Teaser
Recent advances in tissue engineering have enabled the development of microscale biomimetic ‘organ on a chip’ tissue models which have the potential to make an important impact on the various stages of drug discovery and toxicity testing.
Developing biologically relevant models of human tissues and organs is an important enabling step for disease modeling and drug discovery. Recent advances in tissue engineering, biomaterials and microfluidics have led to the development of microscale functional units of such models also referred to as ‘organs on a chip’. In this review, we provide an overview of key enabling technologies and highlight the wealth of recent work regarding on-chip tissue models. In addition, we discuss the current challenges and future directions of organ-on-chip development.
Keywords: Organs on chip, tissue engineering, microfluidics, biomaterials, in vitro models, tissue models
Introduction
Scientists in academia and industry rely heavily on in vivo animal models and in vitro cell culture platforms to investigate biological processes and develop therapeutic strategies. Although these approaches have been informative, and should continue to be so for years to come, they have significant shortcomings [1]. In vivo models can produce integrated multiorgan responses which are impossible to achieve using conventional in vitro models. However, within such a multi-organ system, isolating the salient tissues or cell groups related to a particular physiological or pathophysiological response is difficult and often requires the use of knockouts or transgenic animals, leading to further complications and shortcomings. In addition to the ethics surrounding in vivo model usage, serious concerns exist over their biological relevance to humans [2]. The ability to extrapolate animal model data to human conditions is limited. Current in vitro platforms are useful for studying the molecular basis of physiological and pathological responses. In particular, recent advances in molecular biology enable the interrogation of different signaling pathways to identify the proteins, genes, receptors and ligands involved in physiological and pathological responses. However, in vitro platforms often do not simulate the complex cell–cell and cell–matrix interactions crucial for regulating cell behavior in vivo [3]. The collective limitations associated with current in vivo and in vitro models are exemplified by the significant number of new drug candidates that fail to make it to market owing to low efficiency or severe side effects. These shortcomings together with regulatory restrictions limiting the use of animal models [4] have generated substantial interest in developing human-based tissue-like constructs for disease modeling and drug and chemical testing.
Recent developments in stem cell research, regenerative medicine, biomaterials, tissue engineering and microfluidics could be integrated into new three-dimensional (3D) in vitro models closely mimicking human organs and tissues. A prerequisite for industrial use of such models is scalability. Organ(s)-on-chip devices could provide not only the biological relevance but also the requisite high throughput applications. Herein, we provide an overview of the key microengineering technologies enabling the development of organ-on-chip devices. We then review recent work on exemplar biomimetic micro-organs, their physiological relevance and underlying technologies. We also offer our perspective on exciting future directions and the challenges that remain for creating miniaturized, functional and responsive integrated models of human tissues.
Enabling technologies
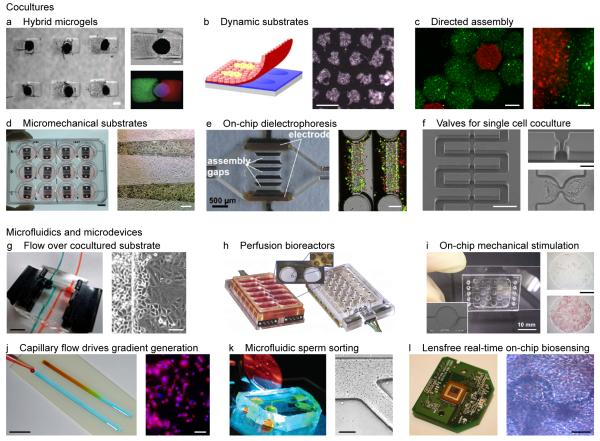
Microfabrication, microfluidics and microelectronics are enabling microengineering technologies used in the development of physiologically relevant in vitro tissue models (Figure 1). Microfabrication approaches such as photolithography, soft lithography, microcontact printing and micromolding are enabling more-complex tissue cultures to be patterned on-chip. Extensive recent reviews outline a host of techniques that hold great promise for synthesizing tailored cellular microenvironments with high spatiotemporal control over the mechanical and biochemical cues to regulate cellular behavior [5-11]. A recent example is the work of Qi et al. who used micromolding and photolithography to encapsulate individual embryoid bodies (EBs) within hybrid microgels to regulate differentiation (Fig. 1a) [12]. Various approaches have been used to engineer patterned co-cultures [13,14], a requirement for developing the complex tissues that comprise organ models. Micrometer-resolution cell positioning within co-cultures has been achieved by the directed assembly on a free-surface (Fig. 1c) [15], micromechanical substrates (Fig. 1d) [16], on-chip dielectrophoresis (Fig. 1e) [17], and direct-write printing [18]. Single-cell co-cultures have been achieved using an array of valves along a microfluidic channel (Fig. 1f) [19]. A common issue associated with patterned tissue culture is removing and transferring the engineered tissue from the substrate used for patterning. This issue was addressed by Tsuda et al., who produced their co-cultures on patterned thermoresponsive substrates (Fig. 1b) [20]. By reducing the temperature, the substrate released the co-culture. A second type of tissue transfer approach was pursued by Kobayashi et al. [21] and Nagamine et al. [22], who exploited the preference of cells to bind to certain gels rather than the substrate for patterning. Also, Tekin et al. [23,24] have used temperature-dependent shape-changing microwells to generate tissue structures of one or multiple cell types either in the form of cell aggregates or cell-laden hydrogels and used the shape changing behavior of this system to retrieve the resulting tissue constructs. Additional examples of such microfabrication techniques applied to specific tissue models are outlined in subsequent sections.
Figure 1. Enabling technologies for on-chip tissue models.
(a) Patterned microenvironment regulates differentiation of embryoid bodies. Scale bars 300 μm. Source: Qi et al. [12], copyright (2010) John Wiley & Sons.[5. AU: Do you have publishers permission to use? Please confirm for all parts of this Figure and ensure all publishers have granted permissionzs] (b) Dynamic substrate enables co-culture harvesting. Scale bar 1 mm. Source: Tsuda et al. [20], copyright 2006 with permission from Elsevier. (c) Directed assembly of cell-laden hydrogels for generating hierarchical tissue constructs. Scale bars 500 μm (left), 100 μm (right). Source: Zamanian et al. [15], copyright (2010) John Wiley & Sons. (d) Micromechanical substrates enable micrometer-resolution cell positioning and co-culture. Scale bars 1 cm (left), 250 μm (right). Source: Hui and Bhatia [16], copyright (2007) National Academy of Sciences, USA. (e) Microfluidic cell culture chambers with integrated electrodes to assemble liver sinusoids by dielectrophoresis. Scale bars 500 μm (left), 200 μm (right). Source: Schütte et al. [17], copyright 2011 Springer Science and Business Media, LLC. (f) A microfluidic channel with an array of valves for single cell co-culture. Scale bars 100 μm (left), 20 μm (right). Source: Frimat et al. [19], reproduced with permission from The Royal Society of Chemistry. (g) Flow of growth factor directs the migration of one cell type into the other within a co-culture. Scale bars 5 mm (left), 100 μm (right). Source: Kaji et al. [40], reproduced with permission from The Royal Society of Chemistry. (h) Multiwell plate with an array of perfusion bioreactors for 3D liver tissue culture. Source: Domansky et al. [38], reproduced with permission from The Royal Society of Chemistry. (i) Pneumatic microfluidic chip for the differentiation of stem cells under mechanical stimulation. Scale bars 10 mm (left), 1 mm (right). Source: Sim et al. [39], reproduced with permission from The Royal Society of Chemistry. (j) Capillary-flow-driven gradient generation within a fluid stripe[6. AU: Is this ok or did you mean strip?] for synthesizing gradient biomaterials to regulate cellular behavior such as spreading. Scale bars 1 cm (left), 100 μm (right). Sources: Hancock et al. [114] (left), copyright (2011) John Wiley & Sons; and Hancock et al. [44] (right), copyright 2011 with permission from Elsevier. (k) A microfluidic device for separating motile and non-motile sperm. Scale bar 200 μm. Sources: Cho et al. [42] (left), copyright 2003 American Chemistry Society; and Wu et al. [43] (right), copyright 2006 Springer Science and Business Media, LLC. (l) CMOS chip from webcam used to detect cardiomyocyte beating within a cell-based biosensor. Source: Kim et al. [48], reproduced with permission from The Royal Society of Chemistry.
[7. AU: I am unable to edit the Figure itself. Please change coculture to co-culture (x3) and lensfree to Lens-free]
Microfluidics is the study of systems that manipulate or process small (i.e. nanoliter and below) amounts of fluids within geometries measuring tens to hundreds of microns [25,26]. Extensive reviews exist on the widespread use of microfluidic devices in biomedical engineering, including drug discovery [2,27,28], stem cell biology [29,30], cancer biology and experimental oncology [31], cell culture and processing [1,2,28,32,33], and probing cellular behaviors such as angiogenesis, migration and cell-cell interaction [34-36], even at the single cell level [37]. Microfluidic systems enable enhanced dynamic control over the cellular microenvironment within on-chip tissue models, such as providing nutrients and dissolved gasses (Fig. 1h) [38] and applying mechanical stimulation to cultured tissues (Fig. 1i) [39]. In a recent example, microchannels were immobilized onto co-cultured substrates to flow growth factors over the cells to promote directed migration (Fig. 1g) [40]. Microfluidic devices have also been developed to mimic in vivo processes, such as identifying smells [41] and sorting motile and non-motile sperm (Fig. 1k) [42,43]. Microfluidics, combined with microfabrication, enables the synthesis of more-complex tissue constructs and biomaterials [5]. In a recent example, capillary flow within fluid stripes[1. AU: Ok? Or is strips better?] was used to generate prepolymer solution gradients that were crosslinked to form gradient biomaterials (Fig. 1j) [44].
Incorporating stimulation and sensing technologies on-chip and interfacing these with cultured cells is another important step for the real-time manipulation and detection of cellular behavior. Electrogenic cells such as heart and brain cells can be cultured directly atop complementary semiconductor–metal–oxide (CMOS) chips in vitro for pinpoint bidirectional connectivity between cells and the chip [45]. On-chip microelectrodes have been used to stimulate myotubes of skeletal muscle [22] and interconnected cardiac cells [46]. Two-way microelectrode contacts have been used to excite and detect the signals from interconnected neurons non-invasively [47]. A second detection method was developed by Kim et al., who attached a webcam chip to a cell culture chamber to detect cardiomyocyte beating in real-time (Fig. 1l) [48].
Moving forward, the challenge is to develop more-sophisticated microsystems that incorporate multiple tissues and vascular channels with active interfaces between them for transporting fluids, nutrients, immune cells and other regulatory factors [49]. An additional challenge is to apply spatiotemporally controlled mechanical forces to such microsystems to simulate the physiological environment, such as breathing in the lung, shear within blood vessels, peristalsis in the gut and tension in the skin [49,50].
Tissue models on a chip
A host of tissue models have been developed in academia and industry to mimic the sub-systems of organs or biological processes (Table 1 and Figure 2). In the following, we provide an overview of on-chip tissue models, their design, fabrication and physiological properties relevant to drug discovery. In particular, we highlight on-chip tissue models of the intestine, liver, lung and muscle, as well as models of tumors and blood vessels. We also outline recent attempts to combine the tissues of several organs on a single chip. Comprehensive reviews exist for bone, breast, cardiac, corneal, liver and tumor tissue models [51-53] as well as neural networks [54,55]; we expand on this literature by focusing more on liver and tumor tissue models. Other on-chip models that could be used for future studies on drug discovery and toxicity include those for sensory organs such as the nose [41,56,57] and biological processes such as sperm motility [42,43,58] and wound healing [59,60].
Table 1. Engineered on-chip tissue models in academia and industry.
| Tissue model | Type/comments | Academic research | Industrial researchb |
|---|---|---|---|
| Blood vessels | Review | [34] | |
| Live analysis | [96] | Quorum Technologies (Guelph, Canada) |
|
| Capillary growth | [21,89,90,108,109] | ||
| Co-culture with endothelial cells |
[40,91,92,95,110] | ||
| Brain | [47] | ||
| Breast | Review | [53] | |
| Cornea | Review | [53] | |
| Gastrointestinal | [64-66,104] | ||
| Heart | Review | [53] | |
| Example | [46] | ||
| Liver | Review | [53] | |
| Examples | [17,38,72,74,103,111] | CellASIC (Hayward, CA, USA); Hepregen (Medford, MA, USA); Hurel (New Brunswick, NJ, USA); RegeneMed (San Diego, CA, USA) |
|
| Lung | [49,80-82] | ||
| Multi-tissue | [103-105] | ||
| Muscle | Review | [53] | |
| Myotube formation |
[22,97,98,100,101,112,113] | Myomics (providence, RI) |
|
| 3D myotube | [97] | ||
| Stimulus- response |
[22,100,101,112] | ||
| Skin | Hurel (New Brunswick, NJ, USA); MatTek Corp. (Ashland, MA, USA); Phenion (Henkel AG & Co., Düsseldorf, Germany) |
||
| Spleen | In developmenta | [107] | |
| Tumor | [31,53,84,85,103] |
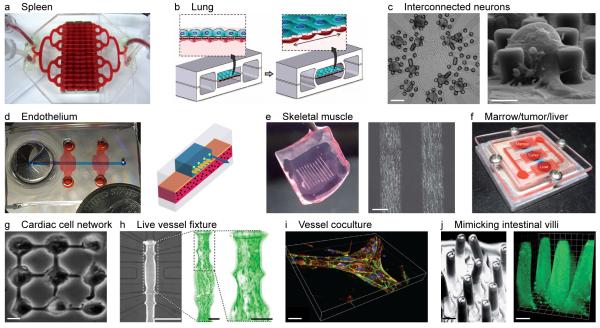
Figure 2. Microscale on-chip tissue models and culture platforms.
(a) Spleen on a chip. Source: Baker [107], reprinted with permission from Macmillan Publishers, Nature, copyright 2011. (b) Lung on a chip with membrane housing epithelium/endothelium separating air flow from fluid flow chambers. Vacuum channels enable membrane stretching to simulate breathing. Source: Huh et al. [49], reprinted with permission from AAAS. (c) Neurons (dark patches) connected by neurites on a chip. Neurons held in place by posts were connected to two-way contacts in the substrate. Scale bars 100 μm (left) and 20 μm (right). Source: Zeck and Fromherz [47], copyright (2001) National Academy of Sciences, USA. (d) Microfluidic endothelium between chemokine and flow channel. Source: Song et al. [95], open access. (e) Fibrin gel with myotube line patterns for use with skeletal muscle-cell-based bioassay system. Scale bar 200 μm. Source: Nagamine et al. [22], reproduced with permission from The Royal Society of Chemistry. (f) Multiple cell types representing liver, bone marrow and tumor cells cultured on-chip and connected via flow channels. Sources: Sung et al. [103] and Baker [107], reprinted with permission from Macmillan Publishers, Nature, copyright 2011. (g) Nine-cell cardiomyocyte network cultured on a chip. Scale bar 10 μm. Source: Kaneko et al. [46], reproduced with permission from The Royal Society of Chemistry. (h) Microfluidic device for on-chip myography with simple resistance vessel fixation. Long-term culture and automated data acquisition of up to ten dose-response sequences. Scale bars 500 μm (left) and 100 μm (middle, right). Source: Günther et al. [96], reproduced with permission from The Royal Society of Chemistry. (i) Branched collagen gel tubes with encapsulated HUVECs labeled for b-catenin (green), actin (red) and nuclei (blue). Scale bar 10 μm. Source: Raghavan et al. [90], the publisher for this copyrighted material is Mary Ann Liebert. (j) Micromolded polydimethylsiloxane (PDMS) (left) and collagen scaffold (right) mimicking human intestinal villi. Scale bars 100 μm. Source: Sung et al. [66], reproduced with permission from The Royal Society of Chemistry.
[8. AU: I am unable to edit the Figure itself. Please change coculture to co-culture]
Intestine on a chip
Orally administered drugs are mainly absorbed in the small intestine, where they must diffuse across a mucous layer covering an epithelial cell layer lining the intestinal wall [61]. Therefore, early in most development processes, drugs and chemicals are first screened against intestinal cells and tissues to assess their absorption, distribution, metabolism, elimination and toxicity (ADMET) [62]. The small intestine epithelium is composed of two main cell types: enterocytes and goblet cells. Enterocytes constitute ~90% of the cell population and are covered with surface-enhancing microvilli. In most areas of the intestine, goblet cells constitute the remaining 10% of the cell population and secrete mucus which coats the epithelial layer and is important for innate immune responses and homeostasis. Coordinated intestinal movements (i.e. peristalsis) have also been shown to have an important role in intestinal biology. Every material absorbed through the intestine into the blood stream must first diffuse across the mucus layer, the enterocytes, the lamina propria and the capillaries formed by endothelial cells. Crossing the epithelial layer, however, has been shown to be the rate-limiting step [63].
Small intestine models destined for absorption and/or metabolism studies should include these key cellular (i.e. enterocytes, goblet and vascular endothelial cells), structural (i.e. villi and mucus) and dynamic (i.e. peristalsis) features. Some of these elements were addressed by Kimura et al. [64], who engineered an intestinal model with a membrane and vascular flow simulating the epithelial barrier and the epithelial-endothelial interface, respectively. Additional important model features include the mucus layer and the digestion process. These were partially addressed by Mahler et al., who developed a microscale cell culture analog of the gastrointestinal (GI) tract incorporating digestion, a mucus layer and physiologically realistic cell populations [65]. To mimic drug transport across the intestinal wall better, additional structural features such as microscopic villi should be incorporated to enhance the intestinal epithelium surface area and to direct mucus. Such structural features have been fabricated in PDMS[2. AU: Please define] and hydrogels seeded with epithelial cells (Fig. 2j) [66], and could be incorporated into future functional tissue models.
Because healthy primary human cells are difficult to obtain, the majority of existing intestinal models were developed using immortalized cells lines. Although such cell lines can serve as informative and convenient surrogates for primary cells, their use limits the extrapolation of results to humans. Existing intestinal models also lack a host of ‘non-GI’ cells, including mast cells which play a key part in the barrier integrity and physiology of the intestinal epithelium [67].
Liver on a chip
Hepatotoxicity is a major side effect of many chemicals and drugs that are administered over prolonged periods of time. Almost half of all drug withdrawals occur as a result of acute liver toxicity [68]. Therefore, efficient, reliable, accurate and inexpensive tools for liver toxicity testing are required. Recent reviews provide an overview of microfluidic devices incorporating liver tissue or cells for metabolism, drug discovery and toxicity studies [53,69]. Existing tools for testing liver toxicity, involving hepatocyte cell lines, two-dimensional (2D) in vitro cultures and animal models, all suffer from major limitations. Cell lines and 2D cultures lack key metabolic activities found in human liver and major interspecies differences limit the predictive value of animal models [70,71]. The key challenge for on-chip liver tissue models is to maintain the metabolic activity and phenotype of the poorly viable cryopreserved human primary hepatocytes. Bioreactors have been developed to meet this challenge, such as the perfused multiwell plate device of Domansky et al. [38], which supported the growth and functional integrity of hepatocytes in 3D culture for up to a week (Fig. 2h), and the multiwell micropatterned co-culture system of Khetani and Bhatia [72] which could maintain phenotypic functions for several weeks. Both groups co-cultured primary hepatocytes with other key liver cells such as sinusoidal endothelial cells containing stellate and Kupffer cells [38] or fibroblasts [72] to improve and maintain primary hepatocyte function considerably. Another desirable feature of liver tissue models is to simulate the morphology of the liver’s functional units (i.e. lobules) to provide the homogenous and heterogeneous cell–cell interactions and cellular matrix support to maintain hepatocyte functionality [72,73]. Toh et al. [74] incorporated this feature into their on-chip liver model that exhibited a reasonable simulation of the liver morphology and functionality for up to 72 hours. In a second example, Schütte et al. used a perfusion system and integrated electrodes to assemble liver sinusoids by dielectrophoresis (Fig. 2g) [17]. A second class of on-chip liver models involves intact tissues extracted from humans or animals and integrated on-chip for the in vitro assessment of metabolism and toxicity [69]. Recently, van Midwoud et al. [75] integrated liver and intestinal slices into different compartments of a microfluidic device with sequential perfusion between the compartments to study interorgan interactions. In addition to reflecting in vivo conditions adequately for investigating liver drug metabolism and toxicity, on-chip models incorporating primary hepatocytes or intact tissue slices must be able to maintain key metabolic activities and normal liver integrity, particularly with regard to repeat dose experiments [69]. This feature was demonstrated by Khetani & Bhatia [72], who performed nine-day[3. AU: Is hyphen ok here? Added to draw a distinction between nine one-day repeat dose exps] repeat dose experiments with their co-culture system to test the chronic toxicity of troglitazone. Finally, an exciting new area is the ability to use in vitro cultures for disease modeling and to monitor disease progression and the impact of treatment in real-time. An example for such an approach is the recent work of Jones et al. [76], who used a fluorescent cell-based reporter system to monitor in real-time the infection of hepatitis C on hematoma cell lines and primary human hepatocytes.
Lung on a chip
The lung epithelium acts like a physical barrier between the host and the environment and responds to a variety of environmental insults such as pathogens and air pollution. The lung’s defense mechanisms include secreting antimicrobial peptides (e.g. defensins) and pro- and anti-inflammatory mediators, and recruiting innate and adaptive immune cells to the site of infection or damage. The efficiency of such protective responses as well as the homeostasis of the airways is underpinned by a variety of lung specialized epithelial cells (e.g. ciliated and mucus producing) and their crosstalk with other cell types such as antigen presenting cells, macrophages and myofibroblasts [77]. The human lung also provides one of the largest vascularized interfaces between the host and the environment, and is therefore considered a convenient port of delivery for local and systemic drugs. Drug delivery studies tend to focus on drug permeability across the lung epithelial barrier. Within this context, most existing lung tissue models are limited to single cell culture representations of the alveolar epithelium comprising epithelial cell lines [78,79], limiting their physiological relevance to the human lung. Certain enhancements have been made to single cell culture models, such as microfluidics and flexible membranes to apply fluid and solid mechanical stresses to single cell alveolar models to simulate the effects of mechanical ventilation and physiologic or pathologic liquid plug flows [80-82].
More relevant lung tissue models destined for disease modeling and drug testing require additional cell types such as airway smooth muscle and immune cells, as well as key structural, functional and mechanical features of the human respiratory epithelium. Toward this goal, Huh et al. (Fig. 2b) [49] co-cultured alveolar epithelial, endothelial and certain immune cells on an artificial membrane and used adjacent vacuum channels to mimic the cyclic stretching during breathing. The inclusion of immune cells recapitulated certain aspects of human lung responses to infection and inflammation, enabling the assessment of the inflammatory response triggered by inhaled pathogens. Despite its many merits, this model used immortalized cell lines rather than human primary cells and did not include several other key structural or immune cells. It is not known whether the anatomical shape of the alveolar space or upper respiratory tract would have any bearing on the dynamics of cell–cell interaction. Future attempts, in addition to inclusion of primary cells, could focus on micropatterning scaffolds to improve the anatomical relevance and function of such tissue models.
Tumor on a chip
Developing anticancer therapies that target rapidly dividing cancer cells but leave normal healthy cells untouched is a grand challenge for cancer research. A recent review contains a comprehensive list of 3D tumor tissue models including multicellular spheroid, hollow fiber and multicellular layer models [53]. Perfusion delivers potential therapeutic agents to such 3D tumor cell cultures, which emulates the heterogeneous blood supply delivered to tumors in vivo [53]. In addition to these approaches, a popular strategy for developing anticancer drugs is HTS in which large combinatorial arrays of drug cocktails are produced and exposed to cells, which are then analyzed to assess the drug effects [31]. Performing HTS within microfluidic devices enables smaller volumes of reagents to be used than in typical multiwell plate experiments. One key engineering challenge is to generate an array of drug concentrations on-chip. To meet this challenge, Jang et al. developed a microfluidic combinatorial system that produced 64 or 100 combinations of two chemical solutions at different concentrations and stored them in isolated chambers [83]. The device inputs could be chosen such that the compositions of the stored mixtures spanned any triangle in the 2D space of chemical concentrations. A second engineering challenge is to expose different combinations or concentrations of drugs to cells cultured on-chip. Two groups recently accomplished this step by exposing human lung cancer cells to different concentrations of the anticancer drug verapamil (VP-16) [84,85]. In one study, the percentage of apoptotic cells in the verapamil-pretreated group was approximately double that of the control group, in agreement with previous flow cytometry analysis [85]. Further HTS advances could be made by using 3D cultures which mimic in vivo conditions better than cell monolayers. For example, Fischbach et al. [86] cultured cancer cells in 3D alginate hydrogels and found that cell signaling was altered.
A second line of cancer research is to test anticancer therapies on-chip to optimize system parameters for different types of cancer cells, while requiring minute amounts of reagents and cells. For example, Jedrych et al. [87] created a microfluidics system for in vitro photodynamic therapy (PDT) [87]. In PDT, light induces a photosensitizer delivered to the carcinoma cells, which in turn reacts with oxygen to produce a chemical toxic to the tumor cells. A key step in developing PDT is to optimize the system parameters such as the photosensitizer concentration for different types of cancer cells. Jedrych et al. [87] demonstrated this step by testing different concentrations of photosensitizer on A549 cancer cells.
Vessels on a chip
Blood vessels are involved in most medical conditions and are thus integral components to organs-on-chip devices [88]. A key engineering challenge is to grow blood vessels and capillaries on-chip. Dike et al. addressed this issue by using a surface patterned with 10 μm wide cell-adhesive lines surrounded by cell-repellent areas [89]. Endothelial cells cultured on the cell-adhesive lines differentiated to form capillary tube-like structures containing central lumens. A different approach was carried out by Kobayashi et al. [21], who patterned endothelial cells along parallel hydrophilic lines and then transferred the cells to a hydrogel scaffold. Once on the scaffold, the cells changed their morphology to form capillary-like tubular structures. The capillary structures were tested by flowing fluorescent solution through them, and also by transplanting them into mice where blood cells were subsequently found in the lumen. To control the endothelial capillary tube formation (tubulogenesis) further, Raghavan et al. cultured endothelial cells within microscale channels filled with collagen hydrogel (Fig. 2i) [90]. The cells were organized into tubes containing lumens with controlled branching and orientation. Future work must be done to generate capillary structures in vitro with similar mechanical properties to those in vivo.
A second major engineering challenge is to reproduce the microenvironment surrounding blood vessel walls, composed predominantly of vascular endothelial cells. A key component of this challenge is to understand how endothelial cells lining the vessel walls interact with other cells types. To address this issue, Chung et al. [91] and Kaji et al. [40,92] developed endothelium models to study the paracrine communication between endothelial and other cell types. Chung et al. found that MTLn3 cancer cells attracted endothelial cells to induce capillary formation, whereas smooth muscle cells suppressed endothelial activity [91]. Kaji et al. used complimentary substrates to co-culture endothelial and HeLa (cervical cancer) cells and found, similarly, that paracrine factors secreted from one cell type and directed via the culture medium to the other cell type affected the migration behavior of both types [40,92]. Research has also been devoted toward understanding how the cyclic stretch due to vessel deformation regulates cell function, and how disruptions in these forces produce diseased states such as hypertension or arteriosclerosis [93]. The effect of mechanical stimulation on endothelial cells in vitro has been previously reviewed [94]. An additional challenge is to apply chemical and mechanical stimuli simultaneously to cells to engineer more biologically relevant models. To address this issue, Song et al. developed a membrane-based microfluidic device that models the adhesion of breast cancer cells on an endothelium (formed on the porous membrane) during intravascular metastasis (Fig. 2d) [95]. They showed that either the addition of the chemokine CXCL12 from the basal side or the elevation of fluidic shear stress could enhance the metastatic cell adhesion. Many additional processes remain to be studied, including the chemical and cellular exchange across vessel walls, regulation of the permeability of surrounding tissues, coagulation of blood, transmigration of leukocytes and the regulation of vessel diameter.
Microfluidic platforms for probing the structure and function of actual blood vessels under physiological conditions also exist. Recently, Günther et al. developed a microfluidic platform enabling on-chip fixation, long-term culture, controlled delivery of drugs and automated acquisition of consecutive dose–response curves of intact mouse mesenteric artery segments (Fig. 2h) [96]. The phenylephrine dose–response relationships produced by the platform were in good agreement with those obtained by conventional pressure myography.
Muscle on a chip
Skeletal muscle is one of the major insulin-target tissues responsible for the maintenance of whole body glucose homeostasis. Defects in the glucose uptake in skeletal muscle contribute to insulin resistance in type 2 diabetes. To investigate insulin- and contraction-induced glucose metabolism, on-chip skeletal tissue muscle models require in-vivo-like structure, electrode stimulation and co-culture with primary human skeletal cells with motor neurons. In-vivo-like structural features include myotube alignment and assembly into sarcomeres. On-chip myotube alignment has been guided by substrate pattern [97,98] and stiffness [99,100] and by electrical stimulation [101]. An important aspect of a muscle tissue model is the ability to control muscle contraction. Such control has been accomplished by Tourovskaia et al. [102] by stimulating myotubes with a laminar stream of agrin, a chemical secreted by neurons in vivo at the location where nerves contact the muscle. A different approach was developed by Kaji et al. [100], who modulated the substrate stiffness with an atelocollagen and electrically stimulated the formed myotubes to demonstrate a positive correlation between the contractility of the myotubes and the glucose uptake. Control of individual myotubes was achieved by Nagamine et al. [22], who integrated a microelectrode array with aligned myotubes on a fibrin gel sheet (Fig. 2e). Future improvements to on-chip muscle tissue models include monitoring the glucose uptake and other metabolism events in real time and incorporating neuromuscular junctions by co-culturing myotubes with neurons and stimulating them to induce muscle contraction.
Multiple organs on a chip
Most existing on-chip tissue models represent a single organ, preventing investigations on a drug’s systemic effect. Microscale cell culture analog systems, also known as ‘animal-on-a-chip’, ‘human-on-a-chip’ or ‘body-on-a-chip’ systems, attempt to improve the prediction of the effects of drugs and toxicity on the whole body. In a typical multiorgan device, multiple cell types representing different organs are cultured in separate chambers on a single chip. The chambers are connected by channels based on their sequence in vivo to assess systematically the effects of drug action and metabolism in different organs. A number of multiorgan devices has been developed with cell lines representing various organs or tissues, including: liver, tumor and bone marrow based on a mathematical pharmacokinetics–pharmacodynamics model and used to test the toxicity of the anticancer drug 5-fluorouracil (Fig. 2f) [103]; intestine, liver and breast cancer cells to evaluate the intestinal absorption, hepatic metabolism and the antitarget cell bioactivity of drugs [104]; liver, lung, kidney and adipose tissue [105]. Despite continued efforts and strong motivations to replace animal testing, these multiorgan systems are still in their infancy. The function of these systems is dictated by the cell types employed, and physiologically relevant cellular responses must be distinguished from artifacts due to in vitro cell culture. Also, barrier tissue analogs must be incorporated into existing multiorgan systems because these tissues can significantly reduce the bioavailability of drugs taken up through inhalation or absorbed through the skin. Finally, it is essential to benchmark the results of the on-chip systems with in vivo data under identical conditions and drug exposure. A more detailed discussion can be found in the literature by Esch et al. [106].
Concluding remarks
With recent advances in tissue engineering, biomaterials and microfluidics, developing functional organs on a chip is becoming a more realistic goal for various diagnostics and drug screening applications. However, barriers still exist before integrated in vitro models can be produced with the necessary capabilities to report and predict chemical metabolism and target interaction over long durations. One of the major challenges to recapitulating the true functional properties of organs is to build systems based on primary human cells rather than cancerous cell lines. An additional challenge is to create in-vivo-like environments with enhanced architectural and cellular complexities. That said, once such complexities are in place, they will pose additional challenges for detecting cellular responses to external stimuli, monitoring the quality of the microenvironment (e.g. metabolites, pH and O2 saturation) and supporting the diverse cellular requirements. Tools to address these issues will require additional advances in biosensing and engineering. In addition, mathematical modeling will guide the design of the next generation of organ-on-chip devices, and nanosensing will enable the real-time and in situ monitoring of the microenvironment and cellular and/or metabolic responses.
Acknowledgements
AMG was supported by an International Scientific Interchange Scheme award (BB/I02643X/1) and a project grant (BB/H011293/1) from the Biotechnology and Biological Sciences Research Council (BBSRC), UK. HK was supported by a JSPS Fellowship for Research Abroad and Grant-in-Aid for Young Scientists (A) (23681027) from the Ministry of Education, Science and Culture, Japan. AK would like to acknowledge funding by the National Institutes of Health (HL092836, DE019024, EB012597, AR057837, DE021468, HL099073, EB008392), the Office of Naval Research, the National Science Foundation and the US Army Corps of Engineers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ziolkowska K, et al. Microfluidic devices as tools for mimicking the in vivo environment. New J. Chem. 2011;35:979–990. [Google Scholar]
- 2.Wu M-H, et al. Microfluidic cell culture systems for drug research. Lab Chip. 2010;10:939–956. doi: 10.1039/b921695b. [DOI] [PubMed] [Google Scholar]
- 3.Guillouzo A, Guguen-Guillouzo C. Evolving concepts in liver tissue modeling and implications for in vitro toxicology. Expert Opin. Drug Met. 2008;4:1279–1294. doi: 10.1517/17425255.4.10.1279. [DOI] [PubMed] [Google Scholar]
- 4.Adler S, et al. Alternative (non-animal) methods for cosmetics testing: current status and future prospects—2010. Arch. Toxicol. 2011;85:367–485. doi: 10.1007/s00204-011-0693-2. [DOI] [PubMed] [Google Scholar]
- 5.Sant S, et al. Biomimetic gradient hydrogels for tissue engineering. Can. J. Chem. Eng. 2010;88:899–911. doi: 10.1002/cjce.20411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stuart MAC, et al. Emerging applications of stimuli-responsive polymer materials. Nat. Mater. 2010;9:101–113. doi: 10.1038/nmat2614. [DOI] [PubMed] [Google Scholar]
- 7.Shi JJ, et al. Nanotechnology in drug delivery and tissue engineering: from discovery to applications. Nano Lett. 2010;10:3223–3230. doi: 10.1021/nl102184c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dvir T, et al. Nanotechnological strategies for engineering complex tissues. Nat. Nano. 2011;6:13–22. doi: 10.1038/nnano.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tay CY, et al. Micro-/nano-engineered cellular responses for soft tissue engineering and biomedical applications. Small. 2011;7:1361–1378. doi: 10.1002/smll.201100046. [DOI] [PubMed] [Google Scholar]
- 10.Khademhosseini A, et al. Microscale technologies for tissue engineering and biology. Proc. Natl Acad. Sci. U. S. A. 2006;103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Ali J, et al. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 12.Qi H, et al. Patterned differentiation of individual embryoid bodies in spatially organized 3D hybrid microgels. Advanced Materials. 2010;22:5276–5281. doi: 10.1002/adma.201002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaji H, et al. Engineering systems for the generation of patterned co-cultures for controlling cell-cell interactions. Biochim. Biophys. Acta - Gen. Subjects. 2011;1810:239–250. doi: 10.1016/j.bbagen.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khademhosseini A, et al. Co-culture of human embryonic stem cells with murine embryonic fibroblasts on microwell-patterned substrates. Biomaterials. 2006;27:5968–5977. doi: 10.1016/j.biomaterials.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 15.Zamanian B, et al. Interface directed self assembly of cell laden microgels. Small. 2010;6:937–944. doi: 10.1002/smll.200902326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hui EE, Bhatia SN. Micromechanical control of cell-cell interactions. Proc. Natl Acad. Sci. U. S. A. 2007;104:5722–5726. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schütte J, et al. ‘Artificial micro organs’—a microfluidic device for dielectrophoretic assembly of liver sinusoids. Biomed. Microdev. 2011;13:493–501. doi: 10.1007/s10544-011-9517-7. [DOI] [PubMed] [Google Scholar]
- 18.Mironov V, et al. Organ printing: tissue spheroids as building blocks. Biomaterials. 2009;30:2164–2174. doi: 10.1016/j.biomaterials.2008.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frimat J-P, et al. A microfluidic array with cellular valving for single cell co-culture. Lab Chip. 2011;11:231–237. doi: 10.1039/c0lc00172d. [DOI] [PubMed] [Google Scholar]
- 20.Tsuda Y, et al. Heterotypic cell interactions on a dually patterned surface. Biochem. Bioph. Res. Co. 2006;348:937–944. doi: 10.1016/j.bbrc.2006.07.138. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi A, et al. In vitro formation of capillary networks using optical lithographic techniques. Biochem. Bioph. Res. Co. 2007;358:692–697. doi: 10.1016/j.bbrc.2007.04.206. [DOI] [PubMed] [Google Scholar]
- 22.Nagamine K, et al. Spatiotemporally controlled contraction of micropatterned skeletal muscle cells on a hydrogel sheet. Lab Chip. 2011;11:513–517. doi: 10.1039/c0lc00364f. [DOI] [PubMed] [Google Scholar]
- 23.Tekin H, et al. Stimuli-responsive microwells for formation and retrieval of cell aggregates. Lab Chip. 2010;10:2411–2418. doi: 10.1039/c004732e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tekin H, et al. Responsive micromolds for sequential patterning of hydrogel microstructures. J. Am. Chem. Soc. 2011;133:12944–12947. doi: 10.1021/ja204266a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Squires TM, Quake SR. Microfluidics: fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005;77:977. [Google Scholar]
- 26.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 27.Dittrich PS, Manz A. Lab-on-a-chip: microfluidics in drug discovery. Nat. Rev. Drug Discov. 2006;5:210–218. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- 28.Yeo LY, et al. Microfluidic devices for bioapplications. Small. 2011;7:12–48. doi: 10.1002/smll.201000946. [DOI] [PubMed] [Google Scholar]
- 29.Gupta K, et al. Lab-on-a-chip devices as an emerging platform for stem cell biology. Lab Chip. 2010;10:2019–2031. doi: 10.1039/c004689b. [DOI] [PubMed] [Google Scholar]
- 30.van Noort D, et al. Stem cells in microfluidics. Biotechnol. Progr. 2009;25:52–60. doi: 10.1002/btpr.171. [DOI] [PubMed] [Google Scholar]
- 31.Wlodkowic D, Cooper JM. Tumors on chips: oncology meets microfluidics. Curr. Opin Chem. Biol. 2010;14:556–567. doi: 10.1016/j.cbpa.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Paguirigan AL, Beebe DJ. From the cellular perspective: exploring differences in the cellular baseline in macroscale and microfluidic cultures. Integr. Biol. 2009;1:182–195. doi: 10.1039/b814565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyvantsson I, Beebe DJ. Cell culture models in microfluidic systems. Annu. Rev. Anal. Chem. 2008;1:423–449. doi: 10.1146/annurev.anchem.1.031207.113042. [DOI] [PubMed] [Google Scholar]
- 34.Chung S, et al. Microfluidic platforms for studies of angiogenesis, cell migration, and cell-cell interactions. Ann. Biomed. Eng. 2010;38:1164–1177. doi: 10.1007/s10439-010-9899-3. [DOI] [PubMed] [Google Scholar]
- 35.Zervantonakis IK, et al. Microfluidic devices for studying heterotypic cell-cell interactions and tissue specimen cultures under controlled microenvironments. Biomicrofluidics. 2011;5:013406. doi: 10.1063/1.3553237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed T, et al. Microfluidics for bacterial chemotaxis. Integr. Biol. 2010;2:604–629. doi: 10.1039/c0ib00049c. [DOI] [PubMed] [Google Scholar]
- 37.Le Gac S, van den Berg A. Single cells as experimentation units in lab-on-a-chip devices. Trends Biotechnol. 2010;28:55–62. doi: 10.1016/j.tibtech.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Domansky K, et al. Perfused multiwell plate for 3D liver tissue engineering. Lab Chip. 2010;10:51–58. doi: 10.1039/b913221j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sim WY, et al. A pneumatic micro cell chip for the differentiation of human mesenchymal stem cells under mechanical stimulation. Lab Chip. 2007;7:1775–1782. doi: 10.1039/b712361m. [DOI] [PubMed] [Google Scholar]
- 40.Kaji H, et al. Directing the flow of medium in controlled cocultures of HeLa cells and human umbilical vein endothelial cells with a microfluidic device. Lab Chip. 2010;10:2374–2379. doi: 10.1039/c004583g. [DOI] [PubMed] [Google Scholar]
- 41.Figueroa XA, et al. Large-scale investigation of the olfactory receptor space using a microfluidic microwell array. Lab Chip. 2010;10:1120–1127. doi: 10.1039/b920585c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho BS, et al. Passively driven integrated microfluidic system for separation of motile sperm. Anal. Chem. 2003;75:1671–1675. doi: 10.1021/ac020579e. [DOI] [PubMed] [Google Scholar]
- 43.Wu J, et al. A surface-modified sperm sorting device with long-term stability. Biomed. Microdev. 2006;8:99–107. doi: 10.1007/s10544-006-7705-7. [DOI] [PubMed] [Google Scholar]
- 44.Hancock MJ, et al. Anisotropic material synthesis by capillary flow in a fluid stripe. Biomaterials. 2011;32:6493–6504. doi: 10.1016/j.biomaterials.2011.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hierlemann A, et al. Growing cells atop microelectronic chips: interfacing electrogenic cells in vitro with CMOS-based microelectrode arrays. Proc. IEEE. 2011;99:252–284. [Google Scholar]
- 46.Kaneko T, et al. An on-chip cardiomyocyte cell network assay for stable drug screening regarding community effect of cell network size. Analyst. 2007;132:892–898. doi: 10.1039/b704961g. [DOI] [PubMed] [Google Scholar]
- 47.Zeck G, Fromherz P. Noninvasive neuroelectronic interfacing with synaptically connected snail neurons immobilized on a semiconductor chip. Proc. Natl Acad. Sci. U. S. A. 2001;98:10457–10462. doi: 10.1073/pnas.181348698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SB, et al. A cell-based biosensor for real-time detection of cardiotoxicity using lensfree imaging. Lab Chip. 2011;11:1801–1807. doi: 10.1039/c1lc20098d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huh D, et al. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ingber DE. Cellular mechanotransduction: putting all the pieces together again. FASEB J. 2006;20:811–827. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 51.Porter JR, et al. Bone tissue engineering: a review in bone biomimetics and drug delivery strategies. Biotechnol. Progr. 2009;25:1539–1560. doi: 10.1002/btpr.246. [DOI] [PubMed] [Google Scholar]
- 52.Janssen FW, et al. Human tissue-engineered bone produced in clinically relevant amounts using a semi-automated perfusion bioreactor system: a preliminary study. J. Tissue Eng. Regen. M. 2010;4:12–24. doi: 10.1002/term.197. [DOI] [PubMed] [Google Scholar]
- 53.Elliott NT, Yuan F. A review of three-dimensional in vitro tissue models for drug discovery and transport studies. J. Pharm. Sci. 2011;100:59–74. doi: 10.1002/jps.22257. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, et al. Microfluidics: a new cosset for neurobiology. Lab Chip. 2009;9:644–652. doi: 10.1039/b813495b. [DOI] [PubMed] [Google Scholar]
- 55.Wheeler BC, Brewer GJ. Designing neural networks in culture. Proc. IEEE. 2010;98:398–406. doi: 10.1109/JPROC.2009.2039029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Misawa N, et al. Highly sensitive and selective odorant sensor using living cells expressing insect olfactory receptors. Proc. Natl Acad. Sci. U. S. A. 2010;107:15340–15344. doi: 10.1073/pnas.1004334107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oh EH, et al. Recent advances in electronic and bioelectronic noses and their biomedical applications. Enzyme Microb. Technol. 2011;48:427–437. doi: 10.1016/j.enzmictec.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Heo YS, et al. Dynamic microfunnel culture enhances mouse embryo development and pregnancy rates. Hum. Reprod. 2010;25:613–622. doi: 10.1093/humrep/dep449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Meer AD, et al. A microfluidic wound-healing assay for quantifying endothelial cell migration. Am. J. Physiol. - Heart and Circ, Physiol. 2010;298:719–725. doi: 10.1152/ajpheart.00933.2009. [DOI] [PubMed] [Google Scholar]
- 60.Nie F-Q, et al. On-chip cell migration assay using microfluidic channels. Biomaterials. 2007;28:4017–4022. doi: 10.1016/j.biomaterials.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 61.Fasinu P, et al. Diverse approaches for the enhancement of oral drug bioavailability. Biopharm. Drug Dispos. 2011;32:185–209. doi: 10.1002/bdd.750. [DOI] [PubMed] [Google Scholar]
- 62.Selick HE, et al. The emerging importance of predictive ADME simulation in drug discovery. Drug Discov. Today. 2002;7:109–116. doi: 10.1016/s1359-6446(01)02100-6. [DOI] [PubMed] [Google Scholar]
- 63.Macpherson A, et al. The mucosal firewalls against commensal intestinal microbes. Semin. Immunopathol. 2009;31:145–149. doi: 10.1007/s00281-009-0174-3. [DOI] [PubMed] [Google Scholar]
- 64.Kimura H, et al. An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab Chip. 2008;8:741–746. doi: 10.1039/b717091b. [DOI] [PubMed] [Google Scholar]
- 65.Mahler GJ, et al. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol. Bioeng. 2009;104:193–205. doi: 10.1002/bit.22366. [DOI] [PubMed] [Google Scholar]
- 66.Sung JH, et al. Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip. 2011;11:389–392. doi: 10.1039/c0lc00273a. [DOI] [PubMed] [Google Scholar]
- 67.Kalesnikoff J, Galli SJ. New developments in mast cell biology. Nat. Immunol. 2008;9:1215–1223. doi: 10.1038/ni.f.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gershell LJ, Atkins JH. A brief history of novel drug discovery technologies. Nat. Rev. Drug Discov. 2003;2:321–327. doi: 10.1038/nrd1064. [DOI] [PubMed] [Google Scholar]
- 69.van Midwoud PM, et al. Microfluidic devices for in vitro studies on liver drug metabolism and toxicity. Integr. Biol. 2011;3:509–521. doi: 10.1039/c0ib00119h. [DOI] [PubMed] [Google Scholar]
- 70.Donato MT, et al. Functional assessment of the quality of human hepatocyte preparations for cell transplantation. Cell Transplant. 2008;17:1211–1219. doi: 10.3727/096368908787236620. [DOI] [PubMed] [Google Scholar]
- 71.Olsavsky Goyak KM, et al. Hepatocyte differentiation. In: Maurel P, editor. Hepatocytes. Vol. 640. Humana Press; 2010. pp. 115–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 73.Gebhardt R, Hovhannisyan A. Organ patterning in the adult stage: the role of Wnt/β-catenin signaling in liver zonation and beyond. Dev. Dyn. 2010;239:45–55. doi: 10.1002/dvdy.22041. [DOI] [PubMed] [Google Scholar]
- 74.Toh Y-C, et al. A microfluidic 3D hepatocyte chip for drug toxicity testing. Lab Chip. 2009;9:2026–2035. doi: 10.1039/b900912d. [DOI] [PubMed] [Google Scholar]
- 75.van Midwoud PM, et al. A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices. Lab Chip. 2010;10:2778–2786. doi: 10.1039/c0lc00043d. [DOI] [PubMed] [Google Scholar]
- 76.Jones CT, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat. Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tam A, et al. The airway epithelium: more than just a structural barrier. Ther. Adv. Respir. Dis. 2011;5:255–273. doi: 10.1177/1753465810396539. [DOI] [PubMed] [Google Scholar]
- 78.Zhang L, et al. Analysis of chemoresistance in lung cancer with a simple microfluidic device. Electrophoresis. 2010;31:3763–3770. doi: 10.1002/elps.201000265. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, et al. High throughput determination of TGFβ1/SMAD3 targets in A549 lung epithelial cells. PLoS ONE. 2011;6:e20319. doi: 10.1371/journal.pone.0020319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Douville NJ, et al. Combination of fluid and solid mechanical stresses contribute to cell death and detachment in a microfluidic alveolar model. Lab Chip. 2011;11:609–619. doi: 10.1039/c0lc00251h. [DOI] [PubMed] [Google Scholar]
- 81.Huh D, et al. Acoustically detectable cellular-level lung injury induced by fluid mechanical stresses in microfluidic airway systems. Proc. Natl Acad. Sci. U. S. A. 2007;104:18886–18891. doi: 10.1073/pnas.0610868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tavana H, et al. Epithelium damage and protection during reopening of occluded airways in a physiologic microfluidic pulmonary airway model. Biomed. Microdev. 2011;13:731–742. doi: 10.1007/s10544-011-9543-5. [DOI] [PubMed] [Google Scholar]
- 83.Jang Y-H, et al. An integrated microfluidic device for two-dimensional combinatorial dilution. Lab Chip. 2011 doi: 10.1039/c1lc20449a. (in press) DOI: 10.1039/c1031lc20449a[4. AU: Please update reference if possible] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang SY, et al. Application of microfluidic gradient chip in the analysis of lung cancer chemotherapy resistance. J. Pharm. Biomed. Anal. 2009;49:806–810. doi: 10.1016/j.jpba.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 85.Zhao L, et al. Chemotherapy resistance research of lung cancer based on micro-fluidic chip system with flow medium. Biomed. Microdev. 2010;12:325–332. doi: 10.1007/s10544-009-9388-3. [DOI] [PubMed] [Google Scholar]
- 86.Fischbach C, et al. Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement. Proc. Natl Acad. Sci. U. S. A. 2009;106:399–404. doi: 10.1073/pnas.0808932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jedrych E, et al. Evaluation of photodynamic therapy (PDT) procedures using microfluidic system. Anal. Chim. Acta. 2011;683:149–155. doi: 10.1016/j.aca.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 88.Phelps EA, García AJ. Engineering more than a cell: vascularization strategies in tissue engineering. Curr. Opin Biotechnol. 2010;21:704–709. doi: 10.1016/j.copbio.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dike LE, et al. Geometric control of switching between growth, apoptosis, and differentiation during angiogenesis using micropatterned substrates. In Vitro Cell. Dev. Biol. — Animal. 1999;35:441–448. doi: 10.1007/s11626-999-0050-4. [DOI] [PubMed] [Google Scholar]
- 90.Raghavan S, et al. Geometrically controlled endothelial tubulogenesis in micropatterned gels. Tissue Eng. Pt. A. 2010;16:2255–2263. doi: 10.1089/ten.tea.2009.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chung S, et al. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip. 2009;9:269–275. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 92.Kaji H, et al. Controlled cocultures of HeLa cells and human umbilical vein endothelial cells on detachable substrates. Lab Chip. 2009;9:427–432. doi: 10.1039/b812510d. [DOI] [PubMed] [Google Scholar]
- 93.Sato M, Ohashi T. Biorheological views of endothelial cell responses to mechanical stimuli. Biorheology. 2005;42:421–441. [PubMed] [Google Scholar]
- 94.Young EWK, Simmons CA. Macro- and microscale fluid flow systems for endothelial cell biology. Lab Chip. 2010;10:143–160. doi: 10.1039/b913390a. [DOI] [PubMed] [Google Scholar]
- 95.Song JW, et al. Microfluidic endothelium for studying the intravascular adhesion of metastatic breast cancer cells. PLoS ONE. 2009;4:e5756. doi: 10.1371/journal.pone.0005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Günther A, et al. A microfluidic platform for probing small artery structure and function. Lab Chip. 2010;10:2341–2349. doi: 10.1039/c004675b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lam MT, et al. Microfeature guided skeletal muscle tissue engineering for highly organized 3-dimensional free-standing constructs. Biomaterials. 2009;30:1150–1155. doi: 10.1016/j.biomaterials.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 98.Shimizu K, et al. Alignment of skeletal muscle myoblasts and myotubes using linear micropatterned surfaces ground with abrasives. Biotechnol. Bioeng. 2009;103:631–638. doi: 10.1002/bit.22268. [DOI] [PubMed] [Google Scholar]
- 99.Serena E, et al. Soft substrates drive optimal differentiation of human healthy and dystrophic myotubes. Integr. Biol. 2010;2:193–201. doi: 10.1039/b921401a. [DOI] [PubMed] [Google Scholar]
- 100.Kaji H, et al. Electrically induced contraction of C2C12 myotubes cultured on a porous membrane-based substrate with muscle tissue-like stiffness. Biomaterials. 2010;31:6981–6986. doi: 10.1016/j.biomaterials.2010.05.071. [DOI] [PubMed] [Google Scholar]
- 101.Ishibashi T, et al. Localized electrical stimulation to C2C12 myotubes cultured on a porous membrane-based substrate. Biomed. Microdev. 2009;11:413–419. doi: 10.1007/s10544-008-9247-7. [DOI] [PubMed] [Google Scholar]
- 102.Tourovskaia A, et al. Localized acetylcholine receptor clustering dynamics in response to microfluidic focal stimulation with agrin. Biophys. J. 2008;95:3009–3016. doi: 10.1529/biophysj.107.128173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sung JH, et al. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab Chip. 2010;10:446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- 104.Imura Y, et al. Micro total bioassay system for ingested substances: assessment of intestinal absorption, hepatic metabolism, and bioactivity. Anal. Chem. 2010;82:9983–9988. doi: 10.1021/ac100806x. [DOI] [PubMed] [Google Scholar]
- 105.Zhang C, et al. Towards a human-on-chip: culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip. 2009;9:3185–3192. doi: 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- 106.Esch M, et al. The role of body-on-a-chip devices in drug and toxicity studies. Annu. Rev. Biomed. Eng. 2010;13:55–72. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]
- 107.Baker M. A living system on a chip. Nature. 2011;471:661–665. doi: 10.1038/471661a. [DOI] [PubMed] [Google Scholar]
- 108.Borenstein J, et al. Functional endothelialized microvascular networks with circular cross-sections in a tissue culture substrate. Biomed. Microdev. 2010;12:71–79. doi: 10.1007/s10544-009-9361-1. [DOI] [PubMed] [Google Scholar]
- 109.Hernández Vera R, et al. Interstitial fluid flow intensity modulates endothelial sprouting in restricted Src-activated cell clusters during capillary morphogenesis. Tissue Eng. Pt. A. 2009;15:175–185. doi: 10.1089/ten.tea.2007.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kelm JM, et al. A novel concept for scaffold-free vessel tissue engineering: self-assembly of microtissue building blocks. J. Biotechnol. 2010;148:46–55. doi: 10.1016/j.jbiotec.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 111.Chao P, et al. Evaluation of a microfluidic based cell culture platform with primary human hepatocytes for the prediction of hepatic clearance in human. Biochem. Pharmacol. 2009;78:625–632. doi: 10.1016/j.bcp.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nedachi T, et al. Contractile C2C12 myotube model for studying exercise-inducible responses in skeletal muscle. Am. J. Physiol.-Endoc. M. 2008;295:1191–1204. doi: 10.1152/ajpendo.90280.2008. [DOI] [PubMed] [Google Scholar]
- 113.Cimetta E, et al. Production of arrays of cardiac and skeletal muscle myofibers by micropatterning techniques on a soft substrate. Biomed. Microdev. 2009;11:389–400. doi: 10.1007/s10544-008-9245-9. [DOI] [PubMed] [Google Scholar]
- 114.Hancock MJ, et al. Surface-tension-driven gradient generation in a fluid stripe for bench-top and microwell applications. Small. 2011;7:892–901. doi: 10.1002/smll.201002088. [DOI] [PMC free article] [PubMed] [Google Scholar]