Abstract
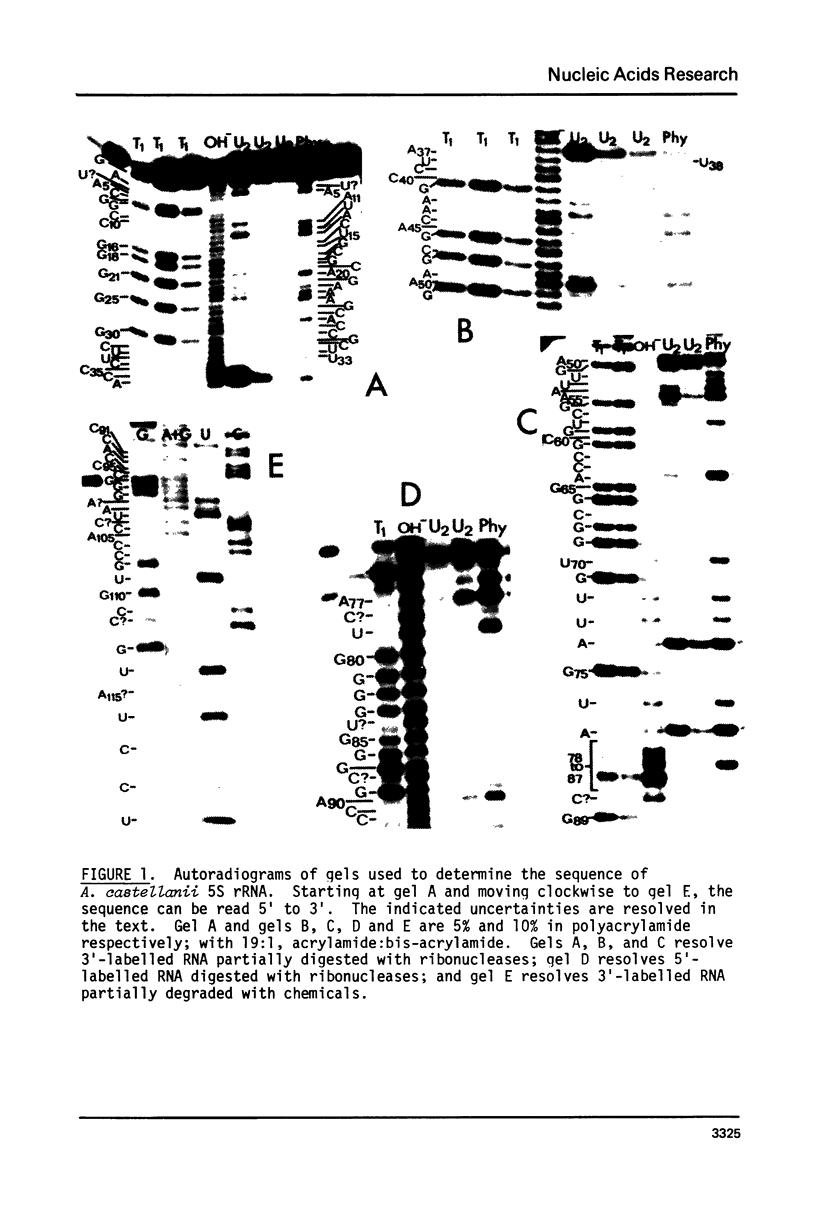
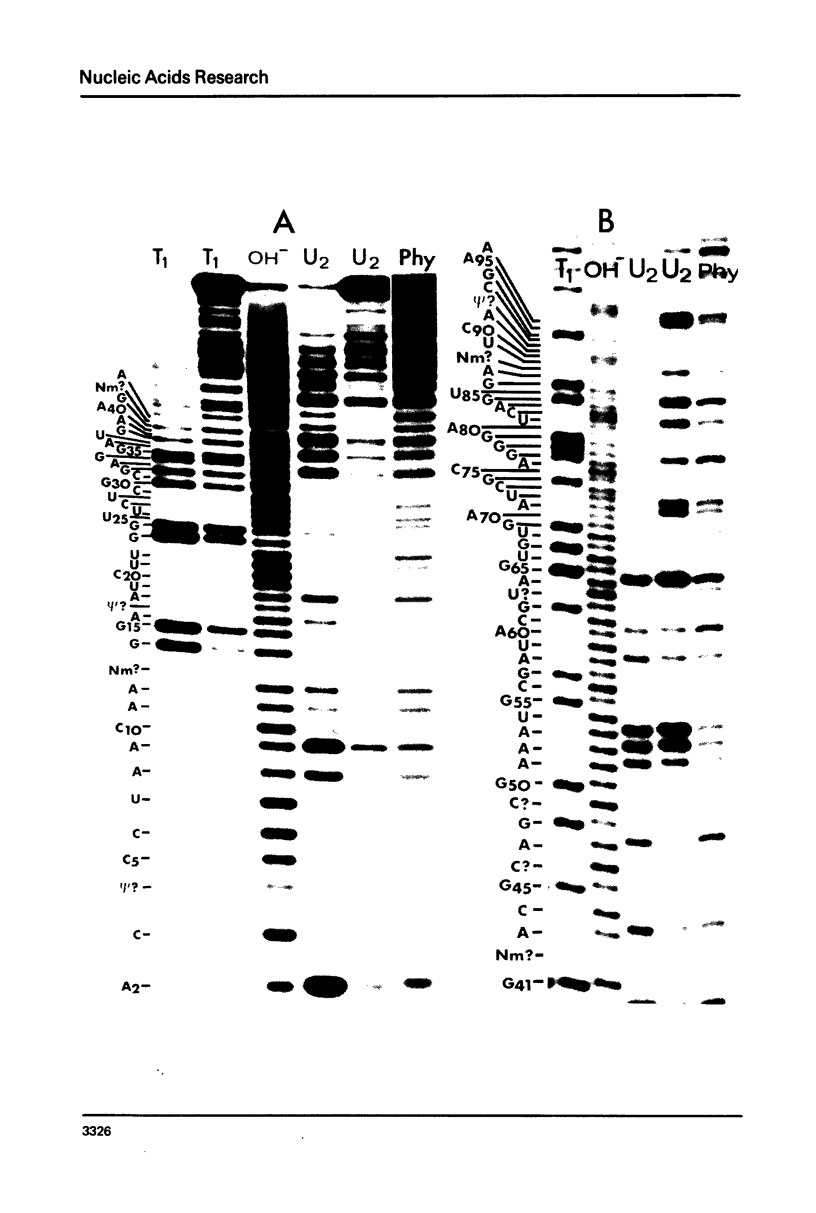
Sequences of 5S and 5.8S rRNAs of the amoeboid protist Acanthamoeba castellanii have been determined by gel sequencing of terminally-labeled RNAs which were partially degraded with chemical reagents or ribonucleases. The sequence of the 5S rRNA is (formula, see text). This sequence is compared to eukaryotic 5S rRNA sequences previously published and fitted to a secondary structure model which incorporates features of several previously proposed models. All reported eukaryotic 5S rRNAs fit this model. The sequence of the 5.8S rRNA is (formula, see text). This sequence does not fit parts of existing secondary structure models for 5.8S rRNA, and we question the significance of such models.
Full text
PDF



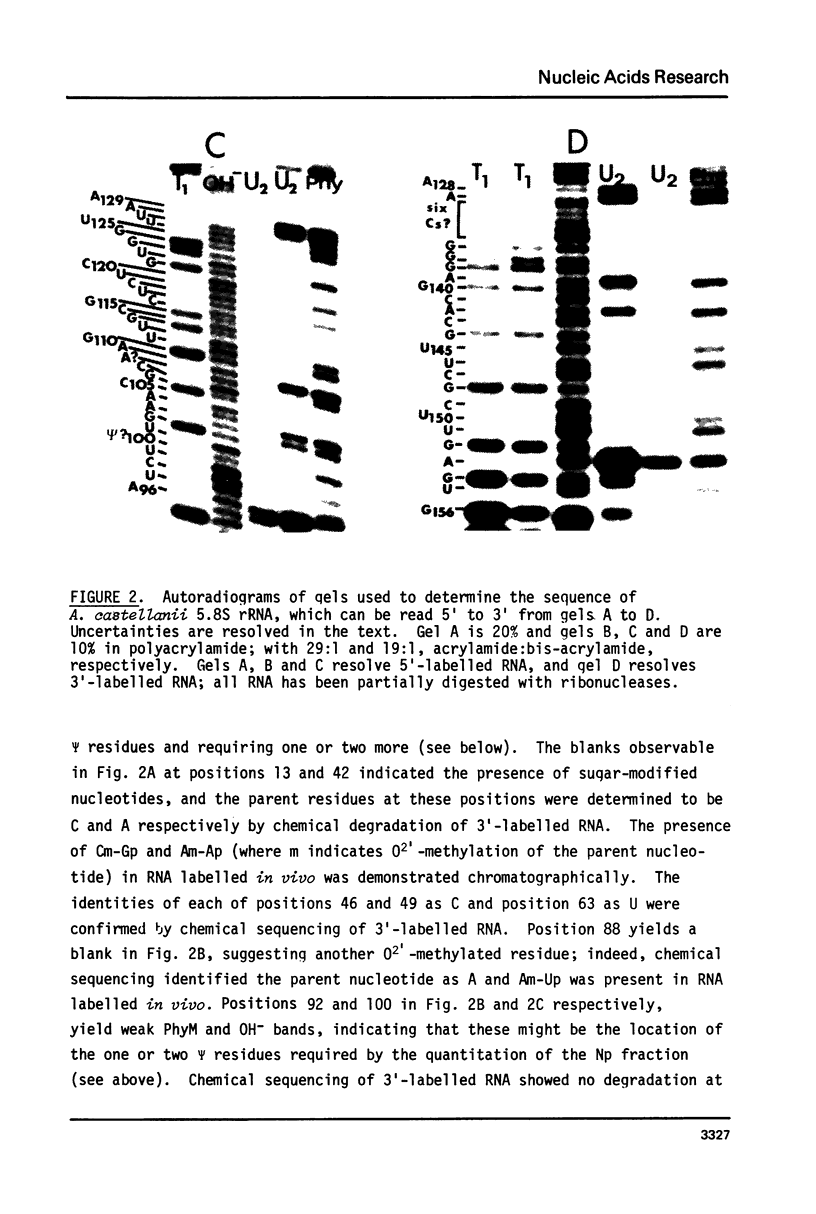

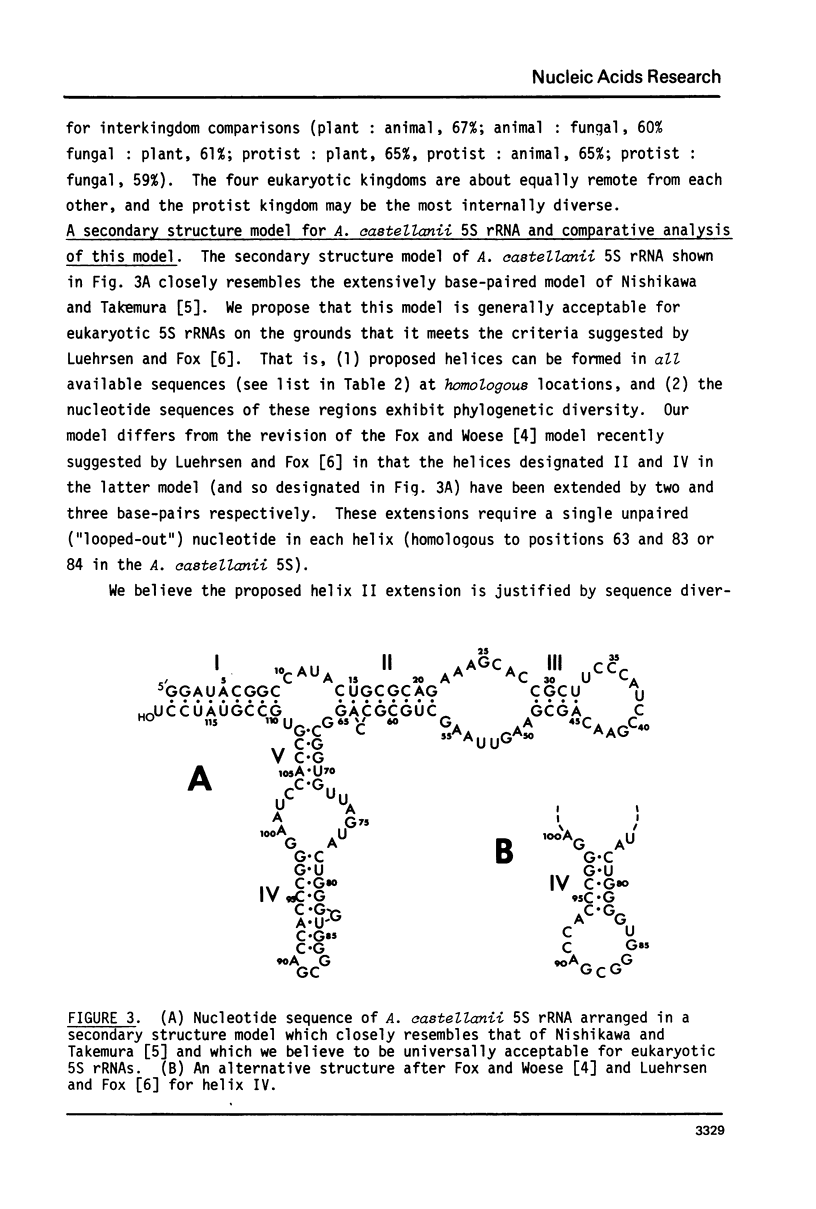


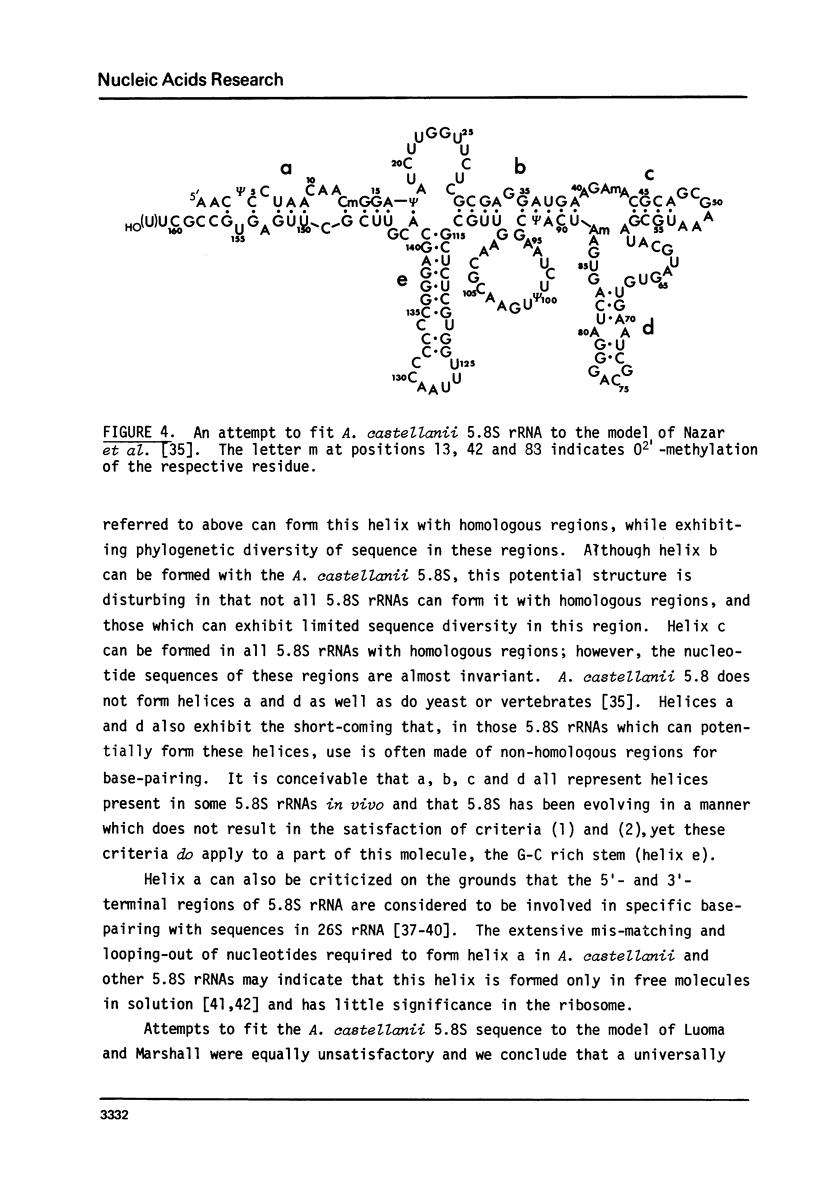


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benhamou J., Jordan B. R. Nucleotide sequence of Drosophila melanogaster 5S RNA: evidence for a general 5S RNA model. FEBS Lett. 1976 Feb 15;62(2):146–149. doi: 10.1016/0014-5793(76)80039-7. [DOI] [PubMed] [Google Scholar]
- Darlix J. L., Rochaix J. D. Nucleotide sequence and structure of cytoplasmic 5S RNA and 5.8S RNA of Chlamydomonas reinhardii. Nucleic Acids Res. 1981 Mar 25;9(6):1291–1299. doi: 10.1093/nar/9.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A. Collection of published 5S and 5.8S RNA sequences and their precursors. Nucleic Acids Res. 1981 Jan 10;9(1):r25–r42. doi: 10.1093/nar/9.1.213-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget B. G., Weissman S. M. Nucleotide sequence of KB cell 5S RNA. Science. 1967 Dec 29;158(3809):1695–1699. doi: 10.1126/science.158.3809.1695. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Gray M. W. The ribosomal RNA of the trypanosomatid protozoan Crithidia fasciculata: physical characteristics and methylated sequences. Can J Biochem. 1979 Jun;57(6):914–926. doi: 10.1139/o79-111. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S., Iwabuchi M. The nucleotide sequence of 5S rRNA from a cellular slime mold Dictyostelium discoideum. Nucleic Acids Res. 1980 Dec 11;8(23):5535–5539. doi: 10.1093/nar/8.23.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. R., Latil-Damotte M., Jourdan R. Coding and spacer sequences in the 5.8S-2S region of Sciara coprophila ribosomal DNA. Nucleic Acids Res. 1980 Aug 25;8(16):3565–3573. doi: 10.1093/nar/8.16.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. M., Cox R. A. The nucleotide sequence at the 3'-end of Neurospora crassa 25S-rRNA and the location of a 5.8S-rRNA binding site. Nucleic Acids Res. 1981 Mar 11;9(5):1111–1121. doi: 10.1093/nar/9.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. M., Maden B. E. Chemical modification studies and the secondary structure of HeLa cell 5.8S rRNA. Nucleic Acids Res. 1980 Oct 10;8(19):4521–4534. doi: 10.1093/nar/8.19.4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Conformation of mammalian 5.8 S ribosomal RNA: S1 nuclease as a probe. FEBS Lett. 1976 Dec 15;72(1):105–110. doi: 10.1016/0014-5793(76)80823-x. [DOI] [PubMed] [Google Scholar]
- Komiya H., Kawakami M., Shimizu N., Takemura S. Nucleotide sequences and evolutional aspect of 5S ribosomal RNAs from Lingula and silkworm. Nucleic Acids Symp Ser. 1980;(8):s119–s122. [PubMed] [Google Scholar]
- Komiya H., Shimizu N., Kawakami M., Takemura S. Nucleotide sequence of 5S ribosomal RNA from Lingula anatina. A study on the molecular evolution of 5S ribosomal RNA from a living fossil. J Biochem. 1980 Nov;88(5):1449–1456. doi: 10.1093/oxfordjournals.jbchem.a133114. [DOI] [PubMed] [Google Scholar]
- LANE B. G., DIEMER J., BLASHKO C. A. END GROUP AND SEDIMENTATION DATA ON FRAGMENTED HIGH MOLECULAR WEIGHT RIBONUCLEATES. Can J Biochem Physiol. 1963 Sep;41:1927–1941. [PubMed] [Google Scholar]
- Lu A. L., Steege D. A., Stafford D. W. Nucleotide sequence of a 5S ribosomal RNA gene in the sea urchin Lytechinus variegatus. Nucleic Acids Res. 1980 Apr 25;8(8):1839–1853. doi: 10.1093/nar/8.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K. R., Fox G. E. Secondary structure of eukaryotic cytoplasmic 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2150–2154. doi: 10.1073/pnas.78.4.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay R. M., Gray M. W., Doolittle W. F. Nucleotide sequence of Crithidia fasciculata cytosol 5S ribosomal ribonucleic acid. Nucleic Acids Res. 1980 Nov 11;8(21):4911–4917. doi: 10.1093/nar/8.21.4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay R. M., Spencer D. F., Doolittle W. F., Gray M. W. Nucleotide sequences of wheat-embryo cytosol 5-S and 5.8-S ribosomal ribonucleic acids. Eur J Biochem. 1980 Dec;112(3):561–576. doi: 10.1111/j.1432-1033.1980.tb06122.x. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O., Busch H. Structural analyses of mammalian ribosomal ribonucleic acid and its precursors. Nucleotide sequence of ribosomal 5.8 S ribonucleic acid. J Biol Chem. 1975 Nov 25;250(22):8591–8597. [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O. Role of the 5'-terminal sequence in the RNA binding site of yeast 5.8 S rRNA. FEBS Lett. 1980 Jun 16;115(1):71–76. doi: 10.1016/0014-5793(80)80729-0. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Nucleotide sequence of 5 S RNA from Torulopsis utilis. FEBS Lett. 1974 Mar 15;40(1):106–109. doi: 10.1016/0014-5793(74)80904-x. [DOI] [PubMed] [Google Scholar]
- Nishimura S., Harada F., Narushima U., Seno T. Purification of methionine-, valine-, phenylalanine- and tyrosine-specific tRNA from Escherichia coli. Biochim Biophys Acta. 1967 Jun 20;142(1):133–148. doi: 10.1016/0005-2787(67)90522-9. [DOI] [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Parish J. H., Kirby K. S. Reagents which reduce interactions between ribosomal RNA and rapidly labelled RNA from rat liver. Biochim Biophys Acta. 1966 Dec 21;129(3):554–562. doi: 10.1016/0005-2787(66)90070-0. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechulla B., Hahn U., McLaughlin L. W., Küntzel H. Nucleotide sequence of 5S ribosomal RNA from Aspergillus nidulans and Neurospora crassa. Nucleic Acids Res. 1981 Mar 25;9(6):1445–1450. doi: 10.1093/nar/9.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg H., Brambilla R., Keith G., Staehelin M. An improved method for the separation and quantitation of the modified nucleosides of transfer RNA. Nucleic Acids Res. 1976 Jan;3(1):285–295. doi: 10.1093/nar/3.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINGH H., LANE B. G. THE ALKALI-STABLE DINUCLEOTIDE SEQUENCES IN 18S+28S RIBONUCLEATES FROM WHEAT GERM. Can J Biochem. 1964 Jul;42:1011–1021. doi: 10.1139/o64-112. [DOI] [PubMed] [Google Scholar]
- Schwartz R. M., Dayhoff M. O. Origins of prokaryotes, eukaryotes, mitochondria, and chloroplasts. Science. 1978 Jan 27;199(4327):395–403. doi: 10.1126/science.202030. [DOI] [PubMed] [Google Scholar]
- Studnicka G. M., Eiserling F. A., Lake J. A. A unique secondary folding pattern for 5S RNA corresponds to the lowest energy homologous secondary structure in 17 different prokaryotes. Nucleic Acids Res. 1981 Apr 24;9(8):1885–1904. doi: 10.1093/nar/9.8.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Whittaker R. H. New concepts of kingdoms or organisms. Evolutionary relations are better represented by new classifications than by the traditional two kingdoms. Science. 1969 Jan 10;163(3863):150–160. doi: 10.1126/science.163.3863.150. [DOI] [PubMed] [Google Scholar]