 and the p-value of either neighboring X is < 0.1 OR the p-value of at least two of three probes (X and its two neighbors) is < .005. A gene was called bound if any of its probes were called bound. The binding strength of a gene is defined as –log10p-value(X□) of its most significantly bound probe. For histone methylation data average probe signals were extracted in 500bp windows as described (Maherali et al., 2007).
Expression Data
RNA was extracted from duplicate samples of V6.5 and E14 ES cells, 1D4 and 2D4 iPS cells, 1A2 and a single sample of 1B3 partially reprogrammed cells, four samples of three different MEF lines, fibroblasts induced to express a single factor and the un-induced control, and analyzed on Affymetrix GeneChip Mouse Genome 430 2.0 arrays at the UCLA microarray core facilities. Quantile normalization was performed using the Affymetrix package (affy) (Bioconductor). To convert probe data into gene expression data, probes ending in “_at” and “_a_at” were averaged for each gene. All gene expression data are provided in Supp Table 4. Analysis combining averaged expression data with binding data was conducted on genes with expression > 200 in ES, iPS, piPS or MEF cells (provided in Supp Table 2). Cluster 3.0 was used for hierarchical clustering and Java Treeview for visualization.
Computational methods
The expected number of binding events (Fig 1B) was computed assuming that binding by each factor is independent of that of the others. Individual binding probabilities were estimated from ChIP-chip data. Specific binding events are then the product of the probabilities (for example p(OSK)=p(Oct4) × p(Sox2) × p(Klf4) × (1-p(c-Myc)). The probability of binding by a specific number of factors is the sum of the probabilities satisfying that condition and the expected number for that condition is this probability multiplied by the number of genes on the array. For Fig 3B, the hamming distance, h, of a gene was defined as the number of mismatches in Oct4, Sox2, c-Myc and Klf4 binding between ES and iPS cells or ES cells and partially reprogrammed cells, ranging from 0 for perfect match in binding to 4 for complete mismatch. The number of factors differentially bound between iPS and partially reprogrammed cells is the average of (h(iPS, ES)-h(piPS,ES)). Significance was determined using the binomial distribution with k equal to the number of times h(piPS,ES) was greater than h(iPS,ES) in a given ES binding cluster, (i.e. p-value = P(X>=k), where X~Bin(n,0.5)). Motif scanning was done using methods developed by Zhou et al (Zhou et al., 2007; see Supplement). Enrichment determined in Fig. 6C is the conditional probability of a gene being 2-fold upregulated in the dox-induced MEF/uninduced MEF given that a gene is 2-fold up in ES/MEF divided by the marginal probability of being 2-fold up in dox-induced MEF/uninduced MEF. Hypergeometric distribution was used to determine significance.
Supplementary Material
Acknowledgements
We thank Stephen Smale, Siavash Kurdistani, Bernadett Papp, Lars Dreier, Mark Chin, Bill Lowry, and Konrad Hochedlinger for critical reading of the manuscript. RS is supported by a training grant from the California Institute of Regenerative Medicine (CIRM), QZ by a NSF grant (DMS-0805491), and KP by the V and Kimmel Scholar Foundations, the NIH Director's Young Innovator Award and a CIRM Young Investigator Award.
Footnotes
Supplemental Data “Supplemental Text and Figures” include a detailed description of the motif scanning method and 10 figures. There are 7 supplemental tables as well as a summary of the tables.
Data access Data are available at GEO under GSE14012.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. Embo J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Wanzel M, Herold S, Eilers M. Transcriptional repression by Myc. Trends Cell Biol. 2003;13:146–150. doi: 10.1016/s0962-8924(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:16438–16443. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.