Abstract
Background
We previously reported that iron chelators inhibit TNFα-mediated induction of VCAM-1 in human dermal microvascular endothelial cells. We hypothesized that iron chelators mediate inhibition of VCAM-1 via inhibition of iron-dependent enzymes such as those involved with oxygen sensing and that similar inhibition may be observed with agents which simulate hypoxia.
Objective
We proposed to examine whether non-metal binding hypoxia mimetics inhibit TNFα-mediated VCAM-1 induction and define the mechanisms by which they mediate their effects on VCAM-1 expression.
Methods
These studies were undertaken in vitro using immortalized dermal endothelial cells, western blot analysis, ELISA, immunofluorescence microscopy, quantitative real-time PCR, and chromatin immunoprecipitation.
Results
Hypoxia and the non-iron binding hypoxia mimetic dimethyl oxallyl glycine (DMOG) inhibited TNFα-mediated induction of VCAM-1. DMOG inhibition of VCAM-1 was dose-dependent, targeted VCAM-1 gene transcription independent of NF-κB nuclear translocation, and blocked TNFα-mediated chromatin modifications of relevant elements of the VCAM-1 promoter. Combined gene silencing of both HIF-1α and HIF-2α using siRNA led to a partial rescue of VCAM expression in hypoxia mimetic-treated cells.
Conclusion
Iron chelators, non-metal binding hypoxia mimetics, and hypoxia all inhibit TNFα-mediated VCAM-1 expression. Inhibition is mediated independent of nuclear translocation of NF-kB, appears to target TNFα-mediated chromatin modifications, and is at least partially dependent upon HIF expression. The absence of complete VCAM-1 expression rescue with HIF silencing implies an important regulatory role for an Fe(II)/α-ketoglutarate dioxygenase distinct from the prolyl and asparagyl hydroxylases that control HIF function. Identification of this dioxygenase may provide a valuable target for modulating inflammation in human tissues.
Introduction
Cytokine inducible cell adhesion molecules (CAMs) on vascular endothelium comprise a tightly regulated family of cell-surface proteins, which mediate leukocyte adhesion to endothelial cells and subsequent diapedesis into the extravascular tissue compartments, including the skin. One specific vascular CAM, vascular cell adhesion molecule (VCAM)-1 is induced by a limited set of cytokines including TNFα and binds to the α4β1integrin, which is present on non-neutrophilic leukocytes [1].
VCAM-1 is robustly expressed on dermal endothelium in numerous inflammatory skin disorders, including psoriasis, atopic dermatitis, and delayed type hypersensitivity, underscoring its integral role in inflammatory processes [2, 3]. Pharmacologic disruption of the α4β1integrin/VCAM-1 interaction blunts inflammatory responses in multiple animal models of cutaneous disease as well as chronic inflammatory states such as rheumatoid arthritis, inflammatory bowel disease, and asthma [4]. The clinical potential of modulating the α4β1integrin/VCAM-1 interaction has been further highlighted by recent clinical trials demonstrating the efficacy of the anti-α4β1integrin monoclonal antibody, natalizumab, in the treatment of relapsing multiple sclerosis [5, 6] as well as Crohn’s disease [7].
Iron chelators potently inhibit TNFα-mediated VCAM-1 protein expression in human dermal endothelial cells (HDMEC) through a marked reduction in VCAM-1 gene transcription [8]. However, iron chelators do not inhibit TNFα-induced NF-kB activation, nuclear translocation, or the ability of nuclear localized nuclear factor kB (NF-kB) complexes to bind to relevant NF-kB binding oligonucleotides, all previously characterized critical regulators of VCAM-1 induction.
One possible target for iron chelators is hypoxia inducible factor (HIF), the central transcription factor implicated in coordinating the cascade of events involved in cellular adaptation to hypoxia [9]. HIF is a heterodimer of a constitutively expressed β subunit and one of three tightly regulated alpha subunits (HIF-1α, 2α, or 3α). HIF-1α and 2α are hypoxia inducible proteins. Under normoxic conditions, prolyl hydroxylases (PHD) hydroxylate both HIF-1α and HIF-2α at specific proline residues enabling ubiquitination and consequent proteasomal degradation. PHDs are oxygen, Fe(II), and α-ketogluterate (α-KG)-dependent and become enzymatically inactive in the absence of any one of these factors [10]. In the absence of any one of the cofactors unhydroxylated HIF-1α/2α escapes destruction and translocates to the nucleus where it affects transcription of hypoxia responsive genes.
It is possible that iron chelators may alter the function of iron- and oxygen-dependent enzymes by sequestering required iron. We therefore hypothesized that iron chelators may mediate anti-inflammatory effects by a novel pathway, functioning as hypoxia mimetics. In this study, we report the abrogation of TNFα-mediated VCAM-1 expression in HDMEC by both hypoxia and a hypoxia mimetic mechanistically distinct from metal chelators, the α-KG antagonist dimethyl oxallyl glycine (DMOG). These agents appear to target TNFα-mediated VCAM-1 gene transcription via inhibition of chromatin modification. Furthermore, using silencing RNA we demonstrate that HIF isoforms are partially responsible for this phenomenon, affirming a novel link between VCAM-1 expression and the hypoxic cellular response pathways, but also implicating other as yet undefined pathways in the transcriptional regulation of this important inflammatory mediator.
Materials and methods
Cell culture
All studies were done in the 5A32 endothelial cell line [11] derived from primary HDMEC immortalized with SV40 Large T which were subcultured as described previously [12]. HDMEC were cultured under continuous reduced oxygen conditions (1%) using a Coy O2 Control Isolation Glovebox (Coy Laboratory Products, Grass Lake, MI). Cell viability was confirmed with trypan blue exclusion.
Reagents and antibodies
DP was obtained from Sigma. DMOG was obtained from Cayman Chemical (Ann Arbor, MI). Recombinant human TNFα and TGFα were purchased from R & D Systems Inc. (Minneapolis, MN). Monoclonal antibodies against VCAM-1 (6G10 or P3C4) and ICAM-1 (P2A4), used in cellular ELISA, were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa (Iowa City, IA). For western blots, Santa Cruz Biotechnology (Santa Cruz, CA) supplied antibodies against VCAM-1 (sc-1809), egr-1 (sc-189), and IRF-1 (sc-497). Novus Biologicals (Littleton, CO) provided antibodies specific for HIF-1α (100-105) and HIF-2α (100-132). Immunocytochemistry involved an anti-p65 antibody (K59165R) from Biodesign International (Saco, Maine).
Cellular ELISA
Expression of VCAM-1 was assessed using a cell-based ELISA system as previous described [13]. All experiments were performed in triplicate, with data from a representative experiment presented. Error bars reflect the average absorbance of 4 wells ± SD.
Western Blot
Cytosolic and nuclear extracts were prepared with Pierce’s NE-PER nuclear extraction kit (Rockford, IL) according to the manufacturer’s protocol. Whole-cell extracts were prepared by cell lysis in a single detergent extraction buffer, and all extracts were electrophoresed by SDS-PAGE under reducing conditions and transferred to nitrocellulose membranes as previously described [11]. Membranes were blocked for 1 hour with TBS-tween/5% non-fat dry milk, and then incubated with primary antibody overnight (egr-1 1:1000, HIF-1α 1:250, HIF-2α 1:500, IRF-1 1:500, VCAM-1 1:200). Membranes were washed with TBS-tween and then incubated with the appropriate HRP-conjugated secondary antibody for 1 hour. After additional washing, immunogen levels were assayed with enhanced chemiluminescence (ECL reagent, GE Healthcare, Piscataway, NJ). Densitometry analysis was performed using GNU Image manipulation software (GIMP).
Quantification of mRNA and hnRNA levels
HDMEC cells were treated with DP or DMOG for 6 hr followed by 4 hr of 1000 U/mL TNFα. Total RNA was isolated with the Qiagen RNeasy Mini Kit (Valencia, CA) with inclusion of a DNase step using Qiagen RNase-free DNase. DNA-free RNA was then reverse transcribed with SuperScript® II Reverse Transcriptase (Invitrogen, Carlsbad, CA) following the random primer protocol. RT-PCR was performed with Sybr green chemistry (Power SYBR® Green PCR Master Mix, Applied Biosystems, Foster City, CA) on a 7500 Fast Real-Time PCR System (Applied Biosystems). The fold difference in mRNA expression relative to untreated cells was calculated employing the ΔΔCT method with 18S as the endogenous control as described by the manufacturer. Real-time-PCR reactions were conducted in triplicate. All experiments were performed at least three times with the data from a representative experiment presented. The error in the calculation of fold difference was determined as described by the manufacturer. hnRNA levels were determined using primers which amplify intronic sequences as described previously [8]. Primer sequences available upon request.
Immunocytochemistry
5A32s were grown on glass cover slips in 12 well plates. Cells were treated with DP or DMOG for 16 hours followed by TNF-α at 1000 U/mL for 30 min. The cover slips were washed in ice-cold PBS and then cells were fixed with 3.7% paraformaldehyde for 7 min followed by permeabilization with 0.1% Triton X-100 in PBS for 5 min. The cover slips were washed again before blocking with 10% FBS in PBS. Next, they were incubated with anti-p65 antibody (1:300) for 30 min. The primary was removed with 3 washes of PBS and then Alexa Fluor 488 goat anti-rabbit (1:3000) (Invitrogen) was added for 30 minutes. After additional washing, the cover slips were mounted on glass slides using Prolong Gold mounting media (Invitrogen) and then fluorescence was visualized with a Leica DMR-E fluorescence microscope equipped with a Hammamatsu Orca camera.
siRNA Studies
HDMEC were reverse transfected with either 50 nM HIF-1α siRNA (Dharmacon, Lafayette, CO), 50 nM HIF-2α siRNA (Dharmacon), or both siRNAs (30 nM of each) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol and plated into either 12 well plates (mRNA quantification) or 10 cm dishes (protein analysis). After 24 hr, the media was changed and DMOG added to the appropriate cells. For mRNA studies, after 6 hr, TNFα was added to the designated cells and all cells were harvested 4 hr later (34 hr after siRNA transfection). RNA was isolated and quantified by RT-PCR as described above. Each siRNA transfection condition was performed in duplicate, and RT-PCR reactions were performed in triplicate. For protein, cells were exposed to DMOG for 6 hr and TNFα for 16 hr (44 hr total from transfection). Cytosolic and nuclear extracts were obtained and western blot performed as described above.
Chromatin Immunoprecipitation (ChIP)
HDMEC cells were grown in 150 mm dishes (3 dishes/treatment, ~4 × 107 cells); at time of experimentation cells were 90-100% confluent. Cells were pretreated with DP (500 μM), DMOG (125 μM), or diluent for the times indicated; TNFα (500 U/mL) was added to control (media-only), diluent-, and hypoxia mimetic-treated cells for an additional 6 hr. Cells were cross-linked using 0.5% formaldehyde for 10 min at room temperature with gentle shaking and chromatin was isolated as previously described [14, 15]. Chromatin was sheared to ~600 bp fragments using the Misonex Sonicator 3000 with cuphorn (Misonex, Farmingdale, NY). Immunoprecipitations were performed overnight at 4°C by adding 5 μg of anti-acetyl Histone H3 (Millipore, Billerica, MA), anti-trimethyl Histone H3, K4 (Millipore), or anti-HA probe (Y-11) (nonspecific, Santa Cruz Biotechnology) to 50 μL of chromatin in 1 mL dilution buffer [14]. Complexes were isolated using Protein A-Sepharose beads (Millipore, 60 μL) and washed as previously described [15]. Following the reversal of the formaldehyde cross-links, DNA was precipitated, extracted, and finally dissolved in 30 μL of water as previously reported [14]. The resulting samples were subjected to RT-PCR (3 μL/well in triplicate) as described above; input (chromatin supernatant retained from nonspecific antibody samples) was diluted 1:30 prior to RT-PCR. DNA was quantified against a standard curve generated from HDMEC genomic DNA. Percent inhibition was determined relative to TNFα in the absence of hypoxia mimetics; statistical analysis was performed using a two-tailed paired Student’s t test. Primer sequences encompassing the VCAM-1 promoter available upon request.
Results
DMOG and Hypoxia inhibit TNFα-mediated expression of VCAM-1 protein in HDMEC
In the presence of an inflammatory stimulus such as TNFα, endothelial VCAM-1 expression rises dramatically in HDMEC (Figure 1a). Our previous studies had demonstrated that iron chelators, such as 2,2′-dipyridyl (DP) and desferoxamine, were potent inhibitors of this induction [8]. Because iron chelators inhibit enzymes involved in the HIF degradation pathway, iron chelators and hypoxia induce a similar constellation of genes in cultured cells. We hypothesized that the mechanism underlying iron chelator-mediated inhibition of VCAM-1 induction involves genes and/or gene products common to the cellular hypoxic response.
Figure 1. Hypoxia and hypoxia mimetics inhibit TNFα-induced VCAM-1.

(a) VCAM-1 expression (by ELISA) on HDMEC, pretreated as indicated with DMOG either 6 or 16 hr prior to TNFα (b) HDMEC pretreated with DMOG or DP for 6 hr followed by TNFα were assessed for VCAM-1 expression by western analysis (c) HDMEC were pretreated with DMOG (500 μM) for times ranging from 6 hr to 2 hr after (−2 hr) TNFα stimulation and assayed for VCAM-1 (d) Pretreatment with FG-4497 prior to TNFα stimulation (e) HDMEC were stimulated with TNFα, hypoxia alone (24 hr), hypoxia (8 hr) followed by TNFα under normoxic conditions, hypoxia (8 hr) followed by TNFα under continuous hypoxia, and TNFα under hypoxia without hypoxia pretreatment. Values above bands represent the relative density as compared to lane 2 adjusted for protein loading. (All TNFα 1000 U/mL × 16 hours – representative experiment of three experiments)
Cellular responses to hypoxia are primarily under the control of the HIF family of transcription factors. HIF stability is regulated by PHD, and in the absence of oxygen, Fe(II), or α-KG, HIF escapes proteasomal degradation and becomes transcriptionally active. To elucidate the possible role of hypoxia inducible pathways in the regulation of VCAM-1 regulation, we examined the effects of an unrelated, non-metal binding hypoxia mimetic known to stabilize HIF, the α-KG antagonist DMOG.
HDMEC exposed to DMOG prior to TNFα administration showed a dose-dependent blockade of TNFα-induced VCAM-1 expression with marked inhibition at doses as low as 250 μM. At doses of ≥500 μM, VCAM-1 expression was reduced to levels observed in unstimulated cells (Figure 1a and b). Preincubation (6 hr) with DMOG prior to cytokine stimulation maximized the inhibitory effect, but submaximal inhibition was observed even without preincubation (Figure 1c). Partial inhibition was observed even if DMOG was added even two hours after TNFα stimulation was initiated (Figure 1 c – (−2 hr)). We also tested a proprietary, small molecule hypoxia mimetic, FG-4497 (FibroGen, South San Francisco, CA), which is a PHD inhibitor 10-100 fold more potent than DMOG and observed a similar dose-dependent inhibition of VCAM-1 induction (Figure 1d).
If the observed effects of iron chelators and α-KG antagonists were related to their capacity to inhibit oxygen-dependent enzymes, then similar inhibition should be observed in hypoxia-treated cells. As shown in Figure 1e, hypoxia (1% O2) markedly blunted TNFα-induced VCAM-1 in HDMEC. This inhibition required continuous exposure to hypoxia both before and during cytokine stimulation, consistent with involvement of an oxygen labile factor unstable in the presence of oxygen.
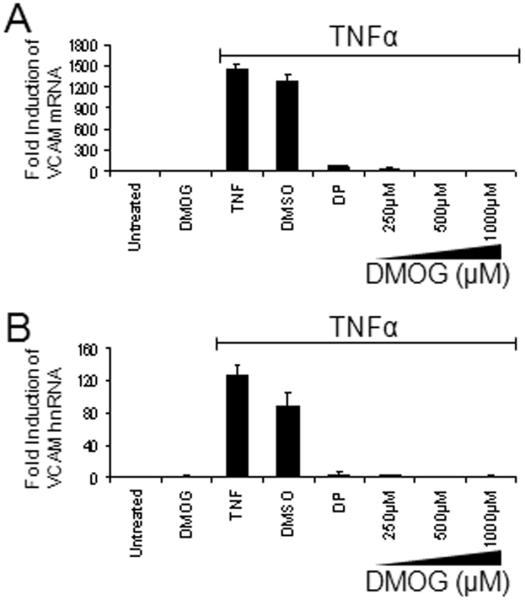
Hypoxia mimetics block TNFα induction of VCAM-1 mRNA and VCAM-1 gene transcription
To further define the mechanisms involved in the inhibition of TNFα-mediated VCAM-1 protein expression, we used quantitative reverse transcription real-time PCR (RT-PCR) to measure VCAM-1 mRNA levels in HDMEC pretreated with hypoxia mimetics. VCAM-1 mRNA levels rose abruptly four hours after TNFα stimulation. DMOG at all doses tested dramatically blocked TNFα induction of VCAM-1 mRNA suggesting that inhibition occurred prior to VCAM-1 translation (Figure 2a). To examine whether DMOG targets VCAM-1 gene transcription, we measured the expression of unspliced heterogenous nuclear RNA (hnRNA) directly by amplifying an intronic sequence within the VCAM-1 gene [8]. Again, dose-dependent inhibition of TNFα-mediated induction of hnRNA was observed (Figure 2b). FG-4497 produced comparable dose-dependent inhibition when employed in analogous experiments (Supplemental Figure 1). These results provide cogent evidence that hypoxia mimetics disrupt TNFα-mediated VCAM-1 gene transcription.
Figure 2. DMOG inhibits TNFα induction of VCAM-1 at a transcriptional level.
HDMEC were pretreated with increasing concentrations of DMOG 6 hr prior to stimulation with TNFα (1000 U/mL × 4 hr) and VCAM-1 (a) mRNA and (b) hnRNA were quantitated by real-time PCR.
TNFα-mediated NF-κB nuclear translocation is preserved in the presence of hypoxia mimetics
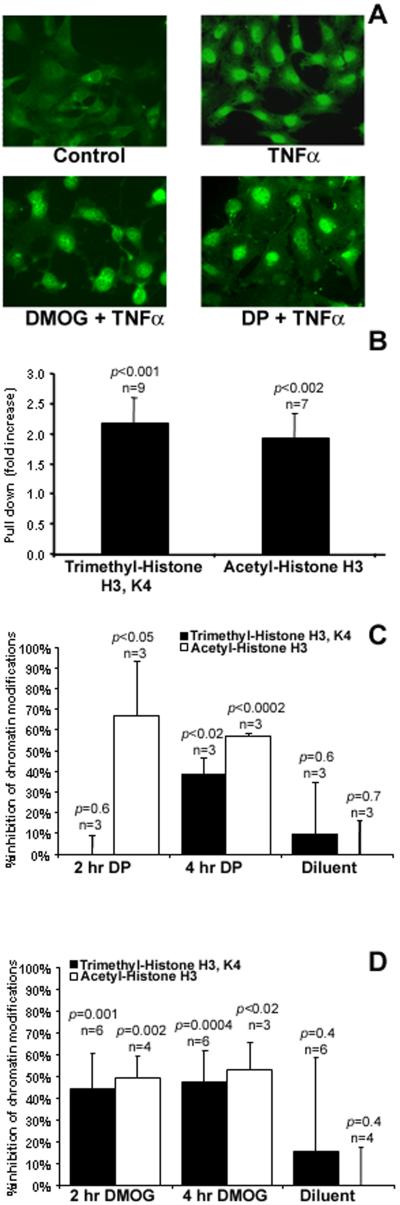
In order to further define the molecular mechanisms involved, we examined whether DMOG affected NF-κB activation and nuclear translocation. As reported previously, iron chelators failed to alter expression of p65 expression or influence the kinetics or magnitude of NF-κB activation or nuclear translocation [8]. Here we further demonstrated, using immunofluorescent staining for p65 in TNFα-treated HDMEC, that NF-κB nuclear translocation proceeds unabated in the presence of DMOG (Figure 3a). Thus, despite not binding iron, the effect of DMOG on VCAM-1 gene expression is comparable to iron chelators. Additionally, DMOG appears to have similar if not identical effects on NF-kB activation and nuclear translocation, all providing evidence that iron chelators and α-KG antagonists share similar mechanisms in modulating TNFα induction of VCAM-1.
Figure 3. DMOG does not inhibit TNFα-mediated nuclear translocation of NFκB, but inhibits TNFα-mediated chromatin modification.
(a) HDMEC were treated with TNFα (1000 U/mL × 30 min), then fixed and stained with antibodies against the p65 domain of NFκB. Bright nuclear NFκB staining persisted in DMOG (500 μM) and DP (125 μM) pretreated cells. (b) HDMEC treated with TNFα (500 U/mL × 6 hr) were evaluated by ChIP as described in materials and methods. TNFα treatment induced trimethylation and acetylation of histone H3 associated with VCAM-1 promoter elements. Pretreated with (c) DP (125 μM) or (d) DMOG (500 μM) as indicated inhibited TNFα-stimulated histone H3 modification. Diluent controls were PBS or DMSO for DMOG- and ethanol for DP-treated cells. (p-values calculated using paired Student’s two-tailed t-test).
NF-κB activation is an absolute prerequisite for TNFα-mediated VCAM-1 transcription in HDMEC [16]. However, optimal VCAM-1 gene induction requires the expression of interferon regulatory factor 1 (IRF-1), another TNFα inducible gene. IRF-1 has been shown to bind to the VCAM-1 promoter and augment transactivation synergistically with NF-κB [17]. In order to address its possible relevance to this phenomenon, we examined IRF-1 expression and nuclear translocation in hypoxia mimetic-pretreated, TNFα-stimulated HDMEC. We observed essentially no effect on IRF-1 expression or nuclear localization under conditions which completely abrogated VCAM-1 induction (Supplemental Figure 2a). These data supported the presence of a target other than IRF-1 expression as the primary mechanism for inhibition of VCAM-1 induction.
TNFα mediates VCAM-1 gene promoter modification which is inhibited by hypoxia mimetics
Because hypoxia mimetics blocked VCAM-1 gene transcription without affecting NF-kB activation and nuclear translocation, we hypothesized that the effects of these agents may target TNFα-induced chromatin modification. Under baseline conditions, promoter sequences in genes such as VCAM-1 are inaccessible to relevant transcription factors due to their tight association with histone proteins. Activation events not only induce the expression and nuclear localization of relevant transcription factors such as NF-kB, but also induce the modification of histones associated with key regulatory DNA sequences associated with specific promoter elements. We hypothesized that hypoxia mimetics such as iron chelators and α-KG antagonists such as DMOG might inhibit VCAM-1 induction by blocking TNFα-mediated chromatin modifications required to allow for transcription factor accessibility.
In order to address this possibility, we initially examined whether TNFα treatment of HDMEC resulted in chromatin modifications in regions of the VCAM-1 promoter, which we had previously demonstrated to be essential for TNFα-mediated gene activation. Consistent with this hypothesis, HDMEC treated with TNFα demonstrated increased acetylation and trimethylation of H3 histones associated with previously characterized TNFα responsive elements of the VCAM-1 promoter [18] as evidenced by increased pull-down of modified chromatin in TNFα treated cells. (Figure 3b). Control ChIP assays using rabbit anti-HA antibody failed to pull down chromatin (<5% of that observed with specific modified histone antibody). Furthermore, pretreatment of HDMEC with either DP or DMOG prior to TNFα inhibited both H3 histone acetylation and trimethylation (Figures 3c and d), consistent with a histone modification target for hypoxia mimetic agents. These data support the hypothesis that chromatin modification is a target for hypoxia mimetic-mediated inhibition of VCAM-1 gene expression.
The effect of hypoxia mimetics is pathway specific
Hypoxia is known to globally suppress metabolism as part of the cell’s energy conservation program [19, 20]. To investigate the possibility that VCAM-1 inhibition by hypoxia mimetics is due to this non-specific hypoxia effect, we examined the expression of other cytokine inducible genes in HDMEC. Transforming growth factor-α (TGFα) induces de novo synthesis of the transcription factor early growth response protein-1 (egr-1) in HDMEC through a signaling cascade that is distinct from the TNFα/VCAM-1 pathway. The egr-1 promoter lacks NF-κB and IRF-1 consensus binding elements, p65 does not translocate to the nucleus in HDMEC treated with TGFα, and TGFα fails to induce IRF-1 in HDMEC (data not shown). We treated HDMEC with or without hypoxia mimetics prior to TGFα stimulation and assayed for egr-1 expression. Egr-1 was rapidly and robustly expressed even in the presence of DMOG and DP (Supplemental Figure 2b). These data demonstrate that hypoxia mimetics do not globally inhibit all cytokine-induced cellular transcription and translation.
HIF stabilization is necessary, but not sufficient for VCAM-1 transcriptional inhibition
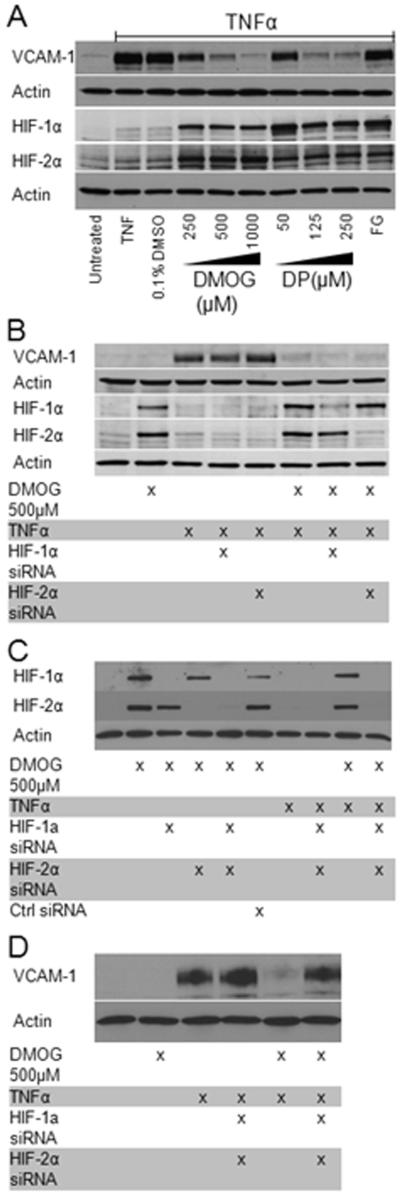
The best characterized targets for oxygen regulated transcriptional mechanisms are hypoxia inducible factors (HIF) 1α and 2α. HIF-1α and HIF-2α are transcription factors that rapidly accumulate in cells exposed to hypoxia or hypoxia mimetics, raising the possibility that these proteins may be involved in the transcriptional repression of VCAM reported above. HIF-1α and 2α are unambiguously demonstrable in nuclear extracts from HDMEC incubated with DP or DMOG at the doses employed in this study (Figure 4a), and the HIF signal persists up to 40 hr if hypoxia mimetic exposure is maintained (data not shown).
Figure 4. HIF stabilization is necessary but not sufficient for inhibition of VCAM-1 protein expression.
(a) HDMEC were pretreated with increasing doses of DMOG and DP (6 hr) prior to TNFα (1000 U/mL × 16 hr). Cytosolic and nuclear extracts were assayed for VCAM and HIF expression, respectively, by western blot. (b) Either HIF-1α or Hif-2α was effectively silenced in separate siRNA experiments (see materials and methods). The knockdown of each isoform independently did not influence DMOG (500 μM) inhibition of VCAM-1. (c) HDMEC treated with both HIF-1α and HIF-2α siRNAs exhibited near complete ablation of both isoforms (last lane). In these double knockdown experiments, partial rescue of VCAM-1 expression was observed (last lane in (d)).
It is possible that high nuclear concentrations of HIF might be impeding TNFα-mediated VCAM-1 transcription by competing (either directly or indirectly through one or more of the HIF target genes) with NF-κB for critical coactivators or other elements of the general transcriptional machinery necessary for chromatin accessibility. However, in all of our studies of hypoxia mimetic dose responses, we failed to observe a strong correlation between HIF stabilization and VCAM-1 inhibition. In particular we observed that HIF protein levels are not increased by escalating the DMOG dose above 250 μM while VCAM-1 inhibition is maximal at higher DMOG concentrations. Similarly, HIF stabilization is most prominent at 50 μM of DP, but VCAM-1 expression is still significant at this modest dosage (Figure 4a). Most strikingly, the more potent PHD inhibitor FG-4497 was capable of inducing HIF expression at low concentrations (5-20 μM) without having any effect on VCAM-1 induction (Figure 4a, last lane).
To further scrutinize the role of HIF in hypoxia mimetic inhibition of VCAM-1 induction, we used an siRNA approach to specifically block the expression of either HIF-1α or 2α independently or together. Knockdown of either HIF-1α or HIF-2α prior to DMOG treatment was straightforward and effective at blunting HIF protein expression but had no effect on TNFα-mediated induction of VCAM-1 (Figure 4b).
Since HIF-1α and HIF-2α demonstrate overlapping activities, we reasoned that knockdown of both proteins might be required to see an effect on VCAM-1 induction. However, treatment with combinations of HIF-1α and HIF-2α siRNAs demonstrated unanticipated effects making complete knockdown technically very difficult. Simultaneous use of both HIF-1α and HIF-2α siRNAs at concentrations and times previously shown to be sufficient to knock out protein expression when used singly, resulted in rescue of expression of HIF-1α protein (data not shown). This was potentially due to suppressed expression of HIF destabilizing enzymes such as the factor inhibiting HIF (FIH-1) and HIF-PHDs [21] which are under the transcriptional control of HIF, ultimately resulting in increased HIF-1α protein stability. Eventually, we were able to define conditions where combinations of HIF-1α and HIF-2α siRNAs markedly decreased protein levels of both HIF isoforms. HIF gene silencing was highly effective in reducing HIF protein and mRNA levels in DMOG treated HDMEC. DMOG alone suppressed HIF-1α mRNA levels 60-70%, but abundant HIF protein persisted in the absence of siRNA. The addition of HIF siRNA to DMOG-treated HDMEC led to 95% reductions in the mRNA of both HIF isoforms with commensurate decreases in detectable protein (Figure 4c and 5a).
If HIF represented the mechanistic link behind our observed phenomenon, then one would predict that TNFα-mediated VCAM-1 expression, normally inhibited by DMOG, could be rescued by transfecting HIF siRNA prior to DMOG administration. When both HIF-1α and HIF-2α were silenced in the same cells, partial rescue of TNFα-induced VCAM protein expression was observed (Figure 4d). This was paralleled by partial rescue at the mRNA level (Figure 5b). These results suggest a redundancy in function between the two major HIF isoforms in HDMEC and that HIF plays a necessary but not sufficient role in mediating hypoxia mimetic inhibition of TNFα-stimulated VCAM-1.
Figure 5. HIF stabilization is necessary but not sufficient for VCAM-1 transcriptional inhibition.
Total RNA was isolated from HDMEC in siRNA experiments paralleling those illustrated in Figure 4c-d (see materials and methods). HIF and VCAM mRNA expression was determined by quantitative real-time PCR. (a) Dual treatment with HIF-1α and HIF-2α siRNA resulted in >95% reduction in mRNA levels of both isoforms. (b) DMOG (500 μM) pretreatment prior to TNFα (1000 U/mL × 16 hr) reduced VCAM induction by 86% (*p<0.05). Dual HIF1α/2α knockdown prior to addition of DMOG increased VCAM expression 2.4 fold (*p<0.05), but VCAM mRNA levels remained less than half of that measured in TNFα- (1000 U/mL × 16 hr) stimulated, HIF 1α/2α silenced HDMEC not exposed to DMOG (*p<0.05). (p-values calculated using paired Student’s two-tailed t-test).
Discussion
Using both hypoxia and pharmacological agents that simulate the hypoxic cellular response, we report the inhibition of TNFα-mediated VCAM-1 gene expression. This work is an extension of our previous studies that examined the effects of pharmacological agents on CAM induction, initially with the hypothesis that these agents acted through their effects on oxygen radicals. While this may still be the case with select agents, many inhibitory agents were also potent metal chelators, and antioxidants that lacked metal chelating properties were inefficient CAM inhibitors [8]. Metals such as iron may serve as catalysts for generation of free radicals and metal chelating agents could blunt this process. We hypothesized that metal chelating agents, particularly iron binders, may have effects on VCAM-1 induction by affecting crosstalk between TNFα signaling pathways and pathways induced by hypoxia and hypoxia mimetics.
Consistent with our hypothesis, DMOG and FG-4497, non-metal binding hypoxia mimetics, blocked TNFα-mediated VCAM-1 induction. These data suggested that previous studies demonstrating inhibition of VCAM-1 induction by iron chelating agents might have actually targeted pathways regulated by hypoxia [8]. Our studies demonstrating a comparable effect with non-metal binding hypoxia mimetics as well as hypoxia alone supported this possibility.
In our model, we detected no effect of hypoxia mimetics alone on NF-kB activation in endothelial cells as measured by p65 nuclear translocation or on NF-kB-mediated gene expression as measured by induction of VCAM-1 by hypoxia or hypoxia mimetics alone. Our observation of hypoxia mimetic-mediated inhibition of VCAM-1, whose TNFα-induced expression is dependent on NF-κB transactivation, clearly showed that hypoxia and hypoxia mimetics can, in some cell types and contexts, have profound suppressive effects on NF-kB mediated gene expression. These effects on NF-kB-dependent gene induction are not entirely consistent with previous studies showing that, under certain circumstances, hypoxia actually facilitates NF-kB activation. This discrepancy may be due to the models used or to alternative interpretation(s) of the data generated.
Previous studies have examined effects of hypoxia on the expression of select NF-kB subunits or regulatory elements, noting that hypoxia may change the expression of select proteins in specific cells such as neutrophils [22, 23]. In addition, the inhibitor of κB kinase β (IKKβ) possesses a putative prolyl hydroxylation domain, and it has been proposed, though not directly demonstrated, that hypoxia suppresses prolyl hydroxylation of IKKβ, activating the IKK complex and potentiating NFκB activity [24].
Several other studies have examined the effects of hypoxia on NF-κB-dependent genes, including VCAM-1. Yamaji-Kegan et al. found that hypoxia induces VCAM-1 expression in whole mouse lung, but provided no evidence of a direct effect of hypoxia on endothelial cell VCAM-1 induction [25]. Conversely, Yamashita et al. provided evidence that HIF-2α directly regulates VCAM-1 transcription in endothelial cells [26], although the levels of mRNA induction documented were in the 2-5 fold range in contrast to the over thousand fold inductions we observed with TNFα in our model. Neither of these studies directly examined TNFα-induced VCAM-1 gene expression, making their observations not entirely relevant to our report. Finally, other models have examined the effect of hypoxia on the expression of NF-kB-dependent transgenes [27]. Understanding tissue specific expression of genes, particularly endothelial cell-associated inducible CAMs such as VCAM-1, likely requires models which can probe tissue specific expression and chromatin remodeling. Transgenes driven by NF-kB outside the context of chromatin packaging may not provide insights relevant to VCAM-1 regulation in endothelial cells [23].
The demonstration that three distinct mechanisms, hypoxia, iron chelation, and α-KG antagonism, all repress VCAM expression strongly implicates an Fe(II)/α-KG dioxygenase, such as the HIF-PHDs, in VCAM regulation. Hypoxia mimetics, such as DMOG and iron chelators, simulate hypoxia through the inhibition of the PHDs involved in the modification and degradation of HIF. A role for HIF is supported by gene silencing experiments which resulted in partial restoration of VCAM-1 induction. Of note, silencing of both transcriptionally active isoforms of HIF was necessary to observe this rescue. Presumably, each isoform can fully compensate for the absence of the other in HDMEC at least vis-à-vis VCAM-1 inhibition.
While it is conceivable that VCAM-1 inhibition is wholly HIF dependent and full rescue would have been observed if it were possible to completely eradicate HIF expression, the persistent, though attenuated, hypoxia mimetic-mediated VCAM inhibition observed in HIF knockdown experiments implies that other regulatory mechanisms in addition to HIF may be relevant. HIF expression is necessary but not sufficient for hypoxia mimetic-mediated inhibition of VCAM-1 induction. This contention is further strengthened by the observation that hypoxia mimetics induced robust HIF expression at doses lower than those required to inhibit TNFα-mediated VCAM-1 induction.
There is some evidence that HIF-PHDs may have alternative targets. Cummins et al. reported a role for the HIF-PHDs and specifically PHD1 in the regulation of NF-κB mRNA expression [27]. In HeLa cells both pharmacologic inhibition and gene silencing of PHD1 increased NF-κB activity as assessed by reporter gene assays and increased expression of cyclooxygenase-2. Silencing of HIF-1α failed to alter this potentiation of NF-κB, but HIF independency was not definitively established as HIF-2α was not concomitantly silenced [27]. HIF-2α is induced by hypoxia and is transcriptionally active in HeLa cells [28]. Our studies should also highlight a particular caution when attempting to manipulate HIF-1α or HIF-2α individually since they have substantial functional overlap and it is highly likely that suppression of either one singly will have profound effects on the other.
The possibility that HIF-PHDs regulate cellular pathways via hydroxylation of non-HIF substrates remains likely, but is not well characterized. Additional candidate Fe(II)/α-KG-dependent enzymes exist that share these requisite cofactors and participate in transcriptional regulation. The Fe(II)/α-KG-dependent asparagyl hydroxylase, FIH-1 modifies HIF domains that interact with the transcriptional coactivator p300; inhibition of FIH-1enhances HIF transcriptional activity by increasing its affinity for p300. FIH-1 has been more recently shown to hydroxylate the NF-κB precursor p105 and the NF-κB cytosolic sequestrant, IκBα [29], but the functional consequence of these posttranslational modifications has yet to be established.
In addition to enzymes associated with HIF, there is a large superfamily of Fe(II)/α-KG-dependent dioxygenases, called cupins, whose biochemical functions have been described in plants, bacteria, and yeast [30, 31]. In humans, histone lysyl demethylase activity has been ascribed to members of the jumonji subgroup of cupins. These proteins possess the jumonji C (JmjC) catalytic domain and are closely related phylogenetically to FIH-1. The few described JmjC domain-containing histone demethylases are capable of specifically demethylating the lysine residues, H3K4, H3K9, H3K27, and H3K36 [32]. These proteins are inhibited by α-KG antagonists and iron chelators [33]. Therefore, a class of histone demethylases that are regulated by hypoxia and hypoxia mimetics represents an intriguing discovery that may provide a mechanism for the effects of pan-hydroxylase inhibitors described in this paper.
Indeed, the role of jumonji proteins in transcriptional upregulation has been substantiated by siRNA experiments with the H3K9 demethylase, JHDM2A. Knockdown of JHDM2A blocked androgen-mediated H3K9 demethylation associated with androgen target genes resulting in a concurrent and marked reduction in gene expression [34]. Within skin, the corepressor hairless has a cupin metalloenzyme domain and has been implicated in chromatin remodeling related to vitamin D responsiveness [35].
These observations raise fundamental questions regarding the effect of oxygen tension on inflammation. Until recently, evidence supported almost exclusively the pro-inflammatory role for hypoxia. However, our studies as well as other recent publications suggest that the role for hypoxia may be much more nuanced with both pro- and anti-inflammatory influences [36, 37]. While hypoxia may augment pro-inflammatory responses mediated via innate immune elements such as neutrophils [38], hypoxia may inhibit pro-inflammatory functions of T cells [39]. This is consistent with previous observations from our laboratory showing that hypoxia mimetics blunt interferon signaling by downregulating the interferon gamma receptor and interferon-induced signaling [11].
Thus, depending upon context, duration, and other modifying factors, low oxygen tension may have both pro- and anti-inflammatory influences. Extending our understanding of the role of oxygen tension in the regulation of inflammation will require improved characterization of the likely extensive targets of oxygen regulated genes. It may also require redefining what hypoxia really means since oxygen tensions in tissues may show tremendous variability, even under non-pathological conditions [37, 40, 41]. Our data would suggest that variations in oxygen tensions across various vascular beds; arterial, venous, or lymphatics may either favor or blunt expression of specific genes such as VCAM-1.
In summary, we have expanded upon our prior report that iron chelators inhibit TNFα-mediated induction of VCAM-1 expression in HDMEC by linking this observation to a more general inhibition of VCAM-1 by hypoxia and hypoxia mimetics. Contrary to the prevailing paradigm of hypoxia as an NF-κB activator and inflammatory stimulant, we provide strong evidence that hypoxia and hypoxia mimetics block the expression of an indispensable participant in inflammatory responses. Using gene silencing experiments, we demonstrated that this inhibition is partially mediated by HIF. However, HIF does not appear to be fully responsible for this phenomenon implicating the participation of an Fe (II)/α-KG dependent dioxygenase that is possibly distinct from the HIF-PHDs. Although specific targets have yet to be defined, the discovery of this enzyme may provide not only a more complete understanding of the cellular hypoxic response but also a novel target for anti-inflammatory agents.
Supplemental Figure 1: FG-4497 inhibits TNFα-induced VCAM-1 in a concentration dependent manner: (a) VCAM-1 expression (by ELISA) on HDMEC, pretreated as indicated with FG-4497 either 6 or 16 hr prior to TNFα. (b) HDMEC pretreated with DMOG or DP for 6 hr, or FG-4497 for 6 or 16 hr, followed by TNFα ( 1000 U/mL × 4 hr) were assessed for VCAM-1 expression by western analysis. FG-4497 inhibits TNFα induction of VCAM-1 at a transcriptional level. HDMEC were pretreated with DP (125 μM), DMOG (1 mM) or increasing concentrations of FG-4497 6 hr prior to stimulation with TNFα (1000 U/mL × 4 hr) and VCAM-1 (c) mRNA and (d) hnRNA were quantitated by real-time PCR.
Supplemental Figure 2: DMOG treatment does not inhibit induction of IRF-1 or egr-1 by TNFα or TGFα in HDMEC: (a) HDMEC were pretreated with increasing concentrations of DMOG 6 hr prior to stimulation with TNFα (1000 U/mL × 16 hr). Induction of IRF-1 was not affected by hypoxia mimetics. (b) HDMEC were pretreated with increasing concentrations of DMOG 6 hr prior to stimulation with TGFα (50 ng/mL × 4 hr). Induction of the TGFα-responsive gene egr-1 was maintained indicating a degree of specificity to the hypoxia mimetic-effect on VCAM.
Supplementary Material
Supplemental Figure 1: FG-4497 inhibits TNFα-induced VCAM-1 in a concentration dependent manner: (a) VCAM-1 expression (by ELISA) on HDMEC, pretreated as indicated with FG-4497 either 6 or 16 hr prior to TNFα. (b) HDMEC pretreated with DMOG or DP for 6 hr, or FG-4497 for 6 or 16 hr, followed by TNFα ( 1000 U/mL × 4 hr) were assessed for VCAM-1 expression by western analysis. FG-4497 inhibits TNFα induction of VCAM-1 at a transcriptional level. HDMEC were pretreated with DP (125 μM), DMOG (1 mM) or increasing concentrations of FG-4497 6 hr prior to stimulation with TNFα (1000 U/mL × 4 hr) and VCAM-1 (c) mRNA and (d) hnRNA were quantitated by real-time PCR.
Supplemental Figure 2: DMOG treatment does not inhibit induction of IRF-1 or egr-1 by TNFα or TGFα in HDMEC: (a) HDMEC were pretreated with increasing concentrations of DMOG 6 hr prior to stimulation with TNFα (1000 U/mL × 16 hr). Induction of IRF-1 was not affected by hypoxia mimetics. (b) HDMEC were pretreated with increasing concentrations of DMOG 6 hr prior to stimulation with TGFα (50 ng/mL × 4 hr). Induction of the TGFα-responsive gene egr-1 was maintained indicating a degree of specificity to the hypoxia mimetic-effect on VCAM.
Acknowledgments
Funding sources
This work was supported by NIH/NIAMS (T32 AR007587) and VA Merit Award.
Abbreviations
- α-KG
α-ketoglutarate
- CAM
cell adhesion molecule
- DMOG
dimethyl oxallyl glycine
- DP
2,2′-dipyridyl
- egr-1
early growth response protein 1
- FIH1
factor inhibiting HIF
- HDMEC
human dermal endothelial cells
- HIF
hypoxia inducible factor(s)
- hnRNA
heterogenous nuclear RNA
- IRF-1
interferon regulatory factor 1
- PHD
prolyl hydroxylase
- RT-PCR
reverse transcription real-time polymerase chain reaction
- TGFα
transforming growth factor-α
- VCAM
vascular cell adhesion molecule
Footnotes
Conflict of Interest
The authors have no conflict of interest to declare
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Elices MJ, Osborn L, Takada Y, Crouse C, Luhowskyj S, Hemler ME, et al. VCAM-1 on activated endothelium interacts with the leukocyte integrin VLA-4 at a site distinct from the VLA-4/fibronectin binding site. Cell. 1990;60:577–584. doi: 10.1016/0092-8674(90)90661-w. [DOI] [PubMed] [Google Scholar]
- [2].Das PK, de Boer OJ, Visser A, Verhagen CE, Bos JD, Pals ST. Differential expression of ICAM-1, E-selectin and VCAM-1 by endothelial cells in psoriasis and contact dermatitis. Acta Derm Venereol Suppl (Stockh) 1994;186:21–22. [PubMed] [Google Scholar]
- [3].Foster CA. VCAM-1/alpha 4-integrin adhesion pathway: therapeutic target for allergic inflammatory disorders. J Allergy Clin Immunol. 1996;98:S270–277. doi: 10.1016/s0091-6749(96)70075-1. [DOI] [PubMed] [Google Scholar]
- [4].Barbadillo C, G-Arroyo A, Salas C, Mulero J, Sanchez-Madrid F, Andreu JL. Anti-integrin immunotherapy in rheumatoid arthritis: protective effect of anti-alpha 4 antibody in adjuvant arthritis. Springer Semin Immunopathol. 1995;16:427–436. doi: 10.1007/BF00196098. [DOI] [PubMed] [Google Scholar]
- [5].Miller DH, Khan OA, Sheremata WA, Blumhardt LD, Rice GPA, Libonati MA, et al. A Controlled Trial of Natalizumab for Relapsing Multiple Sclerosis. N Engl J Med. 2003;348:15–23. doi: 10.1056/NEJMoa020696. [DOI] [PubMed] [Google Scholar]
- [6].Polman CH, O’Connor PW, Havrdova E, Hutchinson M, Kappos L, Miller DH, et al. A Randomized, Placebo-Controlled Trial of Natalizumab for Relapsing Multiple Sclerosis. N Engl J Med. 2006;354:899–910. doi: 10.1056/NEJMoa044397. [DOI] [PubMed] [Google Scholar]
- [7].MacDonald JK, McDonald JW. Natalizumab for induction of remission in Crohn’s disease. Cochrane Database Syst Rev. 2006;3:CD006097. doi: 10.1002/14651858.CD006097. [DOI] [PubMed] [Google Scholar]
- [8].Koo SW, Casper KA, Otto KB, Gira AK, Swerlick RA. Iron chelators inhibit VCAM-1 expression in human dermal microvascular endothelial cells. J Invest Dermatol. 2003;120:871–879. doi: 10.1046/j.1523-1747.2003.12144.x. see comment. [DOI] [PubMed] [Google Scholar]
- [9].Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annual Review of Cell & Developmental Biology. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- [10].Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 107:43–54. doi: 10.1016/s0092-8674(01)00507-4. see comment. [DOI] [PubMed] [Google Scholar]
- [11].Gira AK, Kowalczyk AP, Feng Y, Swerlick RA. Iron chelators and hypoxia mimetics inhibit IFNgamma-mediated Jak-STAT signaling. J Invest Dermatol. 2009;129:723–729. doi: 10.1038/jid.2008.269. [DOI] [PubMed] [Google Scholar]
- [12].Swerlick RA, Lee KH, Wick TM, Lawley TJ. Human dermal microvascular endothelial but not human umbilical vein endothelial cells express CD36 in vivo and in vitro. J Immunol. 1992;148:78–83. [PubMed] [Google Scholar]
- [13].Swerlick RA, Garcia-Gonzalez E, Kubota Y, Xu YL, Lawley TJ. Studies of the modulation of MHC antigen and cell adhesion molecule expression on human dermal microvascular endothelial cells. J Invest Dermatol. 1991;97:190–196. doi: 10.1111/1523-1747.ep12479643. [DOI] [PubMed] [Google Scholar]
- [14].Moreno CS, Beresford GW, Louis-Plence P, Morris AC, Boss JM. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- [15].Beresford GW, Boss JM. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat Immunol. 2001;2:652–657. doi: 10.1038/89810. [DOI] [PubMed] [Google Scholar]
- [16].Marui N, Offermann MK, Swerlick R, Kunsch C, Rosen CA, Ahmad M, et al. Vascular cell adhesion molecule-1 (VCAM-1) gene transcription and expression are regulated through an antioxidant-sensitive mechanism in human vascular endothelial cells. J Clin Invest. 1993;92:1866–1874. doi: 10.1172/JCI116778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Neish AS, Read MA, Thanos D, Pine R, Maniatis T, Collins T. Endothelial interferon regulatory factor 1 cooperates with NF-kappa B as a transcriptional activator of vascular cell adhesion molecule 1. Mol Cell Biol. 1995;15:2558–2569. doi: 10.1128/mcb.15.5.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gille J, Swerlick RA, Lawley TJ, Caughman SW. Differential regulation of vascular cell adhesion molecule-1 gene transcription by tumor necrosis factor alpha and interleukin-1 alpha in dermal microvascular endothelial cells. Blood. 1996;87:211–217. [PubMed] [Google Scholar]
- [19].Aragones J, Fraisl P, Baes M, Carmeliet P. Oxygen sensors at the crossroad of metabolism. Cell Metabolism. 2009;9:11–22. doi: 10.1016/j.cmet.2008.10.001. [DOI] [PubMed] [Google Scholar]
- [20].Johnson AB, Denko N, Barton MC. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutation Research. 2008;640:174–179. doi: 10.1016/j.mrfmmm.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hampton-Smith RJ, Peet DJ. From polyps to people: a highly familiar response to hypoxia. Ann N Y Acad Sci. 2009;1177:19–29. doi: 10.1111/j.1749-6632.2009.05035.x. [DOI] [PubMed] [Google Scholar]
- [22].Walmsley SR, Print C, Farahi N, Peyssonnaux C, Johnson RS, Cramer T, et al. Hypoxia-induced neutrophil survival is mediated by HIF-1alpha-dependent NF-kappaB activity. J Exp Med. 2005;201:105–115. doi: 10.1084/jem.20040624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fitzpatrick SF, Tambuwala MM, Bruning U, Schaible B, Scholz CC, Byrne A, et al. An intact canonical NF-kappaB pathway is required for inflammatory gene expression in response to hypoxia. J Immunol. 2011;186:1091–1096. doi: 10.4049/jimmunol.1002256. [DOI] [PubMed] [Google Scholar]
- [24].Oliver KM, Taylor CT, Cummins EP. Hypoxia. Regulation of NFkappaB signalling during inflammation: the role of hydroxylases. Arthritis Res Ther. 2009;11:215. doi: 10.1186/ar2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yamaji-Kegan K, Su Q, Angelini DJ, Johns RA. IL-4 is proangiogenic in the lung under hypoxic conditions. J Immunol. 2009;182:5469–5476. doi: 10.4049/jimmunol.0713347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yamashita T, Ohneda O, Sakiyama A, Iwata F, Ohneda K, Fujii-Kuriyama Y. The microenvironment for erythropoiesis is regulated by HIF-2alpha through VCAM-1 in endothelial cells. Blood. 2008;112:1482–1492. doi: 10.1182/blood-2007-11-122648. [DOI] [PubMed] [Google Scholar]
- [27].Cummins EP, Berra E, Comerford KM, Ginouves A, Fitzgerald KT, Seeballuck F, et al. Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappaB activity. Proc Natl Acad Sci U S A. 2006;103:18154–18159. doi: 10.1073/pnas.0602235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bracken CP, Fedele AO, Linke S, Balrak W, Lisy K, Whitelaw ML, et al. Cell-specific regulation of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha stabilization and transactivation in a graded oxygen environment. J Biol Chem. 2006;281:22575–22585. doi: 10.1074/jbc.M600288200. [DOI] [PubMed] [Google Scholar]
- [29].Cockman ME, Lancaster DE, Stolze IP, Hewitson KS, McDonough MA, Coleman ML, et al. Posttranslational hydroxylation of ankyrin repeats in IkappaB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH) Proc Natl Acad Sci U S A. 2006;103:14767–14772. doi: 10.1073/pnas.0606877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dunwell JM, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004;65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- [31].Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- [32].Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes & Development. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- [34].Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- [35].Hsieh JC, Slater SA, Whitfield GK, Dawson JL, Hsieh G, Sheedy C, et al. Analysis of hairless corepressor mutants to characterize molecular cooperation with the vitamin D receptor in promoting the mammalian hair cycle. J Cell Biochem. 2010;110:671–686. doi: 10.1002/jcb.22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wilkes DS. Chronic lung allograft rejection and airway microvasculature: is HIF-1 the missing link? J Clin Invest. 2011;121:2155–2157. doi: 10.1172/JCI58329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sica A, Melillo G, Varesio L. Hypoxia: a double-edged sword of immunity. J Mol Med (Berl) 2011;89:657–665. doi: 10.1007/s00109-011-0724-8. [DOI] [PubMed] [Google Scholar]
- [38].Cramer T, Yamanishi Y, Clausen BE, Forster I, Pawlinski R, Mackman N, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. erratum appears in Cell. 2003 May 2;113(3):419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lukashev D, Klebanov B, Kojima H, Grinberg A, Ohta A, Berenfeld L, et al. Cutting edge: hypoxia-inducible factor 1alpha and its activation-inducible short isoform I.1 negatively regulate functions of CD4+ and CD8+ T lymphocytes. J Immunol. 2006;177:4962–4965. doi: 10.4049/jimmunol.177.8.4962. [DOI] [PubMed] [Google Scholar]
- [40].Tsai AG, Johnson PC, Intaglietta M. Is the distribution of tissue pO(2) homogeneous? Antioxid Redox Signal. 2007;9:979–984. doi: 10.1089/ars.2007.1633. [DOI] [PubMed] [Google Scholar]
- [41].Hangai-Hoger N, Cabrales P, Briceno JC, Tsai AG, Intaglietta M. Microlymphatic and tissue oxygen tension in the rat mesentery. Am J Physiol Heart Circ Physiol. 2004;286:H878–883. doi: 10.1152/ajpheart.00913.2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: FG-4497 inhibits TNFα-induced VCAM-1 in a concentration dependent manner: (a) VCAM-1 expression (by ELISA) on HDMEC, pretreated as indicated with FG-4497 either 6 or 16 hr prior to TNFα. (b) HDMEC pretreated with DMOG or DP for 6 hr, or FG-4497 for 6 or 16 hr, followed by TNFα ( 1000 U/mL × 4 hr) were assessed for VCAM-1 expression by western analysis. FG-4497 inhibits TNFα induction of VCAM-1 at a transcriptional level. HDMEC were pretreated with DP (125 μM), DMOG (1 mM) or increasing concentrations of FG-4497 6 hr prior to stimulation with TNFα (1000 U/mL × 4 hr) and VCAM-1 (c) mRNA and (d) hnRNA were quantitated by real-time PCR.
Supplemental Figure 2: DMOG treatment does not inhibit induction of IRF-1 or egr-1 by TNFα or TGFα in HDMEC: (a) HDMEC were pretreated with increasing concentrations of DMOG 6 hr prior to stimulation with TNFα (1000 U/mL × 16 hr). Induction of IRF-1 was not affected by hypoxia mimetics. (b) HDMEC were pretreated with increasing concentrations of DMOG 6 hr prior to stimulation with TGFα (50 ng/mL × 4 hr). Induction of the TGFα-responsive gene egr-1 was maintained indicating a degree of specificity to the hypoxia mimetic-effect on VCAM.