Abstract
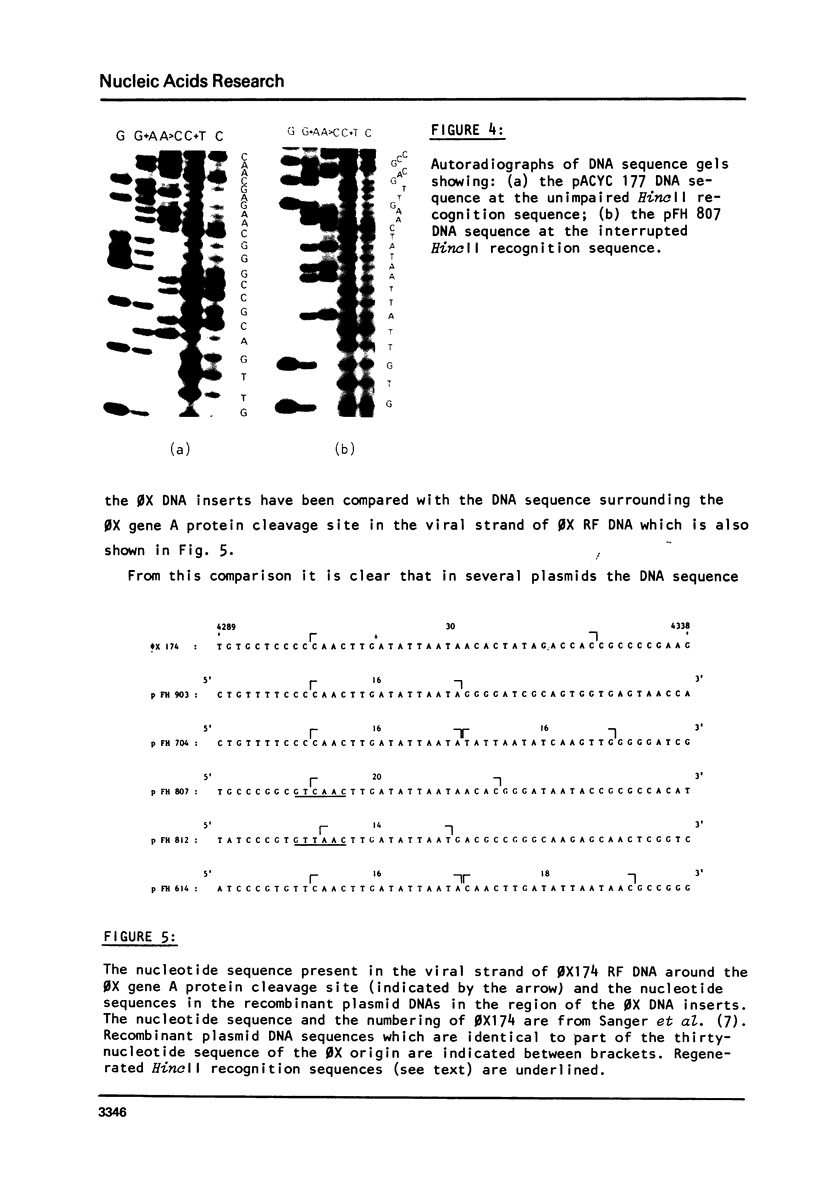
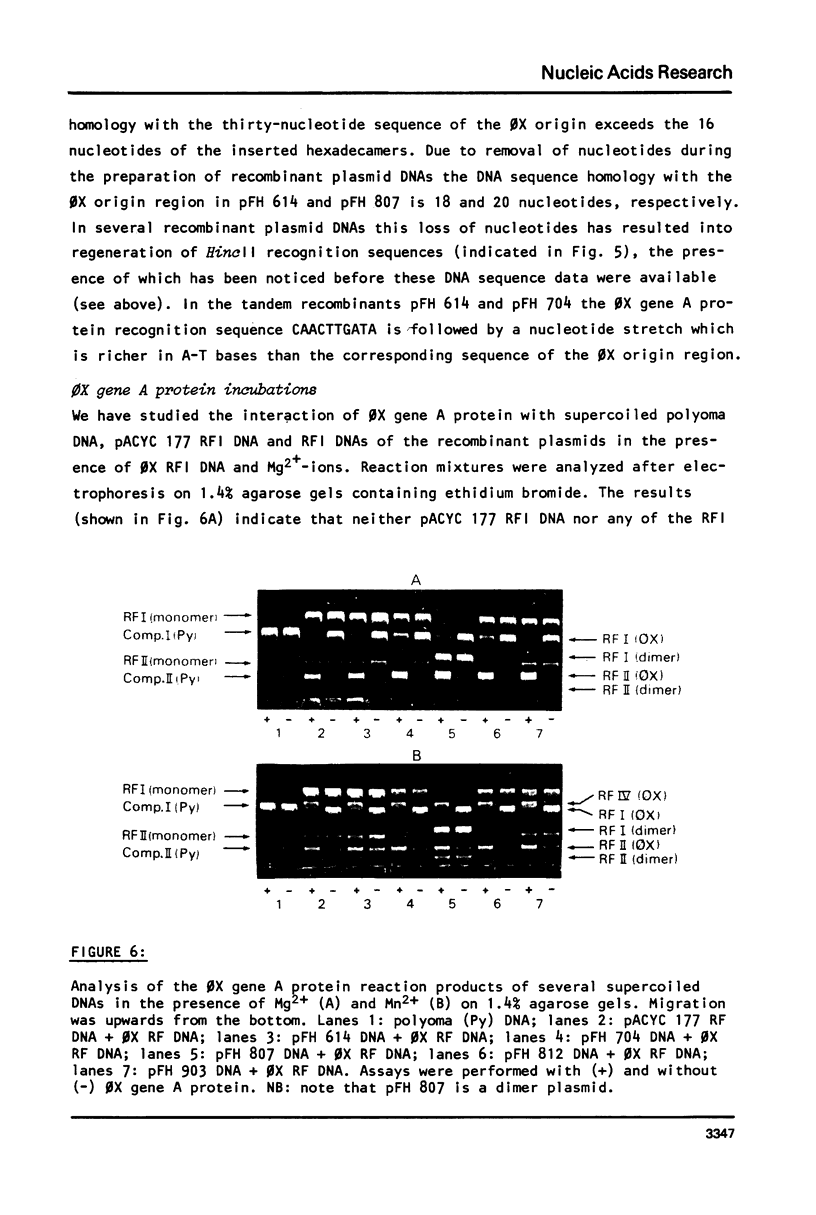
The synthetic DNA fragment (formula, see text) (corresponding to nucleotides 4299-4314 of the phi X DNA sequence) was cloned into either the AmpR gene or the KmR gene of plasmid pACYC 177. The DNA sequence of the KmR gene around the insertion site was determined by nucleotide sequence analysis of the pACYC 177 FnudII restriction DNA fragment N6 (345 b.p.). Of five selected plasmid DNAs, which contained inserted DNA sequences in the antibiotic resistance genes, the nucleotide sequences at and around these insertions were determined. Two recombinant plasmids (pFH 704 and pFH 614) contain the hexadecamer sequence in tandem (tail-to-tail and tail-to-head). In the recombinant plasmids pFH 812, pFH 903 and pFH 807 the DNA sequence homology with the phi X origin region was 14 (No. 4300-4313), 16 (No. 4299-4314) and 20 nucleotides (No. 4299-4318), respectively. None of the supercoiled recombinant plasmid DNAs is nicked upon incubation with phi X gene A protein. Moreover, the recombinant plasmid RFI DNAs cannot act as substitutes for phi X RFI DNA in the in vitro (+) strand synthesizing system. It has been shown earlier that single-stranded DNA, which contains the decamer sequence CAACTTGATA is efficiently nicked by the phi X gene A protein. The present results indicate that for nicking of double-stranded supercoiled DNA nucleotide sequence homology with the phi X origin region of more than 20 nucleotides is required. These results suggest a model for initiation of phi X RF DNA replication, which involves the presence of the recognition sequence CAACTTGATA of the phi X gene A protein as well as a second specific nucleotide sequence which is required for the binding of the phi X gene A protein. This binding causes local unwinding of the DNA double helix and exposure of the recognition sequence in a single-stranded form, which then can be nicked by phi X gene A protein.
Full text
PDF

















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrés I., Slocombe P. M., Cabello F., Timmis J. K., Lurz R., Burkardt H. J., Timmis K. N. Plasmid replication functions. II. Cloning analysis of the repA replication region of antibiotic resistance plasmid R6-5. Mol Gen Genet. 1979 Jan 5;168(1):1–25. doi: 10.1007/BF00267929. [DOI] [PubMed] [Google Scholar]
- Baas P. D., Jansz H. S. Bacteriophage phiX174 DNA synthesis in a replication-deficient host: determination of the origin of phiX DNA replication. J Mol Biol. 1976 Apr 15;102(3):633–656. doi: 10.1016/0022-2836(76)90339-9. [DOI] [PubMed] [Google Scholar]
- Baas P. D., Jansz H. S. PhiX174 replicative form DNA replication, origin and direction. J Mol Biol. 1972 Feb 14;63(3):569–576. doi: 10.1016/0022-2836(72)90448-2. [DOI] [PubMed] [Google Scholar]
- Beck E., Sommer R., Auerswald E. A., Kurz C., Zink B., Osterburg G., Schaller H., Sugimoto K., Sugisaki H., Okamoto T. Nucleotide sequence of bacteriophage fd DNA. Nucleic Acids Res. 1978 Dec;5(12):4495–4503. doi: 10.1093/nar/5.12.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denniston-Thompson K., Moore D. D., Kruger K. E., Furth M. E., Blattner F. R. Physical structure of the replication origin of bacteriophage lambda. Science. 1977 Dec 9;198(4321):1051–1056. doi: 10.1126/science.929187. [DOI] [PubMed] [Google Scholar]
- Duguet M., Yarranton G., Gefter M. The rep protein of Escherichia coli: interaction with DNA and other proteins. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):335–343. doi: 10.1101/sqb.1979.043.01.040. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Griffith J., Kornberg A. phiX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3198–3202. doi: 10.1073/pnas.74.8.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Kornberg A. Purification and characterization of phiX174 gene A protein. A multifunctional enzyme of duplex DNA replication. J Biol Chem. 1979 Jun 25;254(12):5328–5332. [PubMed] [Google Scholar]
- Francke B., Ray D. S. Formation of the parental replicative form DNA of bacteriophage phi-X174 and initial events in its replication. J Mol Biol. 1971 Nov 14;61(3):565–586. doi: 10.1016/0022-2836(71)90065-9. [DOI] [PubMed] [Google Scholar]
- Godson G. N., Barrell B. G., Staden R., Fiddes J. C. Nucleotide sequence of bacteriophage G4 DNA. Nature. 1978 Nov 16;276(5685):236–247. doi: 10.1038/276236a0. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Hayashi M. Template activities of the phi X-174 replicative allomorphic deoxyribonucleic acids. Biochemistry. 1971 Nov;10(23):4212–4218. doi: 10.1021/bi00799a009. [DOI] [PubMed] [Google Scholar]
- Heidekamp F., Langeveld S. A., Baas P. D., Jansz H. S. Studies of the recognition sequence of phi X174 gene A protein. Cleavage site of phi X gene A protein in St-1 RFI DNA. Nucleic Acids Res. 1980 May 10;8(9):2009–2021. doi: 10.1093/nar/8.9.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. L., Ross W., Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980 May 8;285(5760):85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Anderson E. S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975 Apr 2;383(4):457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- Ikeda J. E., Yudelevich A., Hurwitz J. Isolation and characterization of the protein coded by gene A of bacteriophage phiX174 DNA. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2669–2673. doi: 10.1073/pnas.73.8.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob R. J., Lebowitz J., Kleinschmidt A. K. Locating interrupted hydrogen bonding in the secondary structure of PM2 circular DNA by comparative denaturation mapping. J Virol. 1974 Jun;13(6):1176–1185. doi: 10.1128/jvi.13.6.1176-1185.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansz H. S., Pouwels P. H., Schiphorst J. Preparation of double-stranded DNA (replicative form) of bacteriophage phi-X174: a simplified method. Biochim Biophys Acta. 1966 Sep;123(3):626–627. doi: 10.1016/0005-2787(66)90233-4. [DOI] [PubMed] [Google Scholar]
- Langeveld S. A., van Arkel G. A., Weisbeek P. J. Improved method for the isolation of the A and A* proteins of bacteriophage phi X174. FEBS Lett. 1980 Jun 2;114(2):269–272. doi: 10.1016/0014-5793(80)81131-8. [DOI] [PubMed] [Google Scholar]
- Langeveld S. A., van Mansfeld A. D., Baas P. D., Jansz H. S., van Arkel G. A., Weisbeek P. J. Nucleotide sequence of the origin of replication in bacteriophage phiX174 RF DNA. Nature. 1978 Feb 2;271(5644):417–420. doi: 10.1038/271417a0. [DOI] [PubMed] [Google Scholar]
- Levine A. J., Sinsheimer R. L. The process of infection with bacteriophage phi-X174. XIX. Isolation and characterization of a chloramphenicol-resistant protein from phi-X-infected cells. J Mol Biol. 1968 Mar 28;32(3):567–578. doi: 10.1016/0022-2836(68)90343-4. [DOI] [PubMed] [Google Scholar]
- Luck G., Zimmer C. Conformational aspects and reactivity of DNA. Effects of manganese and magnesium ions on interaction with DNA. Eur J Biochem. 1972 Sep 25;29(3):528–536. doi: 10.1111/j.1432-1033.1972.tb02018.x. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Marians K. J., Ikeda J. E., Schlagman S., Hurwitz J. Role of DNA gyrase in phiX replicative-form replication in vitro. Proc Natl Acad Sci U S A. 1977 May;74(5):1965–1968. doi: 10.1073/pnas.74.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pribnow D. Bacteriophage T7 early promoters: nucleotide sequences of two RNA polymerase binding sites. J Mol Biol. 1975 Dec 15;99(3):419–443. doi: 10.1016/s0022-2836(75)80136-7. [DOI] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Zipursky S. L., Reinberg D., Hurwitz J. In vitro DNA replication of recombinant plasmid DNAs containing the origin of progeny replicative form DNA synthesis of phage phi X174. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5182–5186. doi: 10.1073/pnas.77.9.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mansfeld A. D., Langeveld S. A., Baas P. D., Jansz H. S., van der Marel G. A., Veeneman G. H., van Boom J. H. Recognition sequence of bacteriophage phi X174 gene A protein--an initiator of DNA replication. Nature. 1980 Dec 11;288(5791):561–566. doi: 10.1038/288561a0. [DOI] [PubMed] [Google Scholar]
- van Mansfeld A. D., Langeveld S. A., Weisbeek P. J., Baas P. D., van Arkel G. A., Jansz H. S. Cleavage site of phiX174 gene-A protein in phiX and G4 RFI DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):331–334. doi: 10.1101/sqb.1979.043.01.039. [DOI] [PubMed] [Google Scholar]