Abstract
The presence of the cellular prion protein (PrPC) on the cell surface is critical for the neurotoxicity of prions. Although a number of biological activities have been attributed to PrPC, a definitive demonstration of its physiological function remains elusive. In this review, we will discuss some of the proposed functions of PrPC, focusing on recently suggested roles in cell adhesion, regulation of ionic currents at the cell membrane, and neuroprotection. We will also discuss recent evidence supporting the idea that PrPC may function as a receptor for soluble oligomers of the amyloid β peptide and possibly other toxic protein aggregates. These data suggest surprising new connections between the physiological function of PrPC and its role in neurodegenerative diseases beyond those caused by prions.
Introduction
Prion diseases are a group of rare and invariably fatal neurodegenerative disorders of humans and animals, characterized by dementia, motor dysfunction, and cerebral amyloidosis. These disorders include Creutzfeldt-Jakob disease and kuru in humans, as well as bovine spongiform encephalopathy and scrapie in animals. Prion diseases can arise sporadically, as a result of mutations in the gene encoding the prion protein, or by infection from exogenous sources. A great deal of evidence indicates that the key event underlying all forms of prion diseases is conformational conversion of a normal cell surface glycoprotein called PrPC (cellular prion protein) into an aggregated, β-sheet-rich isoform called PrPSc (scrapie prion protein) (Box 1) [1]. The infectious spread of prions occurs via PrPSc-templated conversion of an endogenous pool of PrPC molecules, a process that has analogues in self-propagating proteins described in other species [2].
BOX 1. One protein, two forms: PrPC and PrPSc.
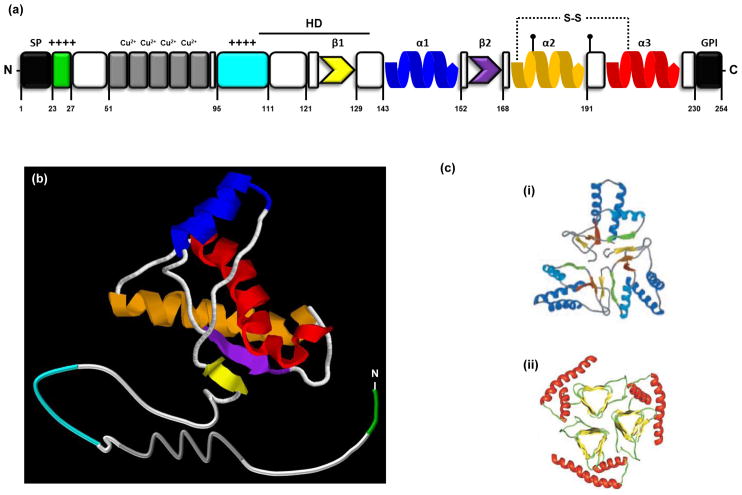
PrPC is the cellular form of the prion protein (Figure Ia). It is an endogenous glycoprotein that is expressed at highest levels in the CNS, and present in a wide range of species from fish to mammals [100–102]. The three-dimensional structure of PrPC includes a disordered N-terminal domain (residues 23–124, numbering for mouse PrP) and a C-terminal globular region (residues 125–228) composed of three α-helices and two short β-strands (Figure Ib) [99, 103]. The N-terminal half encompasses a polybasic region (residues 23–27) and a series of histidine-containing octapeptide repeats (residues 51–90) that can bind metal ions like Cu2+ [104]. The central region encompasses a charged region (residues 105–111) followed by a highly conserved hydrophobic domain (residues 112–130) which serves as a transmembrane anchor in certain situations [69]. During its biosynthesis in the ER, the N-terminal signal peptide (residues 1–22) is removed and a GPI anchor is attached at residue 230 [105]. Two N-linked oligosaccharide chains are also added (at Asn-180 and Asn-196) [106]. Most PrP is found on the cell-surface where it is localized to lipid rafts, although a fraction is endocytosed via clathrin-coated pits [62, 107]. Some of the protein is proteolytically cleaved by cellular proteases near residue 111 to generate N- and C-terminal fragments called N1 and C1, respectively [108, 109].
PrPSc is the infectious isoform of the prion protein. It has the same amino acid sequence as PrPC, but has a higher content of β-sheet structure (Figure Ic), and is relatively resistant to protease digestion [110]. PrPSc acts as a molecular template by physically interacting with PrPC and converting the latter to more molecules of PrPSc. It is this process which accounts for the self-propagating nature of infectious prions [1]. A prion-like propagation mechanism has been described for several proteins in yeast and fungi (reviewed in [111, 112]). Furthemore, such a propagation mechanism may play a role in the neuroanatomical spread of protein aggregates in other neurodegenerative diseases like Alzheimer’s, Huntington’s, and Parkinson’s diseases (see [113] for a recent review).
Despite compelling evidence for this mechanism of prion propagation, our current understanding of the primary causes of neurodegeneration in prion diseases is still limited. Several kinds of experiments suggest that PrPSc, or other pathological PrP conformers, require physiologically active PrPC on the cell membrane in order to exert their toxicity [3]. Interestingly, recent evidence indicates that PrPC may also mediate the toxicity of amyloid beta (Aβ) oligomers that are associated with Alzheimer’s disease (AD) [4], and possibly other β-rich protein aggregates [5]. Therefore, defining the physiological activity of PrPC is not only crucial for understanding the pathogenesis of prion diseases, but may also provide fundamental insights into the pathogenesis of other neurodegenerative disorders caused by misfolded protein assemblies. Here, we review recent findings that have provided fresh insights into potential physiological activities of PrPC, and how these might be subverted to produce pathological effects.
A dual role for PrPC in prion diseases
It is widely agreed that PrPC plays a crucial role in the pathogenesis of prion diseases by virtue of its ability to serve as substrate for generation of PrPSc. However, a number of reports have underscored the distinction between prion infectivity and prion toxicity, and provided evidence that alterations in the normal function of PrPC may lie at the root of prion-induced neurodegenerative processes. In particular, recent experiments suggest that accumulation of infectivity and neurodegeneration proceed in distinct chronological and mechanistic phases. Infectivity accumulates relatively rapidly, and requires only a minimum expression level of PrPC, while neurodegeneration takes much longer and is directly dependent on the amount of PrPC expressed in the brain [6]. A dramatic demonstration of the dissociation of infectivity and pathology is the observation that genetically depleting neuronal PrPC in mice with established prion infection reverses neuronal loss and progression of clinical signs, despite the continuous production of infectious PrPSc by surrounding astrocytes [7]. Similarly, the absence of endogenous PrPC renders host brain tissue resistant to the toxic effects of PrPSc emanating from implanted graft tissue [8]. Finally, the absence of the glycolipid membrane anchor on PrPC has been shown to cause dramatic changes in the characteristics of scrapie-induced illness [9, 10]. Taken together, these lines of evidence indicate that the toxicity of PrPSc requires the presence of membrane-anchored PrPC at the cell surface, and suggest that the normal, physiological activity of the protein could be subverted to create toxicity.
Previous attempts to understand the function of PrPC
Although PrPC was first identified as a normal cellular protein almost 20 years ago [11], its physiological function has remained uncertain. The creation of mice carrying targeted disruption of the gene encoding PrP represents a possible approach to this problem. Since 1992, several lines of PrP knockout mice have been engineered [12, 13]. These animals display resistance to prion infection, as predicted by the prion hypothesis, since they lack PrPC molecules that serve as substrates for the generation of PrPSc [14]. With only one notable exception, however (due to artifactual upregulation of an adjacent gene [15]), PrP knock-out mice display no major developmental or anatomical abnormalities, and lead a normal lifespan [12]. However, some of these animals have been reported to exhibit subtle phenotypic abnormalities at the behavioral and cellular levels (reviewed in [16]), for example alterations in olfactory function [17] or myelination [18]. As yet, the molecular basis for these defects is unclear.
PrPC and cell adhesion: new insights from fish
Several of the proposed functions of PrPC are related to the localization of the protein on the cell surface. Previous work has suggested that PrPC could act as a cellular adhesion molecule, participating in several interrelated processes including neurite outgrowth, neuronal survival, and neuronal differentiation (reviewed in [19]). Consistent with this idea, PrPC has been found to associate with several surface proteins including laminin [20], the 37 kDa laminin receptor precursor (LRP)/67 kDa laminin receptor [21, 22], and the neural cell adhesion molecule (NCAM) [23] (Figure 1a, b). The latter was recruited by PrPC into lipid rafts, producing activation of fyn kinase and promoting cell adhesion and neurite outgrowth [24]. PrPC was also found to bind to the contactin-associated protein (Caspr), a neurite outgrowth-inhibitory factor [25]. This interaction prevented Reelin-mediated shedding of Caspr from the cell surface, which resulted in increased surface levels of Caspr and potent inhibition of neurite outgrowth in cultured cerebellar neurons.
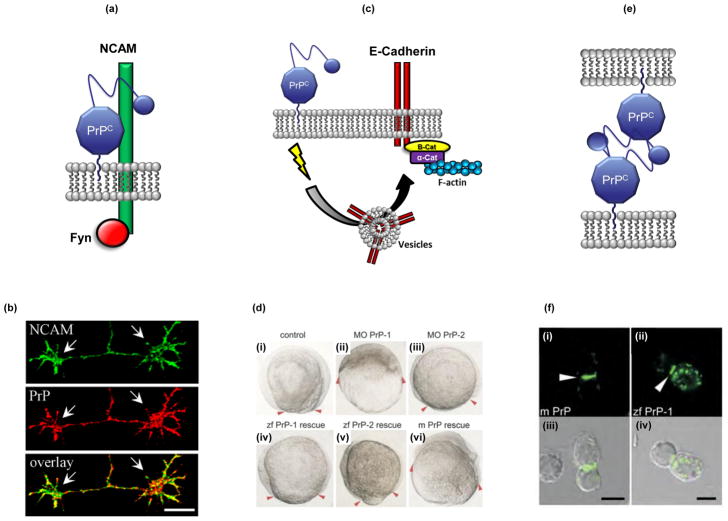
FIGURE 1. A role for PrPC in cell adhesion.
The figure provides three examples of how PrPC could participate in cellular adhesion. (a) Schematic illustration of PrPC interaction with neural cell adhesion molecule (NCAM), which results in activation of Fyn kinase and enhancement of neurite outgrowth in cultured hippocampal neurons (not shown) [24]. (b) PrPC partially co-localizes with NCAM along neurites and in growth cones (arrows) of cultured hippocampal neurons [22]. Bar, 10 μm. (c) Schematic model, based on experiments performed in zebrafish [26], illustrating how PrPC could modulate cell adhesion by influencing delivery of E-cadherin to the plasma membrane. Homodimers of E-cadherin can be anchored to F-actin via α and β catenins [127]; thus, PrPC may indirectly participate in rearrangement of the actin cytoskeleton [26]. The precise mechanism by which PrPC on the plasma membrane regulates the delivery of intracellular vesicles is currently unknown, but may involve activation of a signal transduction cascade involving Src-family tyrosine kinases. (d) Morpholino knockdown of zebrafish PrP-1 causes gastrulation arrest, which can be rescued by re-introduction of several kinds of PrP molecules [26]. (i) Control embryos at the 1–4 cell stage display normal progression of the blastodermal margin (red arrowheads), while morphant (MO) embryos knocked down for zebrafish PrP-1 (ii), but not PrP-2 (iii), are severely arrested. Gastrulation arrest caused by depletion of PrP-1 can be rescued by microinjection of RNA encoding zebrafish (zf) PrP-1 (iv), zebrafish PrP-2 (v) or mouse PrP (m PrP) (vi), suggesting that a biological function of the PrP molecule is conserved from fish to mammals. (e) Model for how PrPC molecules on adjacent cells engage in homophilic interactions that promote cell adhesion, based on experiments in cultured cells [26]. (f) Green fluorescent protein (GFP)-tagged forms of mouse PrPC (i) or zebrafish PrP-1 (ii) expressed in non-adhesive Drosophila S2 cells accumulate at cell junctions (arrowheads). Panels (i) and (ii) show fluorescence images, and panels (iii) and (iv) show the corresponding phase-contrast images. Scale bars, 5 μm. Reproduced, with permission, from [24] (panel b) and [26] (panels d and f).
Compelling evidence for an involvement of PrPC in cell adhesion has emerged recently from a niche of the animal kingdom that was not previously a focus of prion research: the zebrafish Danio rerio [26]. The zebrafish genome contains two PrP genes, called PrP-1 and PrP-2, both encoding glycosylphosphatidylinositol (GPI)-anchored proteins structurally similar to mammalian PrP. The two proteins differ in their spatiotemporal expression patterns, with PrP-1 being expressed primarily at cell contacts during early embryogenesis, while PrP-2 is up-regulated later in the developing nervous system where it is more widely distributed on the cell membrane [26]. Interestingly, experimental depletion of PrP-1 caused gastrulation arrest, a phenotype that was partially rescued by re-introduction of PrP-1, PrP-2 or mouse PrP [26], suggesting that a basic biological activity of PrPC is highly conserved from fish to mammals (Figure 1c, d). Further analysis revealed a role for PrP-1 in modulating cell adhesion by influencing delivery of E-cadherin to the plasma membrane, and by directly serving as a homophilic adhesion molecule (Figures 1e, f). These experiments demonstrate that PrPC function can be studied in zebrafish, a model vertebrate organism that allows direct visualization of developmental events, as well as ease of performing genetic and chemical screens. Importantly, and in contrast to mice, ablation of PrP-1 or PrP-2 produces robust loss-of-function phenotypes that can be dissected in greater detail at the molecular level.
A connection between PrPC, ion channels, and neuronal excitability
Several lines of evidence support the idea that PrPC may play a role in the regulation of ion channels and neuronal excitability. PrPC is localized at synaptic sites where many kinds of ion channels are concentrated, and there is evidence that PrPC is important for normal synaptic development and function [27, 28]. In addition, a variety of electrophysiological abnormalities have been described in cerebellar or hippocampal neurons derived from PrP knock-out mice [29–38].
Perhaps the most compelling evidence for a functional and physical interaction between PrPC and receptors concerns receptors for the excitatory amino acid glutamate. Aberrant activation of the NMDA subtype of ionotropic glutamate receptors is thought to cause nerve cell damage and death in a number of acute and chronic neurological conditions by allowing abnormal entry of Ca2+ ions into neurons, a mechanism referred to as excitotoxicity [39]. In one recent study, hippocampal neurons from Prn-p0/0 mice were found to display enhanced NMDA-induced currents, an effect that was reversed by overexpression of PrPC [40]. Co-immunoprecipitation of PrPC and the NR2D subunit of the NMDA receptor suggested a direct modulation of NMDA receptors by PrPC. Additional evidence connecting PrPC to glutamate-mediated excitotoxicity has emerged from another recent study of transgenic (Tg) mice expressing a PrP form deleted for the central region (residues 105–125, referred to as ΔCR PrP) [41]. A neuropathological hallmark of these mice is massive degeneration of cerebellar granule neurons, which display a unique ultrastructural morphology, typical of non-apoptotic neuronal death occurring after excitotoxic stress [42]. There is also evidence that PrPC modulates the activity of kainate receptors [43] and metabotropic glutamate receptors [44], suggesting that PrPC may serve to suppress neuronal excitability mediated by multiple kinds of glutamate receptors.
Other recent findings suggest the surprising possibility that the PrP molecule itself might form ion channels in the cell membrane, and that the currents associated with these channels might be responsible for certain kinds of PrP-related neurodegeneration. In particular, PrP molecules carrying neurotoxic deletions in the central region (such ΔCR PrP) have been found to induce spontaneous ionic currents in a variety of transfected cell lines [45, 46] (Figure 2a, b). Similar currents were induced by several different point mutations in the central region that are linked to familial prion diseases in humans [45, 46]. These spontaneous ionic currents were silenced by co-expression of wild type (WT) PrP (Figure 2c), parallel to the ability of WT PrP to suppress the neurodegenerative phenotypes of transgenic mice expressing deleted forms of PrP (see below). The biophysical features of the induced currents suggested that they are produced by a non-selective, cation-permeable channel or pore in the cell membrane. Notably, the currents were observed in both neuronal and non-neuronal cell lines derived from a variety of species, ranging from insects to mammals. Therefore, either mutant PrP regulates the activity of an endogenous channel that is widely expressed, or the PrP molecule itself forms a channel or pore in the membrane, independent of other cellular proteins (Figure 2b).
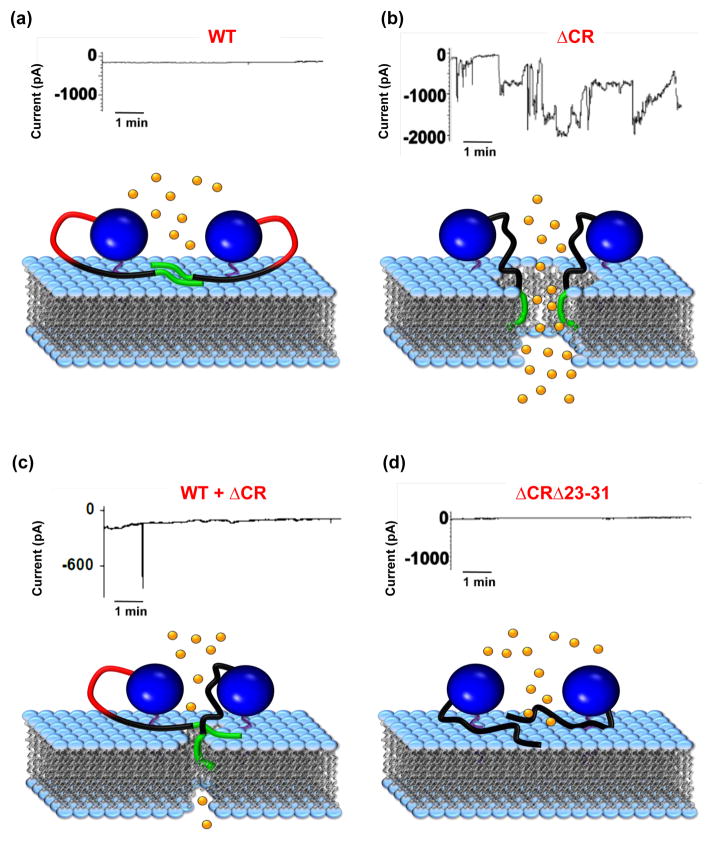
FIGURE 2. Model illustrating how PrP molecules may form ion channels or pores.
The figure illustrates how mutant PrP molecules carrying a deletion (referred to as ΔCR) of residues 105–125 in the central region induce spontaneous ionic currents by the proposed formation of channels or pores in the cell membrane, and how this activity is dependent on residues 23–31 and is suppressed by WT PrP. For each panel, the upper graph shows whole-cell patch-clamp recordings of human embryonic kidney 293 (HEK293) cells expressing the corresponding mouse PrP protein. The schematic below each graph represents one possible model for interpreting the data at the molecular level. In contrast to WT PrP (a), ΔCR PrP (b) induces spontaneous inward currents, which may reach amplitudes of up to several thousand pA [45]. This phenomenon could be explained by hypothesizing that the lack of the central region promotes the aberrant insertion of ΔCR PrP molecules into the plasma membrane, which results in the formation of a channel or pore. (c) The spontaneous ionic currents induced by ΔCR PrP are silenced by co-expression of WT PrP [45, 46]. WT PrP may inhibit formation of the pore by physically interacting with the ΔCR mutant. (d) Deleting residues 23–31 within ΔCR PrP abrogates the ability of this mutant to induce spontaneous currents [46]. One hypothesis to explain this observation is that interaction of ΔCR PrP molecules with the cell membrane requires the polybasic N-terminal region, which has been shown to act as a protein transduction domain (PTD). Whole-cell patch-clamp recordings are reproduced, with permission, from [45] (a–c), and [46] (d). In each cartoon, the blue ball indicates the C-terminal globular domain of PrP (residues 125–231). The N-terminal flexible domain is divided in three segments corresponding to residues 23–31 (green), 32–104 (black) and 105–125 (red).
Neuroprotective effects of PrPC
Despite the fact that propagation of PrPSc is associated with neurodegeneration, several lines of evidence suggest that PrPC may have neuroprotective and pro-survival functions. Overexpression of PrPC has been shown to protect cell lines and primary neurons from several kinds of apoptotic stimuli [47–51]. Moreover, there is evidence that PrPC plays a role in regulating intracellular signaling cascades, including those mediating cellular survival [52]. A direct role for the N-terminal region of PrPC in protecting cells from reactive oxygen species (ROS) induced by serum deprivation has also been recently proposed [53]. Finally, a cytoplasmic co-chaperone molecule called stress-inducible protein 1 (STI1) has been reported to be secreted from cells, where it binds to the central region of PrPC (residues 113–128) and promotes neuronal survival and differentiation [54]. The PrPC-STI1 complex was also recently implicated in self-renewal of neural progenitor/stem cells [55], a role that is consistent with a previously identified function of PrPC in regulating neural precursor proliferation during mammalian neurogenesis [56]. Collectively, these data suggest a role for PrPC, in particular its N-terminal and central regions, in protection from cellular stress, as well as neuronal survival, differentiation and proliferation.
Neuroprotection and neurotoxicity: two sides of the same coin?
In vitro and in vivo studies suggest that, although PrPC possesses neuroprotective activity, deletions and point mutations in the protein can endow it with cytotoxic properties in cultured cells and neurotoxic properties in mice. Interestingly, as will be discussed below, the same domains of PrPC may be responsible for both activities of the molecule, suggesting the intriguing possibility that prions may deliver their toxicity through corruption of the physiological function of PrPC [57].
Previous attempts to determine which parts of PrPC are necessary for prion replication led to the surprising observation that specific deletions encompassing the conserved central region endow the molecule with powerful toxic properties. Expression in transgenic mice on the Prn-p0/0 background of PrP molecules harboring deletion of residues 32–121 or 32–134 led to progressive neurodegeneration, marked by loss of cerebellar granular neurons and white matter vacuolation [58]. Transgenic mice expressing PrP molecules deleted for residues 105–125 (ΔCR) [41] or 94–134 [59] displayed a neonatal lethal phenotype, suggesting that the critical neurotoxicity-determining region encompasses a cluster of positively charged amino acids (residues 105–111), as well as part of the adjacent, central hydrophobic domain (residues 112–130) (Figure 3a; also see Box I). PrP molecules carrying these deletions are also cytotoxic in cell culture assays [60, 61].
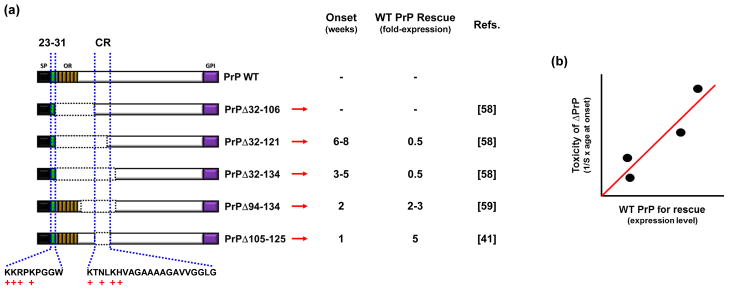
FIGURE 3. Deletions within the central region of PrP are associated with neurotoxicity in transgenic mice.
(a) Deletions up to residue 106 have no adverse effect [58], while deletions extending through residues 121 or 134 cause spontaneous neurodegenerative illness in the absence of endogenous PrPC [58]. Additional deletions (Δ94–134 [59] and Δ105–125 [41]) narrow the critical, neurotoxicity-determining region to a highly conserved segment of 21 amino acids in the central region (residues 105–125). Schematics show the structures of WT mouse PrP and each of the deletion mutants (with the deleted region indicated by dotted lines). The column labeled “Onset” gives the age (in weeks) at which spontaneous neurological illness occurs in mice expressing the corresponding deletion mutant. The column labeled “WT PrP Rescue” gives the fold-expression of WT PrPC needed to prevent the appearance of neurological illness. A minus symbol (“−“) indicates that no spontaneous disease is observed. (b) The graph, which is a representation of the data shown in the first two columns, shows the inverse correlation between age of onset in the different mouse lines and amount of WT PrP expression required for rescuing the neurological phenotype.
Box 1 FIGURE I. Structure of PrPC and PrPSc.
(a) Schematic illustration of PrPC. Residues correspond to the mouse sequence. A signal peptide (SP, residues 1–22), removed during PrP biosynthesis in the ER, precedes a polybasic region (residues 23–27, green) and five histidine-containing octapeptide repeats (residues 51–90, gray) that can bind Cu2+ and other bivalent metal ions. The central part of the molecule includes a positively charged region (residues 95–111, cyan) followed by a highly conserved hydrophobic domain (HD, residues 111–130). The C-terminal part includes two short β-strands (residues 127–129 yellow; and 166–168, purple) and three α-helices (residues 143–152, blue; 171–191, orange; 199–221, red). A C-terminal peptide (residues 231–254) is removed during biosynthesis, followed by attachment of a glycosyl-phosphatidylinositol (GPI) moiety, which anchors the protein to the outer leaflet of the plasma membrane. PrPC also contains two N-linked oligosaccharide chains (at Asn-180 and Asn-196, black lollipops) and a disulfide bond between residues 178 and 231). (b) Three-dimensional structure of PrPC, based on nuclear magnetic resonance (NMR) analysis. The structure of mouse prion protein fragment 121–231 was taken from entry 1XYX of the Protein Data Bank (PDB, http://www.ebi.ac.uk/pdbe), and was modified using the RasMol 7.4 software (www.rasmol.org). Colors correspond to the structural motifs described in (a). (c) Models of aggregated PrPSc. Although a high-resolution structure for PrPSc has not been determined, several different models have been proposed. Two of these are illustrated here. (i) In the first model, derived from a molecular dynamics simulation, the core of the PrPSc aggregate is formed by parallel and antiparallel β-strands, organized in a spiral shape. Reproduced, with permission, from [125]. (ii) In the second model, which is based on electron crystallographic studies, the core is formed by left-handed βhelices. Reproduced, with permission, from [126]. Both pictures show trimers of PrPSc.
A unique feature of transgenic mice expressing toxic PrP deletion mutants, originally described over 10 years ago [58], provides a clue that the neurotoxic and neuroprotective activities of PrPC may be related. In each of these transgenic lines, co-expression of WT PrPC abrogates clinical symptoms and neuropathology, with more toxic mutations requiring higher doses of WT PrPC to rescue the phenotype [41, 58, 59] (Figure 3b). This striking phenomenon suggests that there is a relationship between the physiological activity of PrPC and the toxic effects of the deletion mutants.
Recent transgenic mouse models have implicated another small region in the neurotoxicity of PrP mutants: the N-terminal polybasic domain (residues 23–31) (Figure 3a). This region has been implicated in several aspects of PrPC biology, including regulation of endocytosis [62, 63], and binding to glycosaminoglycans (GAGs) [64]. When residues 23–31 are removed from PrP carrying the neurotoxic deletion Δ32–134, yielding Δ23–134 PrP, the protein is no longer toxic [65]. Moreover, deleting residues 23–31 within ΔCR PrP or a disease-associated PrP point mutant (G130V) completely abrogates the channel-inducing activity and cytotoxicity of the latter mutations [46] (Figure 2d). Interestingly, removal of the 23–31 region greatly diminishes the ability of WT PrP to suppress neurodegeneration in mice expressing a PrP deletion mutant (Δ32-134) [66].
In summary, two different regions of PrP (the central region and the N-terminal polybasic domain) are essential for the neurotoxic effects produced by deleted forms of PrP, as well as for the ability of WT PrPC to suppress these effects. What is the explanation for this correlation? One possibility is that toxic mutations in PrP disrupt a normal physiological function of the protein (e.g., neuroprotection), for example by altering interactions with downstream signaling components, resulting in neurotoxic effects. In this scenario, WT and mutant PrP might compete for binding to cell surface receptors that control cell death or survival via the N-terminal and central domains. Another hypothesis is that PrPC functional activity depends on multimerization via these two domains [57], and incorporation of mutant PrP molecules into the complex somehow results in a neurotoxic signal. Future studies will be necessary to test these and other models, and to determine whether these same two regions of PrP also play a role in the neurotoxicity of infectious prions.
Membrane interactions and PrP toxicity
Most molecules of PrPC are attached to the plasma membrane exclusively via a GPI anchor at the C-terminus, and can be released by phospholipase-mediated cleavage of the anchor [67, 68]. However, there is also evidence that PrPC can associate with the membrane independently of the GPI-anchor, and that the polypeptide chain can permeate the lipid bilayer under some conditions. Such interactions may represent an aspect of the normal cell biology of PrPC, but they may also be the source of cytotoxic effects.
There is extensive evidence that the polypeptide chain of PrPC can span the lipid bilayer under certain circumstances [68]. Two topological variants of PrPC have been described, designated CtmPrP and NtmPrP, with their N- or C-termini, respectively, on the extracellular side of the membrane [69, 70]. Both forms span the membrane via the central, hydrophobic domain (amino acids 112–130). The proportion of CtmPrP synthesized in the endoplasmic reticulum (ER) can be enhanced by mutations in the central, hydrophobic domain as well as the N-terminal signal peptide [69, 70], and expression of these mutants in transgenic mice induces neurodegenerative phenotypes [70, 71]. Interestingly, the phenotype associated with one of these mutants is dependent on the co-expression of WT PrPC, suggesting that CtmPrP subverts the normal function of PrPC function to generate toxicity [72]. Forms of PrP that have been engineered to remain in the cytoplasm by removal of their N- and C-terminal signal peptides also cause neurodegeneration when expressed in transgenic mice [73], but whether such forms exist naturally is unclear. The toxic effects of cytosolic as well as transmembrane forms of PrP may be related to inappropriate interactions between the PrP polypeptide chain and proteins in the cytoplasm [74, 75].
Another potential interaction between the PrP polypeptide chain and the lipid bilayer involves the ability of two different segments of PrP to function as protein transduction domains (PTDs). PTDs are polybasic peptide segments, exemplified by the nine amino acid transactivator of transcription (TAT) peptide derived from the TAT protein of human immunodeficiency virus (HIV-1). These domains are able to penetrate membranes and promote cellular internalization of proteins to which they are fused or covalently attached [76]. Two polybasic domains of PrPC (residues 23–28 and 100–109) resemble prototypic PTDs, and indeed the 23–28 region has been shown to permeabilize membranes of bacteria [77] and to mediate transduction of attached reporter proteins into mammalian cells [78]. It is possible that the PTD activity of the N-terminal domain is related to the ability of PrP molecules with deletions in the central region (like ΔCR) to induce channels or pores in the cell membrane, an effect that is abolished when residues 23–31 are deleted [79]. It will be important to investigate whether the PTD-like domains of PrPC mediate protein transduction events at the cell membrane under normal circumstances.
Intriguing connections between PrPC and Aβ
Recent observations have raised the unexpected possibility that PrPC also plays an important role in the biology of AD. AD is the most prevalent form of dementia in the aging population, and is characterized pathologically by the accumulation of the Aβ peptide, a cleavage product of the amyloid precursor protein (APP) [80, 81]. Considerable evidence indicates that oligomeric assemblies of Aβ, rather than large amyloid fibrils, are the key toxic species in AD, and that these assemblies cause neurodegenerative changes by delivering a toxic signal at synapses [82]. However, the identity of the receptors on neurons that bind Aβ to transduce neurotoxic signals has remained enigmatic.
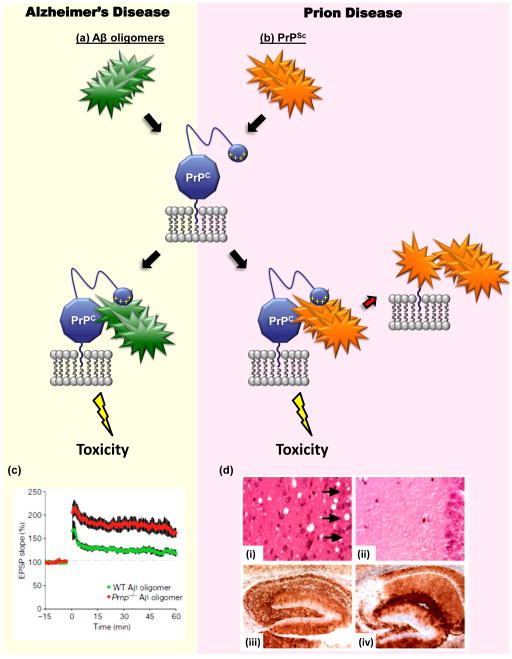
Recent data have suggested that PrPC is a high affinity receptor for Aβ oligomers, and may also mediate the synaptotoxic effects of these assemblies [4] (Figure 4a, b). In the initial study [4], PrPC emerged from an expression cloning screen as a receptor capable of binding Aβ oligomers with nanomolar affinity. Binding was not observed with Aβ monomers or fibrils, suggesting that PrPC was specifically a receptor for oligomers. Importantly, PrPC was also found to be a mediator of Aβ-induced synaptotoxicity. In support of this conclusion, hippocampal slices derived from Prn-p0/0 mice were found to be resistant to Aβ oligomer-induced suppression of long-term potentiation (LTP) (Figure 4c). In a second study from the same group [83], Prn-p0/0 mice were crossed with transgenic mice that model AD, in which mutant forms of APP and presenilin-1 are co-expressed. The results showed that PrPC was required for both the cognitive deficits and reduced survival observed in AD mice, although the presence of PrPC did not influence the rate of Aβ plaque formation in the brain.
FIGURE 4. PrPC may mediate the toxicity of both Aβ oligomers and PrPSc.
The diagram illustrates how cell-surface PrPC, as a consequence of its binding to either Aβ oligomers (green) or PrPSc (orange), may deliver a neurotoxic signal in Alzheimer’s disease (a) and prion diseases (b), respectively. The N-terminal polybasic domain of PrPC (depicted as a small blue ball carrying positive charges) has been shown to participate in binding to both PrPSc [128] and Aβ oligomers [94]. In addition to acting as a transducer of PrPSc-induced neurotoxicty, PrPC also serves as a precursor for the generation of more molecules of PrPSc, which allows prion propagation [1] [(red arrow in part (b)]. (c) PrPC is required for inhibition of hippocampal LTP induced by Aβ oligomers. Field potentials were recorded from the CA1 region of hippocampal slices derived from wild-type mice (green) or Prn-p−/− mice (red), which had both been treated with Aβ oligomers [4]. The red trace is similar to the one recorded from untreated control (Prn-p+/+) slices (not shown). (d) Depletion of neuronal PrPC reverses spongiosis, but not accumulation of PrPSc, in prion-infected mice [7]. Neuronal PrPC expression in mice previously infected with scrapie prions was eliminated by means of Cre-Lox recombination at approximately 12 weeks of age. Hippocampal sections from PrP-expressing mice (i and iii) or PrP-ablated mice (ii and iv) (at 8 and 48 weeks post infection, respectively) were stained with hematoxylin and eosin (i and ii) or an anti-PrP antibody (iii and iv). The severe loss of CA1 to CA3 neurons caused by scrapie infection (arrows) can be rescued by depleting neuronal PrPC, despite continued PrPSc accumulation. Reproduced, with permission, from [4] (c) and [7] (d).
These initial findings motivated other groups to test the link between PrPC and Aβ oligomers. Several studies have confirmed that PrPC plays an essential role in mediating the alterations in synaptic plasticity induced by Aβ oligomers [84], and have provided evidence that immuno-targeting PrPC in vitro or in vivo can rescue Aβ-dependent toxic effects [85–87]. In contrast, however, another study showed that synthetic Aβ oligomers injected intraventricularly into mice impaired consolidation of long-term recognition memory regardless of whether the animals expressed PrPC [88]. In two other recent studies, genetic ablation of PrPC had no effect on Aβ-induced inhibiton of hippocampal LTP in brain slices [89], or learning and memory deficits in a line of AD transgenic mice expressing mutant APP [90]. Using organotypic brain slices, another group reported that PrPC was not required for any of several synaptotoxic effects of Aβ oligomers, including depression of basal synaptic transmission, reduction in the number of dendritic spines, and blockade of LTP [91].
The evident discrepancy that emerges from these studies may, at least in part, be explained by the fact that preparation of synthetic Aβ oligomers is notoriously challenging, and the product obtained can differ from one laboratory to another [92, 93]. The ability of synthetic Aβ oligomers to bind PrPC has been observed in all of the published studies, although it is unclear why only some preparations of Aβ oligomers appear to display PrPC-dependent synaptotoxicity. It is possible that only a specific conformation or size of synthetic oligomers operate through a PrPC-dependent mechanism. The discrepancy between results obtained in vivo by different laboratories may also be due to the use of different transgenic models of AD. It is possible that multiple receptors mediate the toxic effect of Aβ oligomers, and eliminating a PrPC-dependent pathway may produce only a partial rescue of the AD phenotype, or none at all, depending on the mouse model being studied. Given these uncertainties, the role of PrPC in mediating the synaptic toxicity of Aβ clearly requires further clarification.
Two, distinct Aβ oligomer binding sites have been identified on PrPC (residues 95–105 and 23–27), based on deletion analysis, antibody inhibition, and biophysical techniques [4, 94]. It is noteworthy that these two sites are coincident with or lie immediately adjacent to regions that are important determinants of PrPC activity and mutant PrP toxicity (as discussed in previous sections). Moreover, residues 23–31 have been implicated in internalization of PrP via clathrin-mediated endocytosis [63, 95]. Interestingly, another recent study showed that Aβ oligomers can affect the localization of PrPC by increasing the formation of clusters of PrPC on the cell surface [96].
Taken together, these results suggest that the same structural domains involved in the binding to Aβ oligomers may also govern PrPC function, cellular trafficking, and toxicity. Furthermore, a recent study provided evidence that PrPC could mediate the toxicity not only of Aβ oligomers, but also of other β-sheet-rich protein conformers [5]. Therefore, it is possible that oligomeric forms of several different neurotoxic proteins could exert their effects by blocking, enhancing or subverting the normal function of PrPC.
Although considerable attention has been focused on the ability of PrPC to serve as a receptor for Aβ oligomers, there is also evidence that PrPC might play a role in the processing events that generate Aβ from its APP precursor. In two recent studies, PrPC was reported to physically interact with the β-site APP cleaving enzyme 1 (BACE1), an enzyme responsible for cleaving APP and initiating the amyloidogenic cascade [97, 98]. As a result of this interaction, BACE1 activity was inhibited, and the overall production of Aβ peptides decreased.
In conclusion, despite the debate surrounding the recent studies, defining the potential connections between PrPC and Aβ, and determining whether these interactions play a role in pathological conditions is undoubtedly an exciting future line of investigation.
Concluding remarks
The possibility that alterations in the normal function of PrPC play a role in the pathogenesis of prion and other neurodegenerative diseases is sparking renewed efforts to elucidate the cellular and molecular pathways in which this protein is involved. Traditional approaches involving analysis of PrP knock-out mice and biochemical identification of PrPC interacting molecules have been supplemented by the development of new assays for PrP-related functions in cultured cells and transgenic mice, as well as by the use of zebrafish as a genetically tractable model system. Taken together, these studies have uncovered roles for PrPC in cell adhesion, neurite outgrowth, ion channel activity, neuronal excitability, cytotoxicity and cytoprotection.
The disease relevance of these efforts has been heightened by recent reports that PrPC, aside from serving as a precursor of PrPSc and mediating prion-induced neurodegeneration, may also play an important role in the pathology of AD. There is evidence in both prion diseases and AD that the disease phenotype may depend on some alteration in the physiological activity of PrPC. Thus, binding to cell-surface PrPC of either oligomeric Aβ or PrPSc (or other pathogenic PrP aggregates) may initiate toxic signals that lead to neuronal loss and/or synaptic dysfunction, changes that are ameliorated by elimination of PrPC (Figure 4).
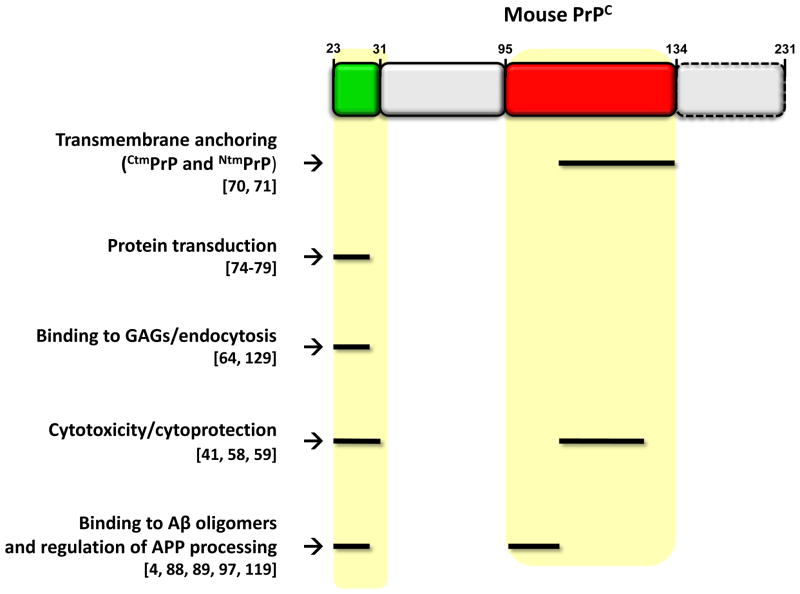
Interestingly, the same domains of PrPC that are critical for its physiological and cellular functions have also been implicated in mediating neurodegenerative processes. Two regions, which lie almost entirely within the unstructured half of the protein [99], seem to be particularly important: the N-terminal polybasic domain (residues 23–31) and the central domain (residues 95–135) (Figure 5). These two regions influence several cell biological features of the protein, including transmembrane anchoring, protein transduction, interaction with GAGs, and endocytic trafficking. Manipulation of these domains creates or alters cytotoxic and cytoprotective activities of the molecule that can be detected in several different assays in vitro and in vivo. They are also responsible for binding to Aβ oligomers, possibly mediating the synaptotoxic effects of the latter, and may regulate the enzymatic processing steps responsible for production of the Aβ peptide.
FIGURE 5. Important functional domains of PrPC.
The figure summarizes the functions attributed to two regions of PrPC (highlighted in yellow shading): the N-terminal polybasic domain (residues 23–31) and the central region (residues 95–135). These two regions influence several cell biological and functional properties of the protein, including transmembrane anchoring (resulting in formation of CtmPrP and NtmPrP) [69, 70], interaction with glycosaminoglycans (GAGs) [64], endocytosis [129], cytotoxicity and cytoprotection [41, 58, 59], binding to Aβ oligomers [4, 88, 89, 119], and regulation of the enzymatic steps involved in the APP processing [97]. In addition, the N-terminal polybasic domain of PrPC (residues 23–28) resembles prototypic protein transduction domains (PTDs) [76–78], short polypeptide segments capable of penetrating cellular membranes. Residues are based on the mouse sequence.
Although several outstanding questions remain (Box 3), these new results highlight a close connection between the normal biology of PrPC and its role in several neurodegenerative diseases. This connection has important therapeutic implications. Compounds that bind to specific functional domains of PrPC and modulate its physiological activity may retard the neurotoxic effects of several kinds of β-rich protein aggregates. Pursuing this novel therapeutic approach will clearly require a deeper understanding of PrPC and its position at the crossroads of physiology and disease.
BOX 2. Outstanding questions.
What is the physiological function of PrPC? A number of functions have been proposed, based largely on subtle abnormalities in PrP knock-out mice or in cells derived from them, including roles in cell adhesion, neurite outgrowth, neuronal excitability, and neuroprotection. However, the underlying molecular mechanisms remain uncertain. A recent addition to the list of experimental models for studying the function of PrPC is the zebrafish, which, unlike the mouse, displays dramatic phenotypes when PrP gene expression is knocked-down. How the different effects of PrP deletion can be reconciled is unclear. PrPC may operate in several, distinct cellular processes, or it may have a more general modulatory role (e.g., protection from cellular stress).
What molecules interact with PrPC? A key to deciphering the function of PrPC is the identification of interacting proteins. A number of candidate interactors have been identified based on co-immunoprecipitation, yeast-two hybrid analysis, and molecular cross-linking [19, 114], but the physiological significance of many of these remains unclear. The interactors include cell surface or secreted molecules involved in adhesion [24, 115, 116]), receptor signaling and trafficking [117], and ion channel/transporter activity [40, 118]. Other proposed PrPC interactors are localized primarily to the cytoplasmic compartment. These might associate with the polypeptide chain of cell-surface PrPC via transmembrane linker proteins, for example as part of a signal transduction complex [119–124]. Intracellular proteins might also bind directly to cytosolic forms of PrP, or the intracellular domains of transmembrane variants (CtmPrP and NtmPrP). The functional activity of PrPC may also depend on interaction with non-protein components, such as lipids of GAGs [64].
How do alterations of the function of PrPC contribute to the neurotoxicity of prions? In addition to its well-known role as a precursor to PrPSc, PrPC is required for transducing prion-related neurotoxic signals. Thus, the normal, physiological function of PrPC may be subverted as part of the pathological process. How this occurs is still poorly understood. PrP molecules missing certain critical regions produce dramatic neurodegenerative phenotypes when expressed in transgenic mice, which may reflect alterations in the normal function of PrPC. It remains to be determined whether the same pathways are affected by infectious PrPSc. If so, then compounds that affect the functional activities of PrPC may represent potent therapeutic agents in prion diseases.
Does PrPC mediate the neurotoxicity of Aβ oligomers in AD, and possibly of β-rich protein assemblies in other neurodegenerative disorders? It is likely that PrPC binds Aβ oligomers, but whether this interaction represents a major pathway for synaptotoxicity is unresolved. Relevant to this issue, it would be useful to know how oligomer binding affects the physiological and cell biological properties of PrPC, for example its localization, processing, and assayable functional activities. Regardless of whether cell-surface PrPC mediates Aβ-induced synaptotoxicity, soluble forms of PrP could have therapeutic benefit by sequestering oligomers in the extracellular space. This effect may extend to β-rich protein oligomers in other neurodegenerative disorders, some of which have also been shown to bind PrPC [5].
Acknowledgments
We thank Edward Malaga Trillo and Roberto Chiesa for critical reading of an earlier draft of the manuscript. The Harris laboratory is supported by grants from the National Institutes of Health (NS052526, NS040975, NS065244, and NS056376), the Dana Foundation, and the Creutzfeldt-Jakob Disease Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuite MF, Serio TR. The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Biol. 2010;11:823–833. doi: 10.1038/nrm3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris DA, True HL. New insights into prion structure and toxicity. Neuron. 2006;50:353–357. doi: 10.1016/j.neuron.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Laurén J, et al. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature. 2009;457:1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Resenberger UK, et al. The cellular prion protein mediates neurotoxic signalling of beta-sheet-rich conformers independent of prion replication. Embo J. 2011;10:2057–70. doi: 10.1038/emboj.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandberg MK, et al. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature. 2011;470:540–542. doi: 10.1038/nature09768. [DOI] [PubMed] [Google Scholar]
- 7.Mallucci G, et al. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 8.Brandner S, et al. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–343. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- 9.Chesebro B, et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308:1435–1439. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- 10.Klingeborn M, et al. Crucial role for prion protein membrane anchoring in the neuroinvasion and neural spread of prion infection. J Virol. 2011;85:1484–1494. doi: 10.1128/JVI.02167-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oesch B, et al. A cellular gene encodes scrapie PrP 27–30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 12.Büeler H, et al. Normal development and behavior of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 13.Manson JC, et al. 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol Neurobiol. 1994;8:121–127. doi: 10.1007/BF02780662. [DOI] [PubMed] [Google Scholar]
- 14.Büeler H, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 15.Moore RC, et al. Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein doppel. J Mol Biol. 1999;292:797–817. doi: 10.1006/jmbi.1999.3108. [DOI] [PubMed] [Google Scholar]
- 16.Steele AD, et al. The prion protein knockout mouse: a phenotype under challenge. Prion. 2007;1:83–93. doi: 10.4161/pri.1.2.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Pichon CE, et al. Olfactory behavior and physiology are disrupted in prion protein knockout mice. Nature neuroscience. 2009;12:60–69. doi: 10.1038/nn.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bremer J, et al. Axonal prion protein is required for peripheral myelin maintenance. Nature neuroscience. 2010;13:310–318. doi: 10.1038/nn.2483. [DOI] [PubMed] [Google Scholar]
- 19.Linden R, et al. Physiology of the prion protein. Physiol Rev. 2008;88:673–728. doi: 10.1152/physrev.00007.2007. [DOI] [PubMed] [Google Scholar]
- 20.Graner E, et al. Cellular prion protein binds laminin and mediates neuritogenesis. Mol Brain Res. 2000;76:85–92. doi: 10.1016/s0169-328x(99)00334-4. [DOI] [PubMed] [Google Scholar]
- 21.Rieger R, et al. The human 37-kDa laminin receptor precursor interacts with the prion protein in eukaryotic cells. Nat Med. 1997;3:1383–1388. doi: 10.1038/nm1297-1383. [DOI] [PubMed] [Google Scholar]
- 22.Gauczynski S, et al. The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. Embo J. 2001;20:5863–5875. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt-Ulms G, et al. Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J Mol Biol. 2001;314:1209–1225. doi: 10.1006/jmbi.2000.5183. [DOI] [PubMed] [Google Scholar]
- 24.Santuccione A, et al. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J Cell Biol. 2005;169:341–354. doi: 10.1083/jcb.200409127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devanathan V, et al. Cellular form of prion protein inhibits Reelin-mediated shedding of Caspr from the neuronal cell surface to potentiate Caspr-mediated inhibition of neurite outgrowth. J Neurosci. 2010;30:9292–9305. doi: 10.1523/JNEUROSCI.5657-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malaga-Trillo E, et al. Regulation of embryonic cell adhesion by the prion protein. PLoS Biol. 2009;7:e55. doi: 10.1371/journal.pbio.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moya KL, et al. Immunolocalization of the cellular prion protein in normal brain. Microsc Res Tech. 2000;50:58–65. doi: 10.1002/1097-0029(20000701)50:1<58::AID-JEMT9>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 28.Kanaani J, et al. Recombinant prion protein induces rapid polarization and development of synapses in embryonic rat hippocampal neurons in vitro. J Neurochem. 2005;95:1373–1386. doi: 10.1111/j.1471-4159.2005.03469.x. [DOI] [PubMed] [Google Scholar]
- 29.Collinge J, et al. Prion protein is necessary for normal synaptic function. Nature. 1994;370:295–297. doi: 10.1038/370295a0. [DOI] [PubMed] [Google Scholar]
- 30.Manson JC, et al. PrP gene dosage and long term potentiation. Neurodegeneration. 1995;4:113–114. doi: 10.1006/neur.1995.0014. [DOI] [PubMed] [Google Scholar]
- 31.Carleton A, et al. Dose-dependent, prion protein (PrP)-mediated facilitation of excitatory synaptic transmission in the mouse hippocampus. Pflugers Arch. 2001;442:223–229. doi: 10.1007/s004240100523. [DOI] [PubMed] [Google Scholar]
- 32.Maglio LE, et al. Hippocampal synaptic plasticity in mice devoid of cellular prion protein. Brain research. 2004;131:58–64. doi: 10.1016/j.molbrainres.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Colling SB, et al. Hippocampal slices from prion protein null mice: disrupted Ca2+-activated K+ currents. Neurosci Lett. 1996;209:49–52. doi: 10.1016/0304-3940(96)12596-9. [DOI] [PubMed] [Google Scholar]
- 34.Herms JW, et al. Prion protein affects Ca2+-activated K+ currents in cerebellar Purkinje cells. Neurobiol Dis. 2001;8:324–330. doi: 10.1006/nbdi.2000.0369. [DOI] [PubMed] [Google Scholar]
- 35.Mallucci GR, et al. Post-natal knockout of prion protein alters hippocampal CA1 properties, but does not result in neurodegeneration. EMBO J. 2002;21:202–210. doi: 10.1093/emboj/21.3.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuhrmann M, et al. Loss of the cellular prion protein affects the Ca2+ homeostasis in hippocampal CA1 neurons. J Neurochem. 2006;98:1876–1885. doi: 10.1111/j.1471-4159.2006.04011.x. [DOI] [PubMed] [Google Scholar]
- 37.Lazzari C, et al. Cellular prion protein is implicated in the regulation of local Ca2+ movements in cerebellar granule neurons. J Neurochem. 2011;116:881–890. doi: 10.1111/j.1471-4159.2010.07015.x. [DOI] [PubMed] [Google Scholar]
- 38.Prestori F, et al. Altered neuron excitability and synaptic plasticity in the cerebellar granular layer of juvenile prion protein knock-out mice with impaired motor control. J Neurosci. 2008;28:7091–7103. doi: 10.1523/JNEUROSCI.0409-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gillessen T, et al. Excitatory amino acid neurotoxicity. Adv Exp Med Biol. 2002;513:3–40. doi: 10.1007/978-1-4615-0123-7_1. [DOI] [PubMed] [Google Scholar]
- 40.Khosravani H, et al. Prion protein attenuates excitotoxicity by inhibiting NMDA receptors. J Cell Biol. 2008;181:551–565. doi: 10.1083/jcb.200711002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li A, et al. Neonatal lethality in transgenic mice expressing prion protein with a deletion of residues 105–125. EMBO J. 2007;26:548–558. doi: 10.1038/sj.emboj.7601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christensen HM, et al. A highly toxic cellular prion protein induces a novel, nonapoptotic form of neuronal death. Am J Pathol. 2010;176:2695–2706. doi: 10.2353/ajpath.2010.091007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rangel A, et al. Enhanced susceptibility of Prnp-deficient mice to kainate-induced seizures, neuronal apoptosis, and death: Role of AMPA/kainate receptors. J Neurosci Res. 2007;85:2741–2755. doi: 10.1002/jnr.21215. [DOI] [PubMed] [Google Scholar]
- 44.Beraldo FH, et al. Metabotropic glutamate receptors transduce signals for neurite outgrowth after binding of the prion protein to laminin gamma1 chain. Faseb J. 2011;25:265–279. doi: 10.1096/fj.10-161653. [DOI] [PubMed] [Google Scholar]
- 45.Solomon IH, et al. Neurotoxic mutants of the prion protein induce spontaneous ionic currents in cultured cells. J Biol Chem. 2010;285:26719–26726. doi: 10.1074/jbc.M110.134619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solomon IH, et al. An N-terminal polybasic domain and cell surface localization are required for mutant prion protein toxicity. J Biol Chem. 2011;286:14724–14736. doi: 10.1074/jbc.M110.214973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shyu WC, et al. Overexpression of PrPC by adenovirus-mediated gene targeting reduces ischemic injury in a stroke rat model. J Neurosci. 2005;25:8967–8977. doi: 10.1523/JNEUROSCI.1115-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milhavet O, Lehmann S. Oxidative stress and the prion protein in transmissible spongiform encephalopathies. Brain Res Rev. 2002;38:328–339. doi: 10.1016/s0165-0173(01)00150-3. [DOI] [PubMed] [Google Scholar]
- 49.Kim BH, et al. The cellular prion protein (PrPC) prevents apoptotic neuronal cell death and mitochondrial dysfunction induced by serum deprivation. Brain Res Mol Brain Res. 2004;124:40–50. doi: 10.1016/j.molbrainres.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Roucou X, et al. Cytosolic prion protein is not toxic and protects against Bax-mediated cell death in human primary neurons. J Biol Chem. 2003;278:40877–40881. doi: 10.1074/jbc.M306177200. [DOI] [PubMed] [Google Scholar]
- 51.Diarra-Mehrpour M, et al. Prion protein prevents human breast carcinoma cell line from tumor necrosis factor alpha-induced cell death. Cancer Res. 2004;64:719–727. doi: 10.1158/0008-5472.can-03-1735. [DOI] [PubMed] [Google Scholar]
- 52.Lo RY, et al. New molecular insights into cellular survival and stress responses: neuroprotective role of cellular prion protein (PrPC) Mol Neurobiol. 2007;35:236–244. doi: 10.1007/s12035-007-8003-y. [DOI] [PubMed] [Google Scholar]
- 53.Haigh CL, et al. Dominant roles of the polybasic proline motif and copper in the PrP23-89-mediated stress protection response. J Cell Sci. 2009;122:1518–1528. doi: 10.1242/jcs.043604. [DOI] [PubMed] [Google Scholar]
- 54.Zanata SM, et al. Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection. EMBO J. 2002;21:3307–3316. doi: 10.1093/emboj/cdf325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santos TG, et al. Enhanced Neural Progenitor/Stem Cells Self-Renewal via the Interaction of Stress-Inducible Protein 1 with the Prion Protein. Stem cells (Dayton, Ohio) 2011;29:1126–1136. doi: 10.1002/stem.664. [DOI] [PubMed] [Google Scholar]
- 56.Steele AD, et al. Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc Natl Acad Sci U S A. 2006;103:3416–3421. doi: 10.1073/pnas.0511290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rambold AS, et al. Stress-protective signalling of prion protein is corrupted by scrapie prions. EMBO J. 2008;27:1974–1984. doi: 10.1038/emboj.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shmerling D, et al. Expression of amino-terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell. 1998;93:203–214. doi: 10.1016/s0092-8674(00)81572-x. [DOI] [PubMed] [Google Scholar]
- 59.Baumann F, et al. Lethal recessive myelin toxicity of prion protein lacking its central domain. EMBO J. 2007;26:538–547. doi: 10.1038/sj.emboj.7601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Massignan T, et al. A novel, drug-based, cellular assay for the activity of neurotoxic mutants of the prion protein. J Biol Chem. 2010;285:7752–7765. doi: 10.1074/jbc.M109.064949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Massignan T, et al. A Drug-Based Cellular Assay (DBCA) for studying cytotoxic and cytoprotective activities of the prion protein: A practical guide. Methods. 2011;53:214–219. doi: 10.1016/j.ymeth.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shyng SL, et al. A glycolipid-anchored prion protein is endocytosed via clathrin-coated pits. J Cell Biol. 1994;125:1239–1250. doi: 10.1083/jcb.125.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sunyach C, et al. The mechanism of internalization of glycosylphosphatidylinositol-anchored prion protein. EMBO J. 2003;22:3591–3601. doi: 10.1093/emboj/cdg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan T, et al. Cell-surface prion protein interacts with glycosaminoglycans. Biochem J. 2002;368:81–90. doi: 10.1042/BJ20020773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Westergard L, et al. A nine amino Acid domain is essential for mutant prion protein toxicity. J Neurosci. 2011;31:14005–14017. doi: 10.1523/JNEUROSCI.1243-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turnbaugh JA, et al. The N-terminal, polybasic region is critical for prion protein neuroprotective activity. PLoS One. 2011;6:e25675. doi: 10.1371/journal.pone.0025675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parkin ET, et al. Dual mechanisms for shedding of the cellular prion protein. J Biol Chem. 2004;279:11170–11178. doi: 10.1074/jbc.M312105200. [DOI] [PubMed] [Google Scholar]
- 68.Harris DA. Trafficking, turnover and membrane topology of PrP. Br Med Bull. 2003;66:71–85. doi: 10.1093/bmb/66.1.71. [DOI] [PubMed] [Google Scholar]
- 69.Hegde RS, et al. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 70.Stewart RS, Harris DA. Mutational analysis of topological determinants in prion protein (PrP) and measurement of transmembrane and cytosolic PrP during prion infection. J Biol Chem. 2003;278:45960–45968. doi: 10.1074/jbc.M307833200. [DOI] [PubMed] [Google Scholar]
- 71.Rane NS, et al. Reduced translocation of nascent prion protein during ER stress contributes to neurodegeneration. Dev Cell. 2008;15:359–370. doi: 10.1016/j.devcel.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stewart RS, et al. Neurodegenerative illness in transgenic mice expressing a transmembrane form of the prion protein. J Neurosci. 2005;25:3469–3477. doi: 10.1523/JNEUROSCI.0105-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma J, et al. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science. 2002;298:1781–1785. doi: 10.1126/science.1073725. [DOI] [PubMed] [Google Scholar]
- 74.Chakrabarti O, Hegde RS. Functional depletion of mahogunin by cytosolically exposed prion protein contributes to neurodegeneration. Cell. 2009;137:1136–1147. doi: 10.1016/j.cell.2009.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rambold AS, et al. Association of Bcl-2 with misfolded prion protein is linked to the toxic potential of cytosolic PrP. Mol Biol Cell. 2006;17:3356–3368. doi: 10.1091/mbc.E06-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gump JM, Dowdy SF. TAT transduction: the molecular mechanism and therapeutic prospects. Trends Mol Med. 2007;13:443–448. doi: 10.1016/j.molmed.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 77.Pasupuleti M, et al. Antimicrobial activity of human prion protein is mediated by its N-terminal region. PLoS One. 2009;4:e7358. doi: 10.1371/journal.pone.0007358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wadia JS, et al. Pathologic prion protein infects cells by lipid-raft dependent macropinocytosis. PLoS One. 2008;3:e3314. doi: 10.1371/journal.pone.0003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Solomon IH, et al. An N-terminal polybasic domain and cell surface localization are required for mutant prion protein toxicity. J Biol Chem. 2011;286:14724–14736. doi: 10.1074/jbc.M110.214973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Selkoe DJ, et al. The role of APP processing and trafficking pathways in the formation of amyloid beta-protein. Ann N Y Acad Sci. 1996;777:57–64. doi: 10.1111/j.1749-6632.1996.tb34401.x. [DOI] [PubMed] [Google Scholar]
- 81.Zhang YW, et al. APP processing in Alzheimer's disease. Mol Brain. 2011;4:3. doi: 10.1186/1756-6606-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walsh DM, Selkoe DJ. A beta oligomers - a decade of discovery. J Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 83.Gimbel DA, et al. Memory impairment in transgenic Alzheimer mice requires cellular prion protein. J Neurosci. 2010;30:6367–6374. doi: 10.1523/JNEUROSCI.0395-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bate C, Williams A. Amyloid-{beta}-induced synapse damage is mediated via cross-linkage of the cellular prion protein. J Biol Chem. 2011 doi: 10.1074/jbc.M111.248724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chung E, et al. Anti-PrPC monoclonal antibody infusion as a novel treatment for cognitive deficits in an Alzheimer's disease model mouse. BMC Neurosci. 2010;11:130. doi: 10.1186/1471-2202-11-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Freir DB, et al. Interaction between prion protein and toxic amyloid beta assemblies can be therapeutically targeted at multiple sites. Nat Commun. 2011;2:336. doi: 10.1038/ncomms1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barry AE, et al. Alzheimer's Disease Brain-Derived Amyloid-{beta}-Mediated Inhibition of LTP In Vivo Is Prevented by Immunotargeting Cellular Prion Protein. J Neurosci. 2011;31:7259–7263. doi: 10.1523/JNEUROSCI.6500-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Balducci C, et al. Synthetic amyloid-beta oligomers impair long-term memory independently of cellular prion protein. Proc Natl Acad Sci U S A. 2010;107:2295–2300. doi: 10.1073/pnas.0911829107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Calella AM, et al. Prion protein and Abeta-related synaptic toxicity impairment. EMBO Mol Med. 2010;2:306–314. doi: 10.1002/emmm.201000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cisse M, et al. Ablation of cellular prion protein does not ameliorate abnormal neural network activity or cognitive dysfunction in the J20 line of human amyloid precursor protein transgenic mice. J Neurosci. 2011;31:10427–10431. doi: 10.1523/JNEUROSCI.1459-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kessels HW, et al. The prion protein as a receptor for amyloid-beta. Nature. 2010;466:E3–4. doi: 10.1038/nature09217. discussion E4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rahimi F, et al. Structure-function relationships of pre-fibrillar protein assemblies in Alzheimer's disease and related disorders. Curr Alzheimer Res. 2008;5:319–341. doi: 10.2174/156720508784533358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Finder VH, Glockshuber R. Amyloid-beta aggregation. Neuro-degenerative diseases. 2007;4:13–27. doi: 10.1159/000100355. [DOI] [PubMed] [Google Scholar]
- 94.Chen S, et al. Interaction between human prion protein and amyloid-beta (Abeta) oligomers: role OF N-terminal residues. J Biol Chem. 2010;285:26377–26383. doi: 10.1074/jbc.M110.145516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taylor DR, et al. Assigning functions to distinct regions of the N-terminus of the prion protein that are involved in its copper-stimulated, clathrin-dependent endocytosis. J Cell Sci. 2005;118:5141–5153. doi: 10.1242/jcs.02627. [DOI] [PubMed] [Google Scholar]
- 96.Caetano FA, et al. Amyloid-beta oligomers increase the localization of prion protein at the cell surface. J Neurochem. 2011;117:538–553. doi: 10.1111/j.1471-4159.2011.07225.x. [DOI] [PubMed] [Google Scholar]
- 97.Parkin ET, et al. Cellular prion protein regulates beta-secretase cleavage of the Alzheimer's amyloid precursor protein. Proc Natl Acad Sci USA. 2007;104:11062–11067. doi: 10.1073/pnas.0609621104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Griffiths HH, et al. Prion Protein Interacts with BACE1 Protein and Differentially Regulates Its Activity toward Wild Type and Swedish Mutant Amyloid Precursor Protein. J Biol Chem. 2011;286:33489–33500. doi: 10.1074/jbc.M111.278556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Riek R, et al. NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 100.Krakauer DC, et al. Phylogenesis of prion protein. Nature. 1996;380:675. doi: 10.1038/380675a0. [DOI] [PubMed] [Google Scholar]
- 101.Bendheim PE, et al. Nearly ubiquitous tissue distribution of the scrapie agent precursor protein. Neurology. 1992;42:149–156. doi: 10.1212/wnl.42.1.149. [DOI] [PubMed] [Google Scholar]
- 102.Brown HR, et al. The mRNA encoding the scrapie agent protein is present in a variety of non-neuronal cells. Acta Neuropathol. 1990;80:1–6. doi: 10.1007/BF00294214. [DOI] [PubMed] [Google Scholar]
- 103.Knaus KJ, et al. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat Struct Biol. 2001;8:770–774. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 104.Stöckel J, et al. Prion protein selectively binds copper(II) ions. Biochemistry. 1998;37:7185–7193. doi: 10.1021/bi972827k. [DOI] [PubMed] [Google Scholar]
- 105.Stahl N, et al. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–249. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 106.Lehmann S, Harris DA. Blockade of glycosylation promotes acquisition of scrapie-like properties by the prion protein in cultured cells. J Biol Chem. 1997;272:21479–21487. doi: 10.1074/jbc.272.34.21479. [DOI] [PubMed] [Google Scholar]
- 107.Gorodinsky A, Harris DA. Glycolipid-anchored proteins in neuroblastoma cells form detergent-resistant complexes without caveolin. J Cell Biol. 1995;129:619–627. doi: 10.1083/jcb.129.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vincent B, et al. The disintegrins ADAM10 and TACE contribute to the constitutive and phorbol ester-regulated normal cleavage of the cellular prion protein. J Biol Chem. 2001;276:37743–37746. doi: 10.1074/jbc.M105677200. [DOI] [PubMed] [Google Scholar]
- 109.Chen SG, et al. Truncated forms of the human prion protein in normal brain and in prion diseases. J Biol Chem. 1995;270:19173–19180. doi: 10.1074/jbc.270.32.19173. [DOI] [PubMed] [Google Scholar]
- 110.Prusiner SB, et al. Purification and structural properties of a major scrapie prion protein. Cell. 1984;38:127–134. doi: 10.1016/0092-8674(84)90533-6. [DOI] [PubMed] [Google Scholar]
- 111.Wickner RB, et al. Prion diseases of yeast: Amyloid structure and biology. Semin Cell Dev Biol. 2011;22:469–475. doi: 10.1016/j.semcdb.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wickner RB, et al. Prion amyloid structure explains templating: how proteins can be genes. FEMS Yeast Res. 2010;10:980–991. doi: 10.1111/j.1567-1364.2010.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 2010;11:155–159. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rutishauser D, et al. The comprehensive native interactome of a fully functional tagged prion protein. PLoS One. 2009;4:e4446. doi: 10.1371/journal.pone.0004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Caughey B, et al. Binding of the protease-sensitive form of PrP (prion protein) to sulfated glycosaminoglycan and congo red. J Virol. 1994;68:2135–2141. doi: 10.1128/jvi.68.4.2135-2141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hundt C, et al. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO J. 2001;20:5876–5886. doi: 10.1093/emboj/20.21.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Taylor DR, Hooper NM. The low-density lipoprotein receptor-related protein 1 (LRP1) mediates the endocytosis of the cellular prion protein. Biochem J. 2007;402:17–23. doi: 10.1042/BJ20061736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kleene R, et al. Prion protein regulates glutamate-dependent lactate transport of astrocytes. J Neurosci. 2007;27:12331–12340. doi: 10.1523/JNEUROSCI.1358-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen S, et al. Prion protein as trans-interacting partner for neurons is involved in neurite outgrowth and neuronal survival. Mol Cell Neurosci. 2003;22:227–233. doi: 10.1016/s1044-7431(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 120.Schneider B, et al. NADPH oxidase and extracellular regulated kinases 1/2 are targets of prion protein signaling in neuronal and nonneuronal cells. Proc Natl Acad Sci USA. 2003;100:13326–13331. doi: 10.1073/pnas.2235648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vassallo N, et al. Activation of phosphatidylinositol 3-kinase by cellular prion protein and its role in cell survival. Biochem Biophys Res Commun. 2005;332:75–82. doi: 10.1016/j.bbrc.2005.04.099. [DOI] [PubMed] [Google Scholar]
- 122.Thellung S, et al. p38 MAP kinase mediates the cell death induced by PrP106–126 in the SH-SY5Y neuroblastoma cells. Neurobiol Dis. 2002;9:69–81. doi: 10.1006/nbdi.2001.0461. [DOI] [PubMed] [Google Scholar]
- 123.Pietri M, et al. Overstimulation of PrPC signaling pathways by prion peptide 106–126 causes oxidative injury of bioaminergic neuronal cells. J Biol Chem. 2006;281:28470–28479. doi: 10.1074/jbc.M602774200. [DOI] [PubMed] [Google Scholar]
- 124.Carimalo J, et al. Activation of the JNK-c-Jun pathway during the early phase of neuronal apoptosis induced by PrP106–126 and prion infection. Eur J Neurosci. 2005;21:2311–2319. doi: 10.1111/j.1460-9568.2005.04080.x. [DOI] [PubMed] [Google Scholar]
- 125.DeMarco ML, Daggett V. From conversion to aggregation: protofibril formation of the prion protein. Proc Natl Acad Sci U S A. 2004;101:2293–2298. doi: 10.1073/pnas.0307178101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Govaerts C, et al. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.D'Souza-Schorey C. Disassembling adherens junctions: breaking up is hard to do. Trends Cell Biol. 2005;15:19–26. doi: 10.1016/j.tcb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 128.Solforosi L, et al. Toward molecular dissection of PrPC-PrPSc interactions. J Biol Chem. 2007;282:7465–7471. doi: 10.1074/jbc.M610051200. [DOI] [PubMed] [Google Scholar]
- 129.Shyng SL, et al. The N-terminal domain of a glycolipid-anchored prion protein is essential for its endocytosis via clathrin-coated pits. J Biol Chem. 1995;270:14793–14800. doi: 10.1074/jbc.270.24.14793. [DOI] [PubMed] [Google Scholar]