Abstract
The vital nature of metal uptake and balance in biology is evident in the highly evolved strategies to facilitate metal homeostasis in all three domains of life. Several decades of study on metals and metalloproteins have revealed numerous essential bio-metal functions. Recent advances in mass spectrometry, x-ray scattering/absorption, and proteomics have exposed a much broader usage of metals in biology than expected. Even elements such as uranium, arsenic, and lead are implicated in biological processes as part of an emerging and expansive view of bio-metals. Here we discuss opportunities and challenges for established and newer approaches to study metalloproteins with a focus on technologies that promise to rapidly expand our knowledge of metalloproteins and metal functions in biology.
Keywords: metallomics, metalloproteomics, metalloproteins, biometals
The elements of biology: metals are fundamental to life processes
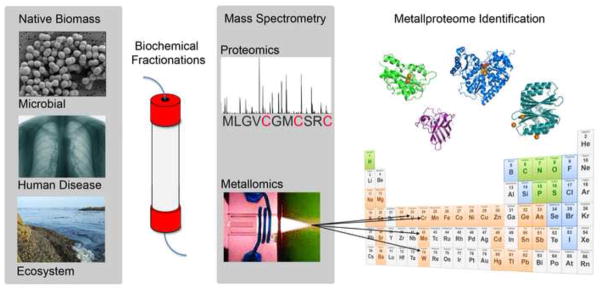
Metals are integral components of nearly all biological processes from the familiar red color of iron-containing blood to the binding of copper as part of the bizarre biology of prions in mammalian brains [1]. Paradoxically, some metals are essential in trace amounts and toxic at elevated levels, some are simply toxic, while others are tolerated at excessively high levels by living cells. Uranium, arsenic, copper, and lead have all recently been implicated in novel biological roles as part of the current expansion of what is known about metal functions in biology [1–3]. The elemental building blocks of life are common knowledge and most students would identify carbon, hydrogen, oxygen and nitrogen as the elements essential to life. While these fundamental components of life are predominate in terms of mass, it is important to note that all life forms have an absolute requirement for any number of metals. When we pause to consider the types of metals in biological systems, most of us would recall the essential minerals, yet we often do not consider how key these ‘trace’ elements are to most biological processes. One might argue that metal uptake, balance, and function are as elementary to life as carbon and nitrogen for many basic and complex biological processes. The expanding numbers and roles of metallic elements implicated in biological functions is striking, and becoming far broader than previously envisioned. Technological advances have given rise to the burgeoning research fields of metallomics and metalloproteomics. One particularly powerful approach highlighted here, combines the simultaneous proteomic and metallomic characterization of fractionated biomass to map entire metalloproteomes (Fig.1). This and other approaches promise a deeper and more comprehensive understanding of the use, balance, and function of metals in cells, ecosystems and human disease. Here we discuss several approaches that promise to be key in defining the largely unexplored landscape of metalloproteins and biological metals in the coming decades.
Figure 1. Diagrammatic representation of state-of-the-art metalloproteomics.
Diverse biological samples can be fractionated using any number of biophysical processes (left), followed by parallel proteomic and metallomic analysis with MS/MS and ICP-MP respectively (middle). This approach reveals the entire native metalloprotein content of any given sample (i.e. microbes [3]). This powerful approach is equally applicable to biological samples from any context.
Bioinformatics of metalloproteins
The exponential growth in available genome sequence information has driven an explosion in bioinformatics in the past decade. Almost 2,000 complete genome sequences are available, and over 11,000 genome projects are in progress (www.genomesonline.org). The amount of genetic data available to researchers will only continue to expand as new sequencing technologies such as semiconductor-based sequencing devices come into production [4]. Currently, many different informatics approaches to identify metalloproteins from primary sequence data have generated metalloprotein database resources [5–7]. However, the accurate prediction of metalloproteins is generally limited to closely homologous well-characterized proteins. It is striking that systems biology and informatics approaches capable of deciphering complex cellular circuitry [8] but remain incapable of predicting metal-binding specificity in proteins on a genome-wide basis. Given that perhaps a third or more of genes in a typical genome sequence correspond to conserved hypothetical or hypothetical proteins, a significant fraction of these genes likely encode novel metal-containing proteins that cannot currently be predicted. Consequently, while several metal-binding motifs have been identified, and some are useful in detecting probable metalloproteins from primary sequence, in most cases computational approaches fail to predict specific protein-bound metals. This limitation is likely due to and incomplete understanding of the complex determinants of metal-binding specificity in proteins rather than a limitation of informatics approaches.
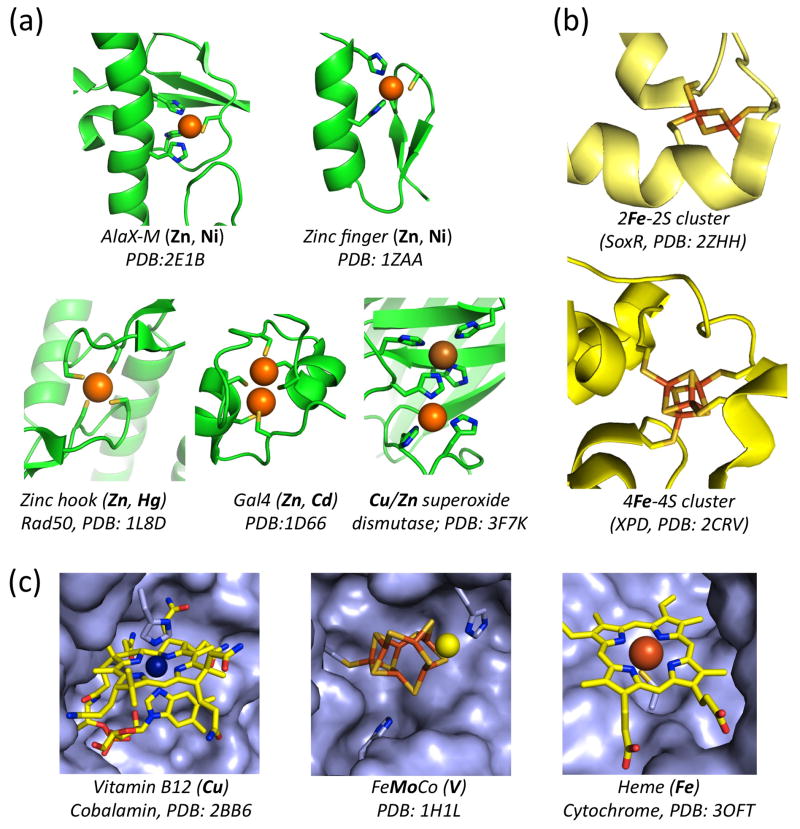
Most metal-binding motifs require only a small number of coordinating amino acids in a specific spatial arrangement (Fig 2). There are nearly infinite combinations of amino acid sequence that can bring several coordinating residues into the proper spatial arrangement for metal binding. This fact results in highly variable sequences at metal binding sites and makes identifying reliable sequence motifs difficult or impossible. Of the many metal binding sites that have been identified nearly all are highly degenerate between species and genera and even between proteins in the same organism [9]. The degenerate regions of metal binding motifs, typically denoted ‘X’, may also play a significant role in determining metal specificity [10,11]. Sequence-based prediction of specific metal binding is further limited by the fact that the same motif may bind more than one type of metal. A prime example is the ubiquitous Cys-X-X-Cys motif, which may coordinate iron-sulfur clusters or zinc, as well as being part of dithiol/disulfide redox sites in metal-free proteins. In addition, metal binding specificity can be dictated by factors other than a protein’s primary sequence, including the cellular context, maturation and ancillary proteins, and metal availability [3,12,13]. One such case is the iron-molybdenum cofactor of nitrogenase, in which a vanadium atom is inserted into the cofactor by alternate ancillary proteins when molybdenum is not available [reviewed in 12]. Another example is the substitution of a tungsten atom for molybdenum in an otherwise identical nitrate reductase [14]. In fact, the processes that regulate specific metal incorporation within a cell are often poorly understood and those that are described are elaborate and often variable between species, metals, proteins, and environments [reviewed in 10]. Consequently, the use of amino acid sequence information is primarily useful for surveying a subset of metalloproteins across genomes but inherently limited in identifying specific metal binding by a given protein [3,15]. While there are some cases, such as the superoxide dismutases where sequence and structure are more reliable predictors of specific metal incorporation [16,17], this is not commonly the case. As informatics approaches incorporate datasets from individual biological systems, three dimensional metalloprotein structures and other non-sequence datasets, their predictive capacity for metal binding will surely continue to improve [18,19]. Informatics approaches will undoubtedly remain an important tool for identifying metalloproteins in diverse genomes, but because of the nature of metal-specificity in proteins, these approaches will require validation by biophysical datasets for the foreseeable future.
Figure 2. A series of exemplary metal binding sites and coordinating side chains from various metalloproteins.
Protein fold names are indicated and examples of metals known to bind each fold listed in bold, PDB file numbers are indicated. (a) Cysteine and histidine coordinated single and double metal binding sites, most commonly Zinc. Top: AlaX-M [44], Zinc-finger [45]; bottom: Zinc-hook [23], Gal4 [46], Cu/Zn superoxide dismutase [47]. (b) Iron-sulfur clusters: 2Fe-2S cluster in SoxR [48] and 4Fe4S-cluster in the XPD helicase [24]. (c) Metal cofactor binding proteins: Vitamin B12 in Transcobalamin [49]; FeMoCo in Nitrogenase [50] and Heme in Cytochrome [51].
Structural Biology of Metalloproteins
Structure databases such as the protein databank (PDB) are the prevailing resources for the properties of metalloproteins at the atomic level. Together with the powerful spectroscopic approaches such as magnetic circular dichroism (MCD) [20], electron paramagnetic resonance (EPR) [21], and Mössbauer spectroscopy [22], a great deal of insight into the coordination and electronic properties of metalloproteins has been realized. Such studies have revealed metalloproteins that directly coordinate metals, bind inorganic metal clusters, and those with prosthetic metal cofactors (Fig. 2A–C respectively). Surprisingly, the determination of atomic resolution structures has lead to the discovery of metal binding in proteins that were previously not known to be metalloproteins [23,24]. However, even these powerful approaches and the resources they generate have inherent limitations for predicting and identifying metal-specific binding in biological systems. Notably, structure databases may be biased toward specific metalloproteins due to several practical considerations of the methods used in crystallography. First, a large fraction of structures in PDB are derived from heterologously expressed recombinant proteins, which often have misincorporated metals that are not representative of the endogenous metalloprotein. Such a case has recently been described for the structure of a recombinantly expressed ‘zinc-protein’ that coordinated a nickel atom when purified from the endogenous microbe [3]. The well-characterized zinc finger folds are known to substitute many different metals for zinc, including; cobalt, cadmium, copper, nickel and iron, which can all coordinated by the same polypeptide [25] (Fig. 2). Secondly, the practice of ‘soaking in’ heavy metals to solve the phase problem in protein crystallography displaces endogenous metals [26]. This process may skew the metalloprotein content of structural databases for folds that readily exchange metals and/or are stable in the apo-forms. Indeed, given that well over a fourth of proteins probably require a metal for normal function, it is striking that so few unique metal-binding folds have been identified. The routine production of recombinant proteins in E.coli and the practice of displacing endogenous metals to solve crystal structures may in part account for the relatively few unique metal-binding folds described to date. Alternatively, the essential nature and primordial origins of metals in biology may have selected a small subset of folds that are readily adapted for diverse metal binding and function. However, a more likely explanation is that there are simply a large number of metal-binding folds that remain uncharacterized. Regardless, while datasets from native biomass are essential to unequivocally determine endogenous metals, structural and spectroscopic approaches for identifying and characterizing metalloproteins remain among the most powerful means to study metalloproteins.
Mass spectrometry of metals and proteins
Spectroscopic and crystallographic approaches are precise methods for characterizing biological metal centers and largely account for what is known about metalloproteins. These approaches typically require large quantities (milligram amounts) of highly purified protein and typically focus on one metalloprotein at a time. The recent development of practical instruments for inductively coupled plasma mass spectrometry (ICP-MS) has made possible rapid and sensitive quantification of metals from complex biological samples. Quantitative ICP-MS metal analysis is a powerful analytical approach for metals in broadly diverse samples such as honeybee venom [27], urine and plasma [28,29], seawater [30], and quantifying toxic metal accumulations in seafood and drinking water [31,32]. As these metallo-approaches become more commonly applied, they will undoubtedly help identify markers of human disease much like the more mature proteomic [33] and metabolomic [34] approaches. The findings of these so-called metallomic studies all reflect the power of ICP-MS to address metals and metal balance issues in widely varied systems from complex ecosystems to human health and disease (Fig. 1).
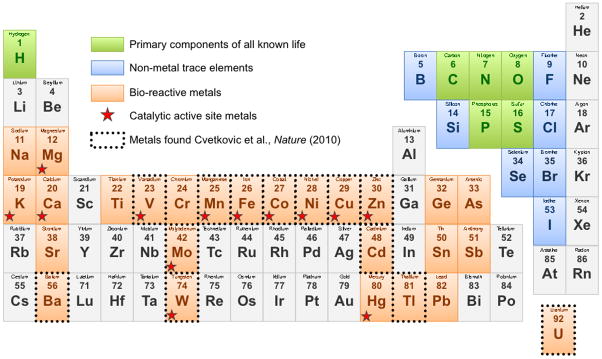
Metallomics studies do not inherently consider the metalloprotein context in biological samples. Metals within protein metal centers often have markedly altered properties that facilitate redox sensing [35], protein stability [24], metal sensing and balance [36], in addition to biochemical catalysis. Recent metalloproteomic studies have combined the sensitive metal detection of ICP-MS with established MS-based proteomics methods to probe fractionated native biomass. These studies have fundamentally changed the scope and depth of our understanding of metalloproteomes [3,15,37]. The approach recently described by Cvetkovic et al combined high-throughput tandem MS/MS based proteomics with ICP-MS analyses and revealed that microbial metalloproteomes are likely twice as extensive than was previously appreciated. The authors surveyed native metalloproteomes from three highly divergent microbial species isolated from particularly divergent environments. The microbes included two archaeal hyperthermophiles, the marine anaerobe Pyrococcus furiosus and the freshwater acidophilic aerobe Sulfolobus solfataricus, and the enteric bacterium Escherichia coli. A remarkable diversity of metalloproteins with twelve various associated metals were detected in the native metalloproteomes of one or more of these species (Fig. 3). Many of the metals bound to proteins were unexpected, because 1) the growth media was not supplemented with the metal and 2) the microbial species was not known to utilize that element. Several trace metals were concentrated by microbes into significant protein-associated quantities irrespective of the supplemented metals. Among these were surprising accumulations of lead, uranium, cadmium, strontium, and arsenic [3] (Fig. 3). While the ‘function’ of these metalloprotein associations are not clear in all cases, the accumulation of these metals into biomass indicates a specific biological interaction whether beneficial or harmful. Importantly, each microbe tested had markedly different metal-use preferences, none of which corresponded to concentration of supplemented metals of in the respective growth mediums. Excluding the predominant iron- and zinc-proteins, P. furiosus incorporated tungsten and nickel into metalloproteins whereas S. solfataricus used molybdenum and barium while the enteric E. coli proteins preferentially bound copper and manganese [3]. Taken together, these data reveal that each microbe has evolved a specific metal-usage profile that is largely maintained irrespective of the available metals. Notably, these observed metal specificities are not reflected in any recognized pattern in metalloprotein amino acid sequence or any other apparent informatics datasets. As part of the metalloproteomic survey, a limited number of native P. furiosus proteins were purified by following metal peaks through multiple chromatography steps. Using this method, a novel metalloprotein was identified as coordinating a nickel atom (PF0086) while the same polypeptide coordinated a Zn atom when expressed in E. coli [3]. These data further illustrate the complexities of specific metal incorporation into proteins within cells and emphasize the importance of factors other than primary amino acid sequence in determining metal-specificity. The comprehensive survey of three native metalloproteomes also expanded the known metallomes for these species and increased the sum of known types of Ni-containing enzymes in biology from 8 to 10 [3]. The metalloproteomics analysis of native biomass from three divergent microbes has brought many unexpected results and solidly established the power of combining biochemical fractionation of native biomass with proteomics and metallomics as an approach at the forefront of biometal research (Fig. 1).
Figure 3. Periodic table emphasizing biologically relevant elements and bio-metals.
The highly abundant building blocks of nature are shown in green, metals described to be associated with metalloproteins are shown in orange and non-metal trace elements in blue. Catalytic metals present in active sites of enzymes are marked with a red star and metals incorporated into microbial biomass in the studies of Cvetkovic et al. (3) are indicated by dotted boarders.
Several other burgeoning approaches for metalloproteomics analyses hold great promise to expand this largely unexplored landscape and further enhance the fields of metallomics and metalloproteomics. Significant advances with metal detection using synchrotron radiation sources reveal the localization and content of metals within cells and have become high throughput in some cases [reviewed in 38,39]. Microsolution isoelectric focusing [40], solid-state NMR [41], and colorimetric reagents [42] among other techniques are poised to contribute to metalloproteomics knowledge bases. Objective methods for observing the impact of metal ions on RNA and protein tertiary structures by small angle x-ray scattering have also been developed [43]. However, many of these techniques lack the breadth of analysis or ease of application of the approach described by Cvetkovic et al. [3]. The development of routine proteomics analysis coupled with practical ICP-MS instrumentation has brought metalloproteomics research into the reach of most institutions and many individual laboratories. Looking ahead, the application of all of these metallo-technologies to bioremediation, bioenergy, carbon cycling, ecology, human health, bio-industrial applications, and the treatment of human disease bring remarkable potential for discovery.
Closing Remarks
The citation of metalloprotein studies has grown steadily and increased ten-fold over the past twenty years reaching nearly 2,500 citations last year. Recent findings have revealed that the sum of these works and our current knowledge is likely limited to far less than half of the metalloproteins that exist [3]. Among known metalloprotein functions are vital roles in the fundamental processes of photosynthesis, electron transport, nitrogen fixation, and oxygen transport in vertebrates among many others. The discovery of such a large number of uncharacterized metalloproteins holds enormous potential for uncovering new biochemistries and novel evolutionary solutions to basic biochemical reactions. The convergence of ‘-omics’ technologies has begun to provide a systems view of microbes, microbial communities, complex organisms, and ecosystems. All known living systems have exploited the unique chemical and physical properties of metals to maintain homeostasis and facilitate life in even the most extreme environments. Considering the ubiquity and indispensable nature of metals to all life, any such ‘systems view’ will be severely lacking without consideration of metal requirements, balance, flux, and functions. As the newer metal-technologies mature to meet established technologies, our ability to rapidly identify and characterize novel metalloprotiens, metal folds, and define metal usage in biological systems will be dramatically expanded.
Metalomics and metalloproteomics will in time find their place with genomics and transcriptomics as key approaches to understanding complex biological systems. The biochemical processing of oxygen, nitrogen, sulfur and carbon are all reliant on metalloproteins and thus an in depth understanding of metalloproteins is fundamental to diverse research areas from climate change, carbon capture, and bioenergy to plant biology and medicine. Future applications of metal-technologies to biological research promise to not only expand our understanding of metal function in microbial physiology, ecology, and human disease, but will almost certainly impact all aspects of biology in the coming decades.
Highlights.
Our knowledge of fundamental metal functions in biology is limited.
Metalloproteins modify and expand metal functions in cells.
Computational approaches are inadequate for identifying metalloproteins.
Structural and spectroscopic characterizations are precise but limited in scope or accuracy.
New mass spectrometry approaches allow for accurate characterization of metalloproteomes.
Acknowledgments
We thank Jill O. Fuss and Barbara K. Burgess for inspiring discussions and acknowledge support of ENIGMA- Ecosystems and Networks Integrated with Genes and Molecular Assemblies, a project supported by the U.S. Department of Energy Office of Science and Office of Biological and Environmental Research under Contract No. DE-AC02-05CH11231.
Abreviations
- ICP-MS
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Brown DR, Qin K, Herms JW, Madlung A, Manson J, Strome R, Fraser PE, Kruck T, von Bohlen A, Schulz-Schaeffer W, et al. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- *2.Wolfe-Simon F, Switzer Blum J, Kulp TR, Gordon GW, Hoeft SE, Pett-Ridge J, Stolz JF, Webb SM, Weber PK, Davies PC, et al. A bacterium that can grow by using arsenic instead of phosphorus. Science. 2011;332:1163–1166. doi: 10.1126/science.1197258. This article asserts a unique and surprising alternative usage of arsenic in microbial nucleic acids and protiens. [DOI] [PubMed] [Google Scholar]
- **3.Cvetkovic A, Menon AL, Thorgersen MP, Scott JW, Poole FL, 2nd, Jenney FE, Jr, Lancaster WA, Praissman JL, Shanmukh S, Vaccaro BJ, et al. Microbial metalloproteomes are largely uncharacterized. Nature. 2010;466:779–782. doi: 10.1038/nature09265. This report presents a novel approach to comprehensively identify the complete metalloproteome from any biological sample and surveys the metalloproteomes of three diverse microbial species. [DOI] [PubMed] [Google Scholar]
- *4.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. This work presents a completely new technology for DNA sequencing with great promise to markedly decrease costs and increase throughput. [DOI] [PubMed] [Google Scholar]
- 5.Castagnetto JM, Hennessy SW, Roberts VA, Getzoff ED, Tainer JA, Pique ME. MDB: the Metalloprotein Database and Browser at The Scripps Research Institute. Nucleic Acids Res. 2002;30:379–382. doi: 10.1093/nar/30.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal-MACiE: a database of metals involved in biological catalysis. Bioinformatics. 2009;25:2088–2089. doi: 10.1093/bioinformatics/btp256. [DOI] [PubMed] [Google Scholar]
- 8.Bonneau R, Facciotti MT, Reiss DJ, Schmid AK, Pan M, Kaur A, Thorsson V, Shannon P, Johnson MH, Bare JC, et al. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007;131:1354–1365. doi: 10.1016/j.cell.2007.10.053. [DOI] [PubMed] [Google Scholar]
- 9.Thilakaraj R, Raghunathan K, Anishetty S, Pennathur G. In silico identification of putative metal binding motifs. Bioinformatics. 2007;23:267–271. doi: 10.1093/bioinformatics/btl617. [DOI] [PubMed] [Google Scholar]
- **10.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. This work provides a concise overview of metal functions usage and metalloprotein/metal specificity in microbial cells. [DOI] [PubMed] [Google Scholar]
- 11.Arnesano F, Banci L, Bertini I, Ciofi-Baffoni S, Molteni E, Huffman DL, O'Halloran TV. Metallochaperones and metal-transporting ATPases: a comparative analysis of sequences and structures. Genome Res. 2002;12:255–271. doi: 10.1101/gr.196802. [DOI] [PubMed] [Google Scholar]
- 12.Rubio LM, Ludden PW. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu Rev Microbiol. 2008;62:93–111. doi: 10.1146/annurev.micro.62.081307.162737. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz G, Mendel RR, Ribbe MW. Molybdenum cofactors, enzymes and pathways. Nature. 2009;460:839–847. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- 14.de Vries S, Momcilovic M, Strampraad MJ, Whitelegge JP, Baghai A, Schroder I. Adaptation to a high-tungsten environment. Pyrobaculum aerophilum contains an active tungsten nitrate reductase. Biochemistry. 2010;49:9911–9921. doi: 10.1021/bi100974v. [DOI] [PubMed] [Google Scholar]
- *15.Lancaster WA, Praissman JL, Poole FL, 2nd, Cvetkovic A, Menon AL, Scott JW, Jenney FE, Jr, Thorgersen MP, Kalisiak E, Apon JV, et al. A computational framework for proteome-wide pursuit and prediction of metalloproteins using ICP-MS and MS/MS data. BMC Bioinformatics. 2011;12:64. doi: 10.1186/1471-2105-12-64. This work describes a computational approach for proteome-wide identification of metal-proten interactions from fractionated biomass using mass spectrometry data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA, Getzoff ED. Nickel superoxide dismutase structure and mechanism. Biochemistry. 2004;43:8038–8047. doi: 10.1021/bi0496081. [DOI] [PubMed] [Google Scholar]
- 17.Borgstahl GE, Parge HE, Hickey MJ, Beyer WF, Jr, Hallewell RA, Tainer JA. The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell. 1992;71:107–118. doi: 10.1016/0092-8674(92)90270-m. [DOI] [PubMed] [Google Scholar]
- 18.Dudev T, Lim C. Metal binding affinity and selectivity in metalloproteins: insights from computational studies. Annu Rev Biophys. 2008;37:97–116. doi: 10.1146/annurev.biophys.37.032807.125811. [DOI] [PubMed] [Google Scholar]
- *19.Maret W. Metalloproteomics, metalloproteomes, and the annotation of metalloproteins. Metallomics. 2010;2:117–125. doi: 10.1039/b915804a. This article provides a tutorial overview of metal usage, localization, and homeostasis in prokaryotes and eukaryotes. [DOI] [PubMed] [Google Scholar]
- *20.Bruckner A. In situ electron paramagnetic resonance: a unique tool for analyzing structure-reactivity relationships in heterogeneous catalysis. Chem Soc Rev. 2010;39:4673–4684. doi: 10.1039/b919541f. This tutorial introduces the technique of in situ electron paramagnetic resonance and its applications to biological catalysis. [DOI] [PubMed] [Google Scholar]
- 21.McMaster J, Oganesyan VS. Magnetic circular dichroism spectroscopy as a probe of the structures of the metal sites in metalloproteins. Curr Opin Struct Biol. 2010;20:615–622. doi: 10.1016/j.sbi.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Papaefthymiou GC. The Mossbauer and magnetic properties of ferritin cores. Biochim Biophys Acta. 2010;1800:886–897. doi: 10.1016/j.bbagen.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 24.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkar B. Metal replacement in DNA-binding zinc finger proteins and its relevance to mutagenicity and carcinogenicity through free radical generation. Nutrition. 1995;11:646–649. [PubMed] [Google Scholar]
- 26.Perry JJ, Yannone SM, Holden LG, Hitomi C, Asaithamby A, Han S, Cooper PK, Chen DJ, Tainer JA. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat Struct Mol Biol. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- 27.Kokot ZJ, Matysiak J. Inductively coupled plasma mass spectrometry determination of metals in honeybee venom. J Pharm Biomed Anal. 2008;48:955–959. doi: 10.1016/j.jpba.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 28.Schramel P, Wendler I, Angerer J. The determination of metals (antimony, bismuth, lead, cadmium, mercury, palladium, platinum, tellurium, thallium, tin and tungsten) in urine samples by inductively coupled plasma-mass spectrometry. Int Arch Occup Environ Health. 1997;69:219–223. doi: 10.1007/s004200050140. [DOI] [PubMed] [Google Scholar]
- 29.Manley SA, Gailer J. Analysis of the plasma metalloproteome by SEC-ICP-AES: bridging proteomics and metabolomics. Expert Rev Proteomics. 2009;6:251–265. doi: 10.1586/epr.09.44. [DOI] [PubMed] [Google Scholar]
- 30.Zhang S, Lu A, Han F, Shan XQ. Preconcentration and determination of trace metals in seawater using a thiol cotton fiber minicolumn coupled with inductively coupled plasma mass spectrometry. Anal Sci. 2005;21:651–654. doi: 10.2116/analsci.21.651. [DOI] [PubMed] [Google Scholar]
- 31.King TJ, Sheridan RS, Rice DH. Analysis of toxic metals in seafood sold in New York state by inductively coupled plasma mass spectrometry and direct combustion analysis. J Food Prot. 2010;73:1715–1720. doi: 10.4315/0362-028x-73.9.1715. [DOI] [PubMed] [Google Scholar]
- 32.Wang JP, Ma XX, Fang GZ, Wang S, Yin HL. Determination of six heavy metals elements in drinking water by inductively coupled plasma mass spectrometry. Guang Pu Xue Yu Guang Pu Fen Xi. 2010;30:2827–2829. [PubMed] [Google Scholar]
- 33.Pendyala G, Trauger SA, Kalisiak E, Ellis RJ, Siuzdak G, Fox HS. Cerebrospinal fluid proteomics reveals potential pathogenic changes in the brains of SIV-infected monkeys. J Proteome Res. 2009;8:2253–2260. doi: 10.1021/pr800854t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wikoff WR, Pendyala G, Siuzdak G, Fox HS. Metabolomic analysis of the cerebrospinal fluid reveals changes in phospholipase expression in the CNS of SIV-infected macaques. J Clin Invest. 2008;118:2661–2669. doi: 10.1172/JCI34138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yannone SM, Burgess BK. Identification of a palindromic sequence that is responsible for the up-regulation of NAPDH-ferredoxin reductase in a ferredoxin I deletion strain of Azotobacter vinelandii. J Biol Chem. 1997;272:14454–14458. doi: 10.1074/jbc.272.22.14454. [DOI] [PubMed] [Google Scholar]
- 36.Rouault TA, Haile DJ, Downey WE, Philpott CC, Tang C, Samaniego F, Chin J, Paul I, Orloff D, Harford JB, et al. An iron-sulfur cluster plays a novel regulatory role in the iron-responsive element binding protein. Biometals. 1992;5:131–140. doi: 10.1007/BF01061319. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Thompson R, Caruso J. Probing the viral metallome: searching for metalloproteins in bacteriophage lambda-- the hunt begins. Metallomics. 2011;3:472–481. doi: 10.1039/c0mt00104j. [DOI] [PubMed] [Google Scholar]
- 38.Ortega R, Deves G, Carmona A. Bio-metals imaging and speciation in cells using proton and synchrotron radiation X-ray microspectroscopy. J R Soc Interface. 2009;6 (Suppl 5):S649–658. doi: 10.1098/rsif.2009.0166.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi WX, Chance MR. Metalloproteomics: forward and reverse approaches in metalloprotein structural and functional characterization. Current Opinion in Chemical Biology. 2011;15:144–148. doi: 10.1016/j.cbpa.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *40.Pioselli B, Munro C, Raab A, Deitrich CL, Songsrirote K, Feldmann J, Thomas-Oates J. Denaturing and non-denaturing microsolution isoelectric focussing to mine the metalloproteome. Metallomics. 2009;1:501–510. doi: 10.1039/b903607e. This work investigates the application of micro-scale isoelectric focusing and mass spectrometry for metalloproteome analysis. [DOI] [PubMed] [Google Scholar]
- *41.Bertini I, Emsley L, Lelli M, Luchinat C, Mao J, Pintacuda G. Ultrafast MAS solid-state NMR permits extensive 13C and 1H detection in paramagnetic metalloproteins. J Am Chem Soc. 2010;132:5558–5559. doi: 10.1021/ja100398q. This article describes new advances in NMR technology that allow observation of the metal coordinating residues in metalloprotiens. [DOI] [PubMed] [Google Scholar]
- 42.Sabel CE, Neureuther JM, Siemann S. A spectrophotometric method for the determination of zinc, copper, and cobalt ions in metalloproteins using Zincon. Anal Biochem. 2010;397:218–226. doi: 10.1016/j.ab.2009.10.037. [DOI] [PubMed] [Google Scholar]
- *43.Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. This article describes a novel approach for using small-angle x-ray scattering to characterize structural dynamics in biomolecules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukunaga R, Yokoyama S. Structure of the AlaX-M trans-editing enzyme from Pyrococcus horikoshii. Acta Crystallogr D Biol Crystallogr. 2007;63:390–400. doi: 10.1107/S090744490605640X. [DOI] [PubMed] [Google Scholar]
- 45.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 46.Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 47.Shin DS, Didonato M, Barondeau DP, Hura GL, Hitomi C, Berglund JA, Getzoff ED, Cary SC, Tainer JA. Superoxide dismutase from the eukaryotic thermophile Alvinella pompejana: structures, stability, mechanism, and insights into amyotrophic lateral sclerosis. J Mol Biol. 2009;385:1534–1555. doi: 10.1016/j.jmb.2008.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe S, Kita A, Kobayashi K, Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc Natl Acad Sci U S A. 2008;105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wuerges J, Garau G, Geremia S, Fedosov SN, Petersen TE, Randaccio L. Structural basis for mammalian vitamin B12 transport by transcobalamin. Proc Natl Acad Sci U S A. 2006;103:4386–4391. doi: 10.1073/pnas.0509099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayer SM, Gormal CA, Smith BE, Lawson DM. Crystallographic analysis of the MoFe protein of nitrogenase from a nifV mutant of Klebsiella pneumoniae identifies citrate as a ligand to the molybdenum of iron molybdenum cofactor (FeMoco) J Biol Chem. 2002;277:35263–35266. doi: 10.1074/jbc.M205888200. [DOI] [PubMed] [Google Scholar]
- 51.Ma M, Bell SG, Yang W, Hao Y, Rees NH, Bartlam M, Zhou W, Wong LL, Rao Z. Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444. Chembiochem. 2011;12:88–99. doi: 10.1002/cbic.201000537. [DOI] [PubMed] [Google Scholar]