Abstract
Wild-type Phospholamban (WT-PLB), a Ca2+-ATPase (SERCA) regulator in the sarcoplasmic reticulum membrane, was studied using TOAC nitroxide spin labeling, magnetically aligned bicelles, and electron paramagnetic resonance (EPR) spectroscopy to ascertain structural and dynamic information. Different structural domains of PLB (transmembrane segment: positions 42 and 45, loop region: position 20, and cytoplasmic domain: position 10) were probed with rigid TOAC spin labels to extract the transmembrane helical tilt and structural dynamic information, which is crucial for understanding the regulatory function of PLB in modulating Ca2+-ATPase activity. Aligned experiments indicate that the transmembrane domain of wild-type PLB has a helical tilt of 13° ± 4° in DMPC/DHPC bicelles. TOAC spin labels placed on the WT-PLB transmembrane domain showed highly restricted motion with more than 100 ns rotational correlation time (τc); whereas the loop, and the cytoplasmic regions each consists of two distinct motional dynamics: one fast component in the sub-nanosecond scale and the other component is slower dynamics in the nanosecond range.
Keywords: Phospholamban (PLB), TOAC nitroxide spin label, Bicelles, Membrane Proteins, Electron Paramagnetic Resonance Spectroscopy
1. Introduction
Phospholamban (PLB) is a 52-residue homopentameric transmembrane protein that regulates the cardiac sarco/endoplasmic reticulum calcium ATPase (SERCA) activity [1, 2]. SERCA is responsible for translocation of calcium ions across the lumen in the sarcoplasmic reticulum (SR) membrane; therefore, controlling the contraction and relaxation cycle of the heart [3, 4]. In its unphosphorylated form, PLB interacts with SERCA with apparent affinity for Ca2+ ions; whereas phosphorylation of PLB at Ser-16 and Thr-17 by cyclic AMP- and Ca2+/calmodulin dependent kinases relieves the PLB-SERCA inhibition [3, 5].
In lipid bilayers, PLB exists mainly in a pentameric form. PLB has 3 main structural features: (i) cytoplasmic region: (residues:1–16, domain Ia), (ii) hinge region: (residues 17–30, domain Ib), and (iii) transmembrane (TM) section (residues 31–52, domain II). Earlier studies of PLB disagree whether the pentameric protein consists of a continuous helix or two helices connected by an unstructured loop or β-sheet region [6, 7]. Two models of PLB have been proposed by the Thomas/Veglia groups (pinwheel model) [8, 9] and the Chou group (bellflower like structural model) [10]. The pinwheel model indicates that the cytoplasmic region of PLB lies on and interacts with the membrane surface, which was also supported by reports from the Middleton and Lorigan groups [11, 12]. However, the bellflower like model, suggests that the cytoplasmic region of the PLB points away from the membrane surface making a bellflower-like structure. In addition, solution and solid-state NMR studies have also been performed on AFA-PLB (fully functional PLB monomer with C36A, C41F, C46A mutations) [13, 14]. The Veglia/Thomas group proposed that the cytosolic segment of AFA-PLB is α-helical and interacts with the membrane surface and is connected to the transmembrane domain through the loop region [13]. Conversely, the Baldus group reports that the cytosolic region of AFA-PLB is completely unstructured and has minimal interaction with the membrane surface [14]. Thus, there is a debate about both the structure and orientation of the cytoplasmic segment of PLB. Even those models, which agree on the α-helical secondary structure of the PLB cytoplasmic domain, disagree on its orientation with respect to the membrane and have reported inconsistent helical tilt angles varying from 20° in bellflower like model [10], to 28° in continuous helix model [6], and 50°–60° in an extended helix-sheet-helix model [7], and ~90° in the pinwheel model [9]. Although all proposed PLB models agree on the α-helical secondary structure of the transmembrane domain, there are also inconsistent reports in the literature about the helical tilt of the transmembrane domain of PLB [6, 9, 15–17]. The helical tilt of the transmembrane domain of PLB varies from 28° to 10°. The purpose of this study is to resolve these inconsistencies with the PLB helix tilt angles using newly developed aligned EPR spectroscopic techniques. The TOAC spin label was used as a probe for EPR measurements because it is a compact probe that is rigidly coupled to the α-carbon and reports more accurately on the position, orientation and backbone dynamics of PLB [18]. SDS-PAGE on TOAC labeled wild-type PLB indicated that this spin label does not significantly alter the pentamer to monomer distribution of wild-type PLB. This is also consistent with a previous study [9]. Previously reported gels suggest point mutations from wild-type PLB to N27A do not significantly alter the pentamer to monomer distribution [19]. Additional biophysical techniques are needed to verify pertinent membrane protein structural information. The second aspect of the project is to compare the dynamics of PLB in different regions (e.g., transmembrane, loop, and cytoplasmic domain) with EPR spectroscopy.
2. Materials and Method
2.1 Peptide Synthesis
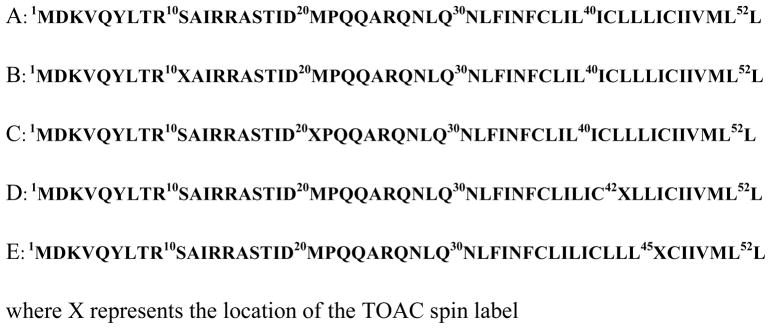
Solid phase peptide synthesis was carried out on a 433A peptide synthesizer (Applied Biosystems Inc., Foster City, CA) using Fmoc-protection chemistry. Amino acid sequences of the phospholamban protein are shown in Figure 1.
Figure 1.
Amino acid sequences of PLB (A) WT-PLB (B) TOAC-10 PLB (C) TOAC-20 PLB (D) TOAC-42 PLB (E) TOAC-45 WT-PLB.
A UV-detector (301 nm wavelength) on the 433A peptide synthesizer was used for monitoring the Fmoc-group removal from the N-terminus of the growing peptide. A modified version of 0.1 mmol Fmoc chemistry protocol (SynthAssist 2.0 software, Applied Biosystems) was used for peptide synthesis. The software modification facilitated optimal peptide synthesis by enabling special functions such as extended double coupling, unnatural amino acid incorporation, and increased reaction times. All amino acids used were in both Fmoc protected and side-chains protected form to minimize unnecessary side chain reactions during peptide synthesis. Solid phase peptide synthesis was used for site-specific TOAC labeling in PLB by functionalizing the amino terminal of the TOAC with the Fmoc group [20]. The synthesis of Fmoc-TOAC was carried out following standard procedures from the literature [20 ]. The synthesis and purification of the full-length PLB provided challenges because of its long amino acid sequence and highly hydrophobic nature. In many cases, TOAC spin-labeling led to peptide truncations. These truncated peptides had similar hydrophobicity, to the full-length peptide, which complicates protein purification and makes the reverse phase HPLC extremely difficult. Therefore, an optimized full-length PLB synthesis took about 10 days for completion. The TOAC labeled PLB was synthesized following standard protocols published in the literature [21–23]. When only HBTU/HOBt coupling agents were used, many truncations that resulted made it difficult to get the full-length TOAC labeled PLB synthesized. The protein synthesis was optimized (i) using a pseudoproline dipeptide Fmoc-Leu-Thr (ψMe,Me-Pro)-OH from Novabiochem, San Diego, CA [24] and (ii) using a superior coupling agent HATU/HOAt [25] to attach the TOAC spin label as well as a few amino acids surrounding it (two amino acids preceding the TOAC and 5–6 amino acids after the TOAC attachment) with extended coupling times. This method ensured the efficient coupling of the TOAC spin probe and also prevented peptide truncation just after the TOAC attachment caused due to low nucleophilicity of the TOAC amino group [21, 26]. The rest of the amino acids were attached using the normal HBTU/HOBt coupling reagent.
About 150 mg of peptide resin was treated with a cleavage mixture (85% TFA, 5% TIS, 5% anisole and 5% distilled water) for 3 hrs and 45 mins. The protein-resin mixture was filtered; resins were rinsed with TFA. The filtrate was concentrated and precipitated in ice-cold methyl tert-butyl ether. The proteins were collected by centrifugation and vacuum dried overnight. The crude proteins were purified on an Amersham Pharmacia Biotech AKTA Explorer 10S HPLC system using reversed phase polymer column (Vydac cat. # 259VHP82215, 8μm, 22 mm × 150 mm). The HPLC solvent system for PLB purification comprised of H2O with 0.1% TFA (Solvent A) and a mixture of 57% isopropyl alcohol (IPA), 38% acetonitrile (MeCN), and 5% water with 0.1% TFA (Solvent B). All TOAC-labeled PLBs were double purified with a slightly modified solvent system in reversed phase C-18 semiprep column (Vydac cat. # 218TP1010, 10μm, 10mm × 250 mm) to ensure the high purity needed for EPR spectroscopy (~95%). The solutions from the corresponding HPLC peaks were lyophilized to yield pure protein. The HPLC purified proteins were treated with a few drops of 10% aq. ammonia (3.5 hr, pH= 9.5) in a 80:20 n-propanol-water mixture to fully retain the radical character of the TOAC nitroxide. This step was necessary since TFA converts nitroxide label into a hydroxylamine form during peptide cleavage and purification process, and the radical character of TOAC is partially lost. The purity of the peptide was confirmed to be ~95% by analytical high performance liquid chromatography (HPLC).
2.2 Sample Preparation
The PLB bicelle samples were prepared from DMPC, DHPC, PEG200-PE, cholesterol and spin-labeled peptide in the molar ratio of 3.5/1/0.035/0.35/0.0035 respectively. The peptide-lipid mixture (250 lipids/PLB) was taken in a pear-shaped flask and dried with nitrogen gas and then desiccated overnight using a high-vacuum pump to remove the residual solvents. The next day, 150 μL of the HEPES buffer (100 mM, pH = 7.0) was added to the lipid peptide mixture to make 25% lipids sample (w/w). The mixture was periodically vortexed and chilled in an ice bath until all of the lipids were completely dissolved. Extreme care was taken in this step to make the sample completely homogeneous and clear sample. Dissolved bicelle samples were sonicated (30 min, Fisher Scientific FS30, Florence, KY) and subjected to four freeze thaw cycles to remove air bubbles and further homogenize the sample. The magnetic alignment of bicelles was achieved by adding 20 mol% of specific lanthanide ions (either Dy3+ or Tm3+) into the DMPC/DHPC bicelles [22, 27]. About 50 μL of sample were drawn into capillary tubes, sealed with cristoseal at the end and placed into a quartz tube (Wilmad, 707-SQ-250M) for EPR spectroscopy.
2.3 EPR Spectroscopy
EPR experiments were performed on a Bruker EMX X-band CW-EPR spectrometer operating at a microwave frequency of 9.5 GHz, 10 mW microwave power, 100 kHz modulation frequency, and 1.0 kHz modulation amplitude. All EPR spectra were recorded at 318 K to achieve optimum alignment. A 42-s field sweep scan with a central field of 3400 G and sweep-width of 100 G were applied.
2.4 EPR Spectral Simulations
The EPR spectra were rigorously analyzed using the nonlinear least-squares (NLSL) analysis program [28, 29]. The EPR spectrum of TOAC-10 PLB, and the randomly dispersed spectra of TOAC-20, TOAC-42 and TOAC-45 PLB were analyzed using the MOMD (macroscopic order, microscopic disorder) model incorporated into the NLSL program. The parallel and perpendicular aligned samples were simulated using NLSL simulations with the introduction of the director tilt angle (Ψ). We assumed that the restoring potential is symmetric to the bilayer normal so that only the c20 axial symmetry term determines the order parameters. The diffusion tilt angle (βd=21°) was taken from the solution structure of PLB and fixed during simulations. The truncated basis sets: Lemax=16, Lomax=15, Kmax=10, and Mmax=10 were used along with the nuclear spin transition index values p1= 2 for the 42-TOAC and 45-TOAC transmembrane spectral simulations. For the cytoplasmic and loop region spectra simulations, these parameters were taken [12, 11, 8, 8, 2] for the T-component and [4, 3, 2, 2, 2] for the R-component, similar to Nesmelov et al. [30]. During simulations, A, g, linewidths and the dynamic parameters were tightly refined using the starting set of hyperfine and g-tensoral parameters from the literature [30]. Each randomly dispersed spectrum was simulated with 40 different orientations (described by the parameter nort in the MOMD simulation) to get full convergence. The rotational correlation time is defined in terms of rotational diffusion coefficients DR, as τc = 1/6DR. For anisotropic motions, the correlation time for unrestricted axial rotation is given by τc(||) = 1/6DR(||) and restricted wobbling motions as τc(⊥) = 1/6DR((⊥) with an associated order parameter S as described by Nesmelov et al. [30].
3. Results and Discussion
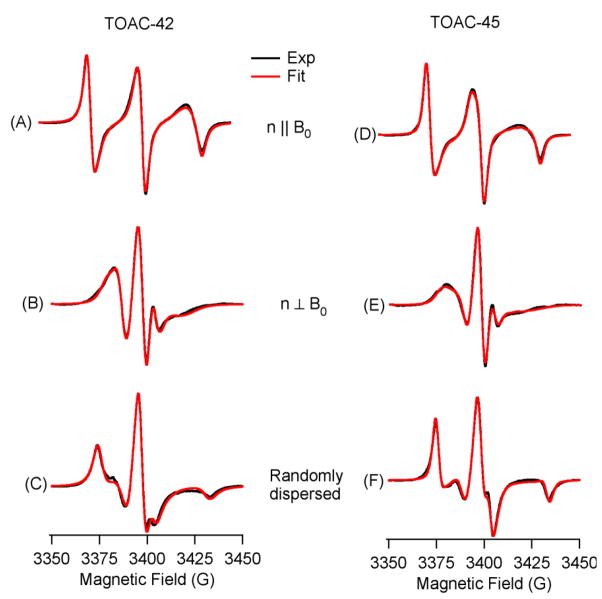
Figure 2 shows CW-EPR spectra of two PLB transmembrane sites labeled with TOAC at positions 42 (left panel) and 45 (right panel) reconstituted into DMPC/DHPC lipid bilcelles at 318 K. The black traces represent experimental EPR spectra and the red traces represent the corresponding best fits from the NLSL simulations. The EPR simulation parameters are given in Table S1. Figures 2(A) and 2(D) represent Tm3+-doped parallel-aligned spectra and Figures 2(B) and 2(E) show Dy3+-doped perpendicular aligned spectra. The parallel and perpendicular alignments are described in the terms of the orientation of the molecular Z-axis of the TOAC spin label (or Z-axis of static magnetic field) with respect to the bilayer normal. Figures 2(C) and 2(F) show a broad unaligned spectrum corresponding to the rigid limit spectra of the TOAC spin label at positions 42 and 45. Orientation-dependent alignment of the TOAC spin probe in the PLB data is clearly visible in the EPR spectra of aligned DMPC/DHPC lipid bicelles, when compared to the unoriented spectra.
Figure 2.
CW-EPR spectra of TOAC-42 PLB [(A), (B), (C)], and TOAC-45 PLB [(D), (E), (F)] in DMPC/DHPC bilcelles at 318 K. (A) and (D) represent parallel aligned (n || Bo) spectra, (B) and (E) represent perpendicular aligned (n ⊥ Bo) spectra, and (C) and (F) represent randomly dispersed spectra. The black traces represent experimental EPR spectra, and red traces are the corresponding fits from NLSL simulations.
In parallel-aligned bicelles, the membrane normal as well as the z-axis of the TOAC nitroxide are nearly parallel to the static magnetic field giving rise to a maximum hyperfine splitting value of 27.2 G and 26.9 G for TOAC-42 and TOAC-45 respectively. For the perpendicular alignment (n ⊥ Bo), the z-axis of the TOAC nitroxide orients nearly perpendicular to the magnetic field thereby averaging the x- and y-nitroxide hyperfine tensors yielding an EPR spectrum with a minimal hyperfine splitting (11.2 G for TOAC-42 and 11.4 G for TOAC-45).
3.1 Transmembrane Helical Tilt Calculation for WT-PLB
The aligned EPR spectra in Fig. 2 can be used to calculate the helical tilt angle of the transmembrane domain of WT-PLB using established procedures in the literature [22, 31, 32]. The details about the matrix algebra required to calculate the angle between the long helical axis vector (h) and the bilayer normal (n), angle φ from the experimentally determined director tilt vector (ZD) and the bilayer normal (angle ξ) of a parallel aligned bicelle sample can be found in Inbaraj et al. [22]. Briefly, equation 1 can be used to calculate the director tilt angle when the magnetic field (Bo) makes an angle (ψ) with the director tilt vector (ZD), the axis of motional averaging.
| (1) |
| (2) |
which finally reduces to,
| (3) |
where, (αn,βn,0) and (αD,βD,0) represent Euler’s angles with respect to the bilayer normal (n) and the director tilt vector (ZD) respectively.
The hyperfine splitting values measured from the aligned PLB spectra at 42- position, were Aexp = 27.2 G for parallel aligned spectrum (Fig. 2(A)), and Aexp = 11.2 G for perpendicularly aligned spectrum (Fig. 2(B)). A|| = 29.8 G and A⊥ = 7.5 G were obtained from the randomly dispersed spectrum [Fig. 2(C)]. Using equation 1, the director tilt angle (ψ), was calculated as 25°± 4° which agrees well with the 27°± 4° director tilt angle obtained from the best-fit NLSL simulations. Using the 25° director tilt angle, the 21° tilt of ZD with respect to the helical axis based upon crystal structure, and unitary transformations involving three Euler angles (α, β, γ), a helix tilt angle of 11°± 4° was calculated. This value agrees well with a helical tilt of 14°± 4° calculated by using the director tilt angle of 27° directly obtained from the NLSL simulations.
Similarly, the hyperfine splitting values measured from aligned TOAC-45 PLB spectra were Aexp = 26.9 G for the parallel aligned spectrum (Fig. 2(D)), and Aexp = 11.37 G for the perpendicularly aligned spectrum (Fig. 2(E)). A|| = 29.68 G and A⊥ = 7.65 G values were obtained directly from the randomly dispersed spectrum (Fig. 2(F)). Using equation 1, the director tilt angle (ψ), was calculated to be 26°± 4° which agreed well with the director tilt angle of 28°± 4° obtained from the best-fit NLSL simulations. The helix tilt angle of 12°± 4° was calculated using the 26° director tilt from aligned spectra [Fig. 2(D), (E), and (F)]. Similarly, the 15°± 4° helical tilt angle was obtained using a 28° director tilt from the NLSL simulations.
By orienting TOAC-labeled PLB reconstituted in DMPC/DHPC bicelles and applying the method used in Inbaraj et al., the transmembrane helical tilt of WT-PLB was calculated to be 13°± 4°(average of four tilt angles for two residues). This helical tilt angle agrees well with the tilt angles reported in previous literature [9, 16, 33]. Torres et al. reported the helix tilt of 10°± 6° for TM-PLB in a hydrated lipid bilayer using ATR-FTIR (Attenuated Total Reflection Fourier Transform Infrared) site-directed dichroism and the restraints obtained from molecular dynamics simulation [33]. Karp et al. performed 13C-CPMAS (cross-polarization magic angle spinning), 2H, and REDOR (Rotational-Echo Double Resonance) solid-state NMR studies on TM-PLB and calculated a helical tilt angle of 11°± 5° [16]. A very important study by Traaseth et al. used 1H/15N PISEMA (Polarization Inversion Spin Exchange at the Magic Angle) solid-state NMR experiments on WT-PLB in oriented lipid bilayer and calculated a 15°± 5° helix tilt angle with respect to the bilayer normal [9]. Thus, utilizing the rigidly bound TOAC spin label and the magnetic alignment techniques the helical tilt of the membrane bound protein can easily be calculated. It is important to verify the structural parameters such as helical tilt with more than one biophysical technique. The helical tilt information can only be ascertained using the aligned techniques discussed in this paper.
3.2 Backbone Dynamics of the Transmembrane Domain of PLB
The rigidly attached TOAC spin label on the PLB transmembrane segment not only enables us to calculate the helical tilt angle, but also provides direct insight into the protein backbone dynamics [18, 22, 34]. The EPR spectra of randomly dispersed WT-PLB at positions 42 and 45 (Figs. 2(C) and 2(F)) produced broadened EPR lineshapes indicating slow motion on the EPR timescale. Simulations of the TOAC EPR spectra using the MOMD method within the NLSL program [28] (see Table S1 for EPR simulation parameters) resulted in a single component fit with very long rotational correlation times. The results show slow uniaxial rotation around the helix axis parallel to the bilayer normal indicated by the τc(||) value (τc(||) = 296 ns for TOAC-45 PLB and τc(||) = 310 ns for TOAC-42 PLB). However, the wobbling motion of the TOAC nitroxide perpendicular to the long helix-axis is quite fast [τc(⊥) = 3 ns for TOAC-45 PLB and τc(⊥) = 8 ns for TOAC-42 PLB]. The molecular order parameters for TOAC-45 PLB and TOAC-42 PLB calculated from the NLSL simulations were 0.84 ± 0.02 and 0.87 ± 0.02 respectively, which indicate a restricted motion of the TOAC nitroxides even at a high temperature of 318 K in the membrane.
Nesmelov et al. have reported the slow rotational motion around the transmembrane helix of PLB monomer [τc(||) = 105 ns] for TOAC-46 AFA-PLB and fast wobbling motion of TOAC nitroxide perpendicular to the helical axis [τc(⊥) = 2 ns] with a 0.93 order parameter value [30]. Our simulation results are similar to Nesmelov et al. given the fact that wild-type PLB has slower dynamics than its monomeric counterpart. Abu-Baker et al. have shown a broad and well-defined powder pattern spectrum for 15N-labeled WT-PLB at position 42 in POPC multilamellar vesicles with a corresponding chemical shift anisotropy (CSA) value of 172.8 ppm [35]. This also indicates that the backbone motion of transmembrane section of WT-PLB is quite immobile on the NMR timescale.
3.3 Conformational Dynamics of the Cytoplasmic Domain of PLB
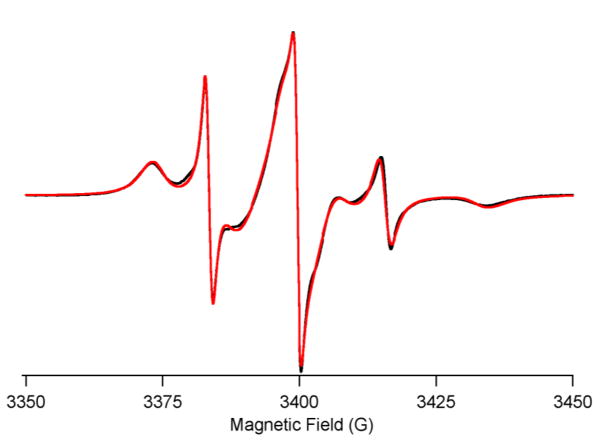
To study the conformational dynamics of the cytoplasmic domain of PLB, SDSL was conducted at Ser-10 with TOAC spin label. The TOAC labeled PLB was reconstituted into DMPC/DHPC bicelles, and the EPR spectrum was recorded at 318 K. The TOAC-10 PLB spectrum (Fig. 3) clearly shows two motional components similar to the EPR spectrum of TOAC-11 AFA PLB (fully functional PLB monomer with C36A, C41F, and C46A mutations and TOAC labeling at position Ala-11) [13, 18, 30].
Figure 3.
CW-EPR spectra of TOAC-10 PLB in DMPC/DHPC bilcelles at 318 K. The black traces represent experimental EPR spectra and red traces are the fits from MOMD simulations.
The EPR spectrum in Fig. 3 was fit best by two-component MOMD simulations with distinct rotational correlation times: τc(T) = 7 ns, for the ordered T-state, and τc(R) = 0.56 ns, for the dynamically disordered R-state. The two-component model gave a better fit than a single component, but a three-component fit did not improve the fit. The order parameter was determined to be 0.62 ± 0.04. The EPR simulation parameters are given in Table S1. The T- and R-components in the EPR spectrum correspond to the two distinct conformational states of the PLB cytoplasmic domain [36]. The T conformer is stabilized through a charged interaction with the membrane surface, whereas the R-conformer points away from the surface and is positioned favorably to interact with its regulatory target SERCA [13, 30]. The T and R components in the EPR spectrum were calculated to be ~85% for the ordered T-component and ~15% for the more dynamic R-component, which is similar to the report of Nesmelov et al. [30].
Nesmelov and co-workers [30] have reported the rotational correlation times of 3.5 ± 0.3 ns and 0.68 ± 0.02 ns for the T and R conformers of the PLB monomer with a mole fraction of 0.84 and 0.16 for T and R conformations respectively. In this paper, our results on the dynamics of the PLB pentamer is 7.0 ± 0.4 ns for the T conformation and 0.56 ± 0.03 ns for the R conformation. The mole fraction of T (0.85 ± 0.02) and R (0.15 ± 0.02) conformations indicate that the pentameric PLB prefers to stay predominantly in the T-conformation similar to the Pinwheel model proposed by Thomas/Veglia groups [8, 9] and supported by the Lorigan [12] and Middleton groups [11]. Veglia and co-workers have also shown almost identical resonance positions of WT-PLB and AFA-PLB in their published (1H/15N]-TROSY-HSQC spectrum for residues 2–31 [9]. The close match between our rotational correlation time for the T and R components and their relative proportion indicates that WT-PLB and AFA-PLB have similar dynamic features in the cytoplasmic domain. Topology mapping studies on the PLB cytoplasmic domain showed that Ser-10 was the most solvent exposed residue, as its TROSY-HSQC spectrum was most effectively quenched by adding Gd3+ ions [9]. However, the Ser-10 residue also retained ~80% of the intensity with 5-doxyl steric acid; another paramagnetic agent that effectively relaxes the hydrophobic residues just below the phosphate head group in the membrane [9]. This indicates that Ser-10 also interacts with the membrane surface, which is consistent with our two-component spectrum of TOAC-10 PLB. Using both uniformly 15N-labeled WT-PLB and site-specifically 15N-labeled PLB (at Ala-11) incorporated in POPC bilayers, Abu-Baker et al., 2007 revealed two distinct 15N-amide backbone dynamics; a relatively narrow isotropic component corresponding to the mobile cytoplasmic backbone component and a broad well-defined powder-like component corresponding to the rigid component [35].
Magnetic alignment studies were also performed on TOAC-10 PLB. However, due to a high backbone dynamics of the cytoplasmic domain and its conformational heterogeneity, we could not obtain well-aligned EPR spectra. The EPR spectra were highly isotropic in nature (data not shown) due to the fast motional averaging of the spin probe on the EPR timescale.
3.4 Loop Region Conformational Dynamics of PLB
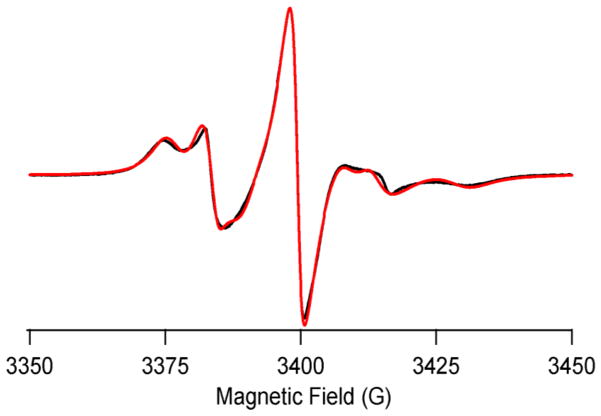
The 20Met→TOAC mutation was made to study the loop region dynamics of PLB. Figure 4 shows the randomly dispersed spectrum at 318 K, which reveals two motional components in the EPR spectrum; a fast motional component (R) represented by a sharp peak and a less dynamic component (T) represented by the broad peak. The two-component MOMD fit (see Table S1 for EPR simulation parameters) to the experimental spectrum shows that the rotational correlation time of the less dynamic T-component is (3.0 ± 0.4) ns and the dynamically disordered R-component is (2.0 ± 0.3) ns. The spectral analysis yielded the distribution of T and R components as ~69% and ~31%, respectively.
Figure 4.
CW-EPR spectra of 20-TOAC PLB in DMPC/DHPC bilcelles at 318 K. The black trace represents experimental EPR spectra and red trace is the fit from MOMD simulations.
4. Conclusions
Rigid TOAC spin labeling and aligned EPR spectroscopic techniques were used to probe the transmembrane helical tilt and dynamics of the wild-type phospholamban. This study reveals that the motional dynamics of transmembrane domain is highly restricted with a rotational correlation time more than 100 ns. Both the loop and the cytoplasmic domain of WT-PLB possess two backbone motions corresponding to the R- and T-conformational states similar to AFA-PLB. The T-component had a few nanosecond rotational correlation times as opposed to a sub-nanosecond rotational correlation time for the dynamically disordered R-component. The major T component corresponds to the cytoplasmic domain of PLB interacting with the membrane surface, while the minor R component (smaller population) is moving faster and does not interact with the membrane. The data agrees with the NMR/EPR pinwheel model discussed in the literature by the Minnesota researchers [37]. Recent work by the Veglia group suggests that the T state is the non-active ground state [36], while the R state is the loss of function state [37]. The function of SERCA can be controlled by enhancing the R state via proper design of mutants [37]. In addition, they report a helical tilt of 13° using solid-state NMR spectroscopy which agrees very well with the reported helical tilt of 13°± 4° for the transmembrane domain calculated using NLSL and aligned EPR spectroscopic techniques. WT-PLB pre-exists in multiple conformations and equilibrated in its two major T and R conformational states when reconstituted in different lipid membrane environments [37]. By fine tuning the PLB conformational equilibria, lipid membrane can be represented as a third element for allosteric control of SERCA[38].
Highlights.
EPR spectroscopic methods developed for determining helical tilt of PLB peptide
Dynamic properties of rigid TOAC spin label attached to PLB measured
Bicelles are an excellent model membrane system
Acknowledgments
This project was supported by a NIGMS/NIH (GM60259-01) grant, and the Ohio Board of Regents (MRI-0722403).
Abbreviations
- PLB
Phospholamban (PLB)
- SERCA
sarco/endoplasmic reticulum Ca2+-ATPase
- DMPC
1,2-Dimyristoyl-sn-Glycero-3-Phosphocholine
- DHPC
1,2-Dihexanoyl-sn-Glycero-3-Phosphocholine (DHPC)
- HPLC
High Performance Liquid Chromatography
- MALDI-TOF
Matrix Assisted Laser Desorption Ionization Time-of- flight Mass Spectrometry
- TOAC
2,2,6,6-tetramethylpiperidine-1oxyl-4-amino-4-carboxylic acid
Footnotes
Supporting Information related to this article can be found online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fuji J, Ueno A, Kitano K, Tanaka S, Kadoma M, Tada M. Characterization of structural unit of phospholamban by amino acid sequencing and electropherotic analysis. Biochim Biophys Res Commn. 1987;138:1044–1050. doi: 10.1016/s0006-291x(86)80387-4. [DOI] [PubMed] [Google Scholar]
- 2.Li HM, Cocco MJ, Steitz TA, Engelman DM. Conversion of phospholamban into a soluble pentameric helical bundle. Biochemistry. 2001;40:6636–6645. doi: 10.1021/bi0026573. [DOI] [PubMed] [Google Scholar]
- 3.Simmerman HKB, Jones LR. Phospholamban: protein structure, mechanism of action, and role in cardiac function. Physiol Rev. 1998;78:921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 4.Kirchberger MA, Tada M, Katz AM. Phospholamban a regulatory protein of the cardiac sarcoplasmic reticulum. Recent Adv Stud Card Struct Metab. 1975;5:103–115. [PubMed] [Google Scholar]
- 5.James P, Inui M, Tada M, Chiesi M, Carafoli E. Nature and site of phospholamban regulation of the calcium pump of sarcoplasmic reticulum. Nature. 1989;342:90–92. doi: 10.1038/342090a0. [DOI] [PubMed] [Google Scholar]
- 6.Arkin IT, Rothman M, Ludlam CFC, Aimoto S, Engelman DM, Rothschild KJ, Smith SO. Structural model of the phospholamban ion-channel complex in phospholipid-membranes. J Biol Chem. 1995;248:824–834. doi: 10.1006/jmbi.1995.0263. [DOI] [PubMed] [Google Scholar]
- 7.Tatulian SA, Jones LR, Reddy LG, Stokes DL, Tamm LK. Secondary structure and orientation of phospholamban reconstituted in supported bilayers from polarized attenuated total-reflection FTIR spectroscopy. Biochemistry. 1995;34:4448–4456. doi: 10.1021/bi00013a038. [DOI] [PubMed] [Google Scholar]
- 8.Robia SL, Flohr NC, Thomas DD. Phospholamban pentamer quaternary conformation determined by in-gel fluorescence anisotropy. Biochemistry. 2005;44:4302–4311. doi: 10.1021/bi0478446. [DOI] [PubMed] [Google Scholar]
- 9.Traaseth NJ, Verardi R, Torgersen KD, Karim CB, Thomas DD, Veglia G. Spectroscopic validation of the pentameric structure of phospholarnban. Proc Natl Acad Sci USA. 2007;104:14676–14681. doi: 10.1073/pnas.0701016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oxenoid K, Chou JJ. The structure of phospholamban pentamer reveals a channel-like architecture in membrane. Proc Natl Acad Sci USA. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clayton JC, Hughes E, Middleton DA. The cytoplasmic domains of phospholamban and phospholemman associate with phospholipid membrane surfaces. Biochemsitry. 2005;44:17016–17026. doi: 10.1021/bi0511383. [DOI] [PubMed] [Google Scholar]
- 12.Abu-Baker S, Lu JX, Chu S, Shetty KK, Gor’kov PL, Lorigan GA. The structural topology of wild-type phospholamban in oriented lipid bilayers using N-15 solid-state NMR spectroscopy. Protein Sci. 2007;16:2345–2349. doi: 10.1110/ps.072977707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zamoon J, Mascioni A, Thomas DD, Veglia G. NMR solution structure and topological orientation of monomeric phospholamban in dodecylphosphocholine micelles. Biophys J. 2003;85:2589–2598. doi: 10.1016/s0006-3495(03)74681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andronesi CA, Becker S, Seidel K, Heise H, Young HS, Baldus M. Detemination of membrane protein structure and dynamics by magic-angle-spininng solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 15.Traaseth NJ, Buffy JJ, Zamoon J, Veglia G. Structural dynamics and topology of phospholamban in oriented lipid bilayers using multidimensional solid-state NMR. Biochemistry. 2006;45:13827–13834. doi: 10.1021/bi0607610. [DOI] [PubMed] [Google Scholar]
- 16.Karp ES, Tiburu EK, Abu-Baker S, Lorigan GA. The structural properties of the transmembrane segment of the integral membrane protein phospholamban utilizing 13C CPMAS, 2H, and REDOR solid-state NMR spectroscopy. BBA-Biomembranes. 2006;1758:772–780. doi: 10.1016/j.bbamem.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Adams PD, Arkin IT, Engelman DM, Brunger AT. Computational searching and mutagenesis suggest a structure for the pentameric transmembrane domain of phospholamban. Nature Struct Biol. 1995;2:154–162. doi: 10.1038/nsb0295-154. [DOI] [PubMed] [Google Scholar]
- 18.Karim CB, Kirby TL, Zhang ZW, Nesmelov Y, Thomas DD. Phospholamban structural dynamics in lipid bilayers probed by a spin label rigidly coupled to the peptide backbone. Proc Natl Acad Sci USA. 2004;101:14437–14442. doi: 10.1073/pnas.0402801101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu S, Coey AT, Lorigan GA. Solid-state 2H and 15N NMR studies of side-chain and backbone dynamics of phospholamban in lipid bilayers: Investigation of the N27A mutation. Biochimica et Biophysica Acta. 2010;1978:210–215. doi: 10.1016/j.bbamem.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchetto R, Schreier S, Nakaie CR. A Novel Spin-Labeled Amino-Acid Derivative For Use in Peptide-Synthesis - (9-Fluroenylmethyloxycarbonyl)-2,2,6,6-Tetramethylpiperidine-N-Oxyl-4-Amino-4-Carboxylic Acid. J Am Chem Soc. 1993;115:11042–11043. [Google Scholar]
- 21.Martin L, Ivancich A, Vita C, Formaggio F, Toniolo C. Solid-phase synthesis of peptides containing the spin-labeled 2,2,6,6-tetramethylpiperidine-1-oxyl-4-amino-4-carboxylic acid (TOAC) J Pept Res. 2001;58:424–432. doi: 10.1034/j.1399-3011.2001.00940.x. [DOI] [PubMed] [Google Scholar]
- 22.Inbaraj JJ, Cardon TB, Laryukhin M, Grosser SM, Lorigan GA. Determining the topology of integral membrane peptides using EPR spectroscopy. J Am Chem Soc. 2006;128:9549–9554. doi: 10.1021/ja0622204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karim CB, Zhang Z, Thomas DD. Synthesis of TOAC spin-labeled proteins and reconstitution in lipid membranes. Nature Protocols. 2007;2:42–49. doi: 10.1038/nprot.2007.2. [DOI] [PubMed] [Google Scholar]
- 24.Abu-Baker S, Lorigan GA. Phospholamban and its phosphorylated form interact differently with lipid bilayers: A 31P, 2H, and 13C solid-state NMR spectroscopic study. Biochemistry. 2006;45:13312–13322. doi: 10.1021/bi0614028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpino LA. 1-Hydroxy-7-Azabenzotriazole - an Efficient Peptide Coupling Additive. J Am Chem Soc. 1993;115:4397–4398. [Google Scholar]
- 26.Carpino LA, Elfaham A, Minor CA, Albericio F. Advantageous Applications of Azabenzotriazole (Triazolopyridine)-Based Coupling Reagents to Solid-Phase Peptide-Synthesis. J Chem Soc, Chem Commun. 1994:201–203. [Google Scholar]
- 27.Cardon TB, Tiburu EK, Padmanabhan A, Howard KP, Lorigan GA. Magnetically aligned phospholipid bilayers at the parallel and perpendicular orientations for X-band spin-label EPR studies. J Am Chem Soc. 2001;123:2913–2914. doi: 10.1021/ja005574i. [DOI] [PubMed] [Google Scholar]
- 28.Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J Magn Reson A. 1996;120:155–189. [Google Scholar]
- 29.Schnedier DJ, Freed JH. Calculating slow motional magnetic resonance spectra: a user’s guide. In: Berliner LJJaR., editor. Spin Labeling: Theory and Application. Biological Magnetic Resonance. Vol. 8. Plenum Press; New York: 1989. pp. 1–76. [Google Scholar]
- 30.Nesmelov YE, Karim CB, Song L, Fajer PG, Thomas DD. Rotational dynamics of phospholamban determined by multifrequency electron paramagnetic resonance. Biophys J. 2007;93:2805–2812. doi: 10.1529/biophysj.107.108910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karp ES, Inbaraj JJ, Laryukhin M, Lorigan GA. Electron paramagnetic resonance studies of an integral membrane peptide inserted into aligned phospholipid bilayer nanotube arrays. J Am Chem Soc. 2006;128:12070–12071. doi: 10.1021/ja064077k. [DOI] [PubMed] [Google Scholar]
- 32.Mayo DJ, Inbaraj JJ, Subbaraman N, Grosser SM, Chan CA, Lorigan GA. Comparing the structural topology of integral and peripheral membrane proteins utilizing electron paramagnetic resonance spectroscopy. J Am Chem Soc. 2008;130:9656–9657. doi: 10.1021/ja803590w. [DOI] [PubMed] [Google Scholar]
- 33.Torres J, Adams PD, Arkin IT. Use of a new label C-13=O-18 in the determination of a structural model of phospholamban in a lipid bilayer. Spatial restraints resolve the ambiguity arising from interpretations of mutagenesis data. Journal of Molecular Biology. 2000;300:677–685. doi: 10.1006/jmbi.2000.3885. [DOI] [PubMed] [Google Scholar]
- 34.Victor KG, Cafiso DS. Location and dynamics of basic peptides at the membrane interface: Electron paramagnetic resonance spectroscopy of tetramethyl-piperidine-N-oxyl-4-amino-4-carboxylic acid-labeled peptides. Biophys J. 2001;81:2241–2250. doi: 10.1016/S0006-3495(01)75871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abu-Baker S, Lu JX, Chu SD, Brinn CC, Makaroff CA, Lorigan GA. Side chain and backbone dynamics of phospholamban in phospholipid bilayers utilizing H-2 and N-15 solid-state NMR spectroscopy. Biochemistry. 2007;46:11695–11706. doi: 10.1021/bi700749q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9101–9106. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gustavsson M, Traaseth NJ, Karim CB, Lockamy EL, Thomas DD, Veglia G. Lipid-Mediated Folding/Unfolding of Phospholamban as a Regulatory Mechanism for the Sarcoplasmic Reticulum Ca2+-ATPase. Journal of Molecular Biology. 2011;408:755–765. doi: 10.1016/j.jmb.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gustavsson M, Traaseth NJ, Veglia G. Probing ground and excited states of phospholamban in model and native lipid membranes by magic angle spinning NMR spectroscopy. Biochimica et Biophysica Acta. 2011 doi: 10.1016/j.bbamem.2011.07.040. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]