Abstract
Trisomy for human chromosome 21 (Hsa21) results in Down syndrome (DS). The finished human genome sequence provides a thorough catalog of the genetic elements whose altered dosage perturbs development and function in DS. However, understanding how small alterations in the steady state transcript levels for <2% of human genes can disrupt development and function of essentially every cell presents a more complicated problem. Mouse models that recapitulate specific aspects of DS have been used to identify changes in brain morphogenesis and function. Here we provide a few examples of how trisomy for specific genes affects the development of the cortex and cerebellum to illustrate how gene dosage effects might contribute to divergence between the trisomic and euploid brains.
Introduction
Trisomy for human chromosome 21 (Hsa21) causes Down syndrome in about 1 of every 800 live births [1]. Individuals with DS show a wide range of effects in many organ systems, some of which are congenital and some of which are progressive. These include cardiac malformations [2], gastrointestinal anomalies [3], craniofacial and skeletal anomalies [4, 5], and contrasting cancer phenotypes which include an increased frequency of childhood leukemia and reduced prevalence of many types of cancer in adults [6, 7]. There is broad variability in both the occurrence (penetrance) and severity (expressivity) of these phenotypes across the DS population. All individuals with DS have some degree of intellectual disability (ID) making this the most frequent genetic cause of ID with intelligence quotients (IQs) that can range from mild to severe; the mean IQ in DS is ~50 [8]. Studies in animal models and in people have uncovered cellular, physiological and functional changes that contribute to the etiology of cognitive deficits and provide the basis for emerging clinical trials aimed at improving cognitive abilities in DS [9–11].
Central nervous system (CNS) abnormalities in DS appear to arise from a combination of abnormal development and functional changes that result directly from overexpression of genes in trisomic cells. Neuropathological changes found during late prenatal development in DS include delayed and disorganized cortical lamination [12], a smaller and hypocellular hippocampal dentate gyrus [13] and hypomorphic cerebellum [14–18]. In the cerebral cortex, pyramidal neurons possess smaller dendritic arborizations [19–21] and there are fewer synapses in the prenatal DS brain [22, 23]. These deficits are followed postnatally by degeneration of cortical pyramidal neurons [24], profound dendrite and synapse abnormalities [19, 21, 24, 25] and reductions in the total number of neurons in the hippocampus and cerebral cortex [13, 26]. Cerebellar development is delayed and ultimately appears to terminate prematurely. These findings indicate that prenatal changes in CNS growth and differentiation underlie many of the pathological abnormalities associated with ID in DS.
Several fundamental cellular growth mechanisms that are critical for development are abnormal in DS. Specific changes have been reported for cell cycle rate and progression in multiple organ systems, suggesting that altered kinetics of cell production may contribute to many phenotypes. DNA synthesis and doubling times are both slower in DS fibroblasts [27, 28] and DS cerebral cortex contains less cyclin-dependent kinase [29] indicating lower replication rates. Recent studies have also shown that proliferation is abnormal in the developing DS forebrain [30] and cerebellum [31]. Many details regarding the molecular pathways underlying these phenotypes and how altered brain formation may lead to ID remain unclear. Here, we consider emerging data from mouse models of DS which demonstrate key deficiencies in neural stem and progenitor cell expansion and changes in the resulting specification of post-mitotic neurons and glial cells in the developing CNS. Characterizing the molecular basis of the abnormalities in the developing DS brain is essential to understanding the etiology of, and developing potential treatments for, the postnatal cognitive disabilities that characterize this syndrome.
Mouse models of Down syndrome and gene effects
Cell intrinsic abnormalities caused by trisomy may be amplified by aberrant cell-cell interactions during development and these cascading effects pose challenges to understanding and ultimately treating the structural and functional defects observed in the brain after birth [32, 33]. The complexity of these relationships in many organ systems cannot be recapitulated in a test tube or cell culture.
Mouse models of DS are trisomic for chromosome regions that show conserved synteny with Hsa21 (Fig. 1) [34]. In comparison to human, orthologous mouse chromosome regions show minor differences in gene content and in the structures of conserved genes, although there are undoubtedly also important differences in gene regulation. However, the occurrence of DS-like outcomes in mice trisomic for orthologs of Hsa21 genes has been established and refined to a great degree using the Ts65Dn mouse model [35]. Increasingly sophisticated mouse models of DS have been developed through cell and chromosome engineering in the last few years, including mice with mosaic trisomy for human chromosome 21 [36] and with trisomy for all mouse orthologs of Hsa21 [37]. The models and their genetic and molecular validation have been reviewed extensively (see, for example, [38–40]).
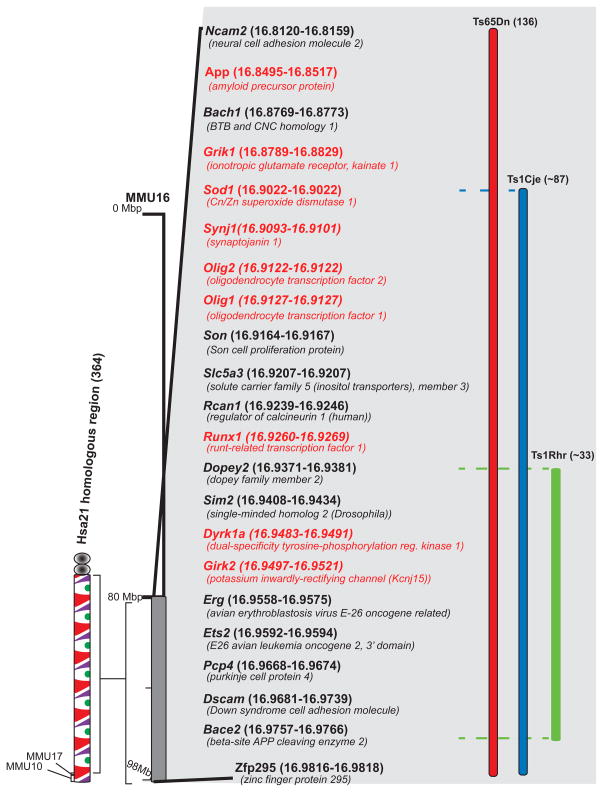
Figure 1. Genetic maps of trisomic mouse models.
The Hsa21 homologous region in the mouse is Mmu16. This region contains orthologs of about half the genes on Hsa21 [41]. The gene content of segmental trisomies occurring as translocations or duplications in the mouse models discussed in this review (ie. Ts65Dn, Ts1Cje and Ts1Rhr) are shown with the approximate number of genes contained in these regions given in parentheses. Some of the individual genes within this region are listed, with the specific genes that are discussed in this review highlighted in red [35, 113, 114]. Gene positions are noted within parentheses; gene labels are not positioned to scale.
Despite significant parallels between DS and mouse models, a mouse is not a human being and the consequence of trisomy in non-human species is not Down syndrome. Nevertheless, fundamental processes of development and function are conserved across mammals and beyond, and dosage changes of the orthologous genes will, in many cases, have comparable effects on outcomes (Figure 2). The proper focus for model studies, then, is on evolutionarily conserved processes and the mechanisms by which they are perturbed. Given a sophisticated understanding of the advantages and limitations of the genetic basis for DS vs. trisomy in the mouse, the utility of the model depends both on the precision with which phenotypes can be determined and the strength of those comparisons to the situation in people. Human phenotypes are normally limiting in this context, as the possibility for intrusive studies in animal models provides for a far more sensitive and comprehensive substrate for assessment.
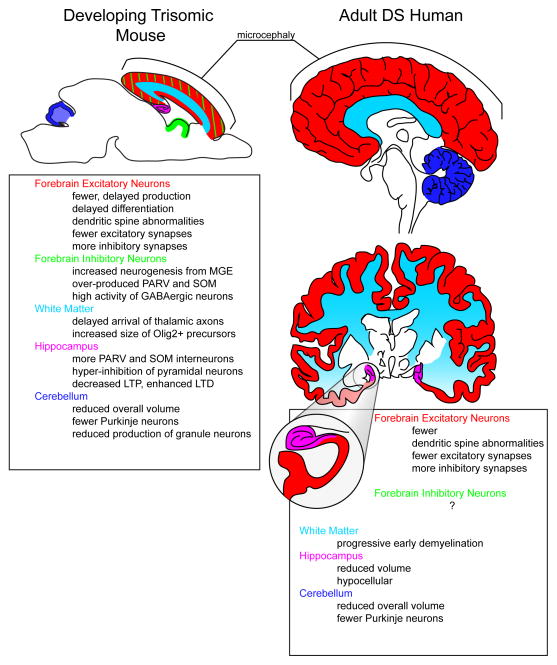
Figure 2. Comparison of altered cellular and anatomical alterations observed in prenatal brain from trisomic mouse models and postnatal human Down syndrome (DS) brain.
Evidence from several trisomic mouse models of DS (left panel) indicate altered production of excitatory and inhibitory forebrain neurons in the neocortex [59, 60], delayed development of the cortical white matter [59], and defective synapse formation/maintenance [58]. In addition, supernumerary interneurons (INs) in the hippocampus result in over-inhibition of pyramidal neurons [58] and may be involved in the observed alterations in synaptic plasticity (ie. long-term potentiation/depression, LTP/LTD) [83, 84]. Defective proliferation of the granule cell precursors also reduces the overall volume and neuronal constituency of the cerebellum [99]. Many of these abnormalities are found during mouse development and presumably underlie morphological and behavioral outcomes in the human DS brain (right panel)(see [12–15, 17–19, 115, 116] for relevant human studies). The magnified area depicts the hippocampus. Abbreviations: MGE, medical ganglionic eminence; PARV, parvalbumin; SOM, somatostatin.
Ts65Dn mice have a number of well-characterized neural phenotypes affecting learning, memory, brain morphology, synaptogenesis, electrophysiology and neurochemical properties of the brain. They undergo neurodegenerative processes affecting structure and function as they age. Many of these effects parallel changes that are observed in the DS brain [41] (Figure 2). A number of trisomic genes have been implicated in various aspects of brain development and function [Box 1], however, such findings are complicated by the fact that behavioral tests with the same name are seldom performed in the same way [41]. Here we consider just three genes - the oligodendrocyte transcription factors (Olig1 and 2), and dual specificity tyrosine-phosphorylation-regulated kinase 1A (Dyrk1a) - as examples of how over-expression due to trisomy can affect function and development of the hippocampus and cortex. We also discuss the anomalous response of trisomic cerebellar precursor neurons to the sonic hedgehog (SHH) growth factor-mediated signaling pathway. This canonical pathway plays a significant role in trisomic mice, but is based on disomic genes. The trisomic gene(s) that cause the aberrant response have not yet been identified.
Box 1. Hsa21 genes implicated in brain structure and function.
Virtually any trisomic gene could potentially affect development or function in the CNS. For example, an enzyme that changes metabolic rates, a growth factor signal that is over-expressed at a critical time in development, or a transcription factor whose increased expression results in increased transcript levels for hundreds of genes [32, 33]. There are many genes on chromosome 21 whose functions in the CNS suggest more specific possible contributions to maldevelopment in DS, and these have been considered at length (see e.g. [117], and a recent meta-analysis of expression data in trisomy [118]). Several of the many genes that have been proposed to affect the CNS when trisomic are mentioned here, however, this is by no means intended as a comprehensive overview of gene dosage effects in DS.
i) Cell surface molecules
A number of these are encoded on Hsa21, including cell adhesion molecules, Neural Cell Adhesion Molecule 2 (NCAM2), Down Syndrome cell adhesion Molecule (DSCAM) and synaptojanin (SYNJ1) which all have potential roles in neuronal development and function. Perhaps one of the most extensively studied cell surface molecules encoded on Hsa21 is APP. Mutations in APP are associated with familial Alzheimer’s Disease (AD), and AD occur in all individuals with trisomy 21. In addition to its role in plaque formation in the aging brain, increased expression of APP has been shown to play a role in the degeneration of cholinergic neurons in the basal forebrain of a mouse model of DS [119], and it likely plays an important role in learning and memory processes [113,.
(ii) Channels and transporters
Multiple ion channels and transporters that are expressed in the brain are encoded on Hsa21. These include GRIK1, an ionotropic glutamate receptor subunit that is implicated in cognition as well as a number of neurological disorders (reviewed in [121]). SLC5A3, solute carrier family 5 (sodium/myo-inositol cotransporter), member 3, is a member of a family of inositol transporters). Inositol transport is an essential function in many cells and tissues, including the brain, but the primary neuronal effects attributed to SLC5A3 are in the peripheral nervous system [122]. GIRK2, a G protein-coupled inwardly rectifying potassium channel, was initially identified because a mutation in this gene (known as the mouse weaver mutation) caused early loss of cerebellar GCPs, resulting in ataxia [123]. Altered Girk2 expression may also contribute to cognitive deficits in DS by interfering with GABAB receptor signaling in the hippocampus[124].
(iii) Transcription factors
Elevated expression of transcription factors would obviously have a direct effect on many dozens or hundreds of additional genes, most of which will be disomic. Thirteen transcription factors and transcription modifiers are encoded on Hsa21. Some of the better studied transcription factors include OLIG1/2, GABP ALPHA, RUNX1, ERG, ETS2, BACH1, SON and NRIP1 (see Figure 1 for gene details).
(iv)Other genes
Some other genes of interest on Hsa21 include Regulator of calcineurin 1 (RCAN1), a negative regulator of calcineurin. Down-regulation of calcineurin by RCAN1 affects calcineurin-dependent gene transcription through the inhibition of NFATc translocation to the nucleus [125]. RCAN1 is highly expressed in cardiac and skeletal muscle, but interactions with DYRK1A in many tissues during development have been posited to perturb Nuclear factor of activated T-cells, cytoplasmic (NFATC) functions and potentially contribute to multiple aspects of DS [125]. Super oxide dismutase1 (SOD1) is another gene that has been extensively researched in DS, and similar to APP, it has been associated with other neurological diseases (including AmyotrophicLateral Sclerosis and Parkinson’s disease [126].
The specification and mapping of forebrain neurons
The function of brain circuits, and the motor and cognitive paradigms they subserve, depends on the proper ratio of excitatory and inhibitory neurons within the network. Fine tuning of this ratio prevents excess excitation and/or inhibition in the neuronal pathways and is important for proper development [42–45]. Excitatory glutamatergic neurons and inhibitory GABAergic neurons converge on the forebrain through a surprisingly diverse but precisely ordered sequence of developmental steps; the faithful execution of each of these steps is necessary for a normally functioning cortex. The proper numbers of neurons must first be generated, specified to the correct cell type, and then migrate to their appropriate position, which is often millimeters to centimeters away from the site of origin.
Excitatory neurons are generated from precursors in the dorsal telencephalic ventricular and subventricular zones (VZ and SVZ) and migrate radially into the neocortical wall [46]. Most inhibitory interneurons are born in the ventral VZ of the ganglionic eminence and populate the cortex and hippocampus via tangential migration [47–51]. Despite their distinct sites of origin, cortical excitatory and inhibitory neurons born on the same embryonic day occupy the same laminar positions within the neocortex [46, 52–56]. Thus, both their allocation within the neocortex and their temporal programs of differentiation must be tightly coordinated. In individuals with DS and in mouse models, deficient production and allocation of neuronal cell types is markedly abnormal in several key regions, including the dorsal and ventral telencephalon and the cerebellum. Together, these studies suggest that cellular defects during the production phase of brain development alter the ratios of different types of neurons and may be key contributors to the eventual cognitive deficits in DS.
The dorsal forebrain phenotype: under-production of excitatory neurons
Due to the delayed development and laminar disorganization evident in late gestational and perinatal DS forebrain, it is now clear that the mechanisms controlling both embryonic production and allocation of neurons is disturbed in DS. Excitatory neurons are particularly affected and synapses with and among these cells are fewer than in euploid brain. How does this arise? Several groups have now quantitatively determined that excitatory neurons are under-produced by the neural stem and progenitor cells in the embryonic VZ in DS fetal brain [30, 57] and in the Ts65Dn mouse [58]. Not only is the cell cycle of VZ cells lengthened, but the commitment of new neurons is also delayed. This combination results in fewer new neurons and a reduction in expansion of the maturing cortical layers. The delay in neuronal differentiation also affects the development of axonal tracts in the white matter [59–61]. This shortfall in excitatory neurons is maintained after birth as neurons of the cerebral cortex begin to establish synapses with both near and distant targets (Figure 3). Since proper synaptic integration of neurons is predicated on their readiness (i.e. timely arrival and proper differentiation state), one major hypothesis from these studies is that the reductions and delays in neurogenesis may prevent or curtail critical windows of development and set the stage for the subsequent defects in synapse formation and consolidation.
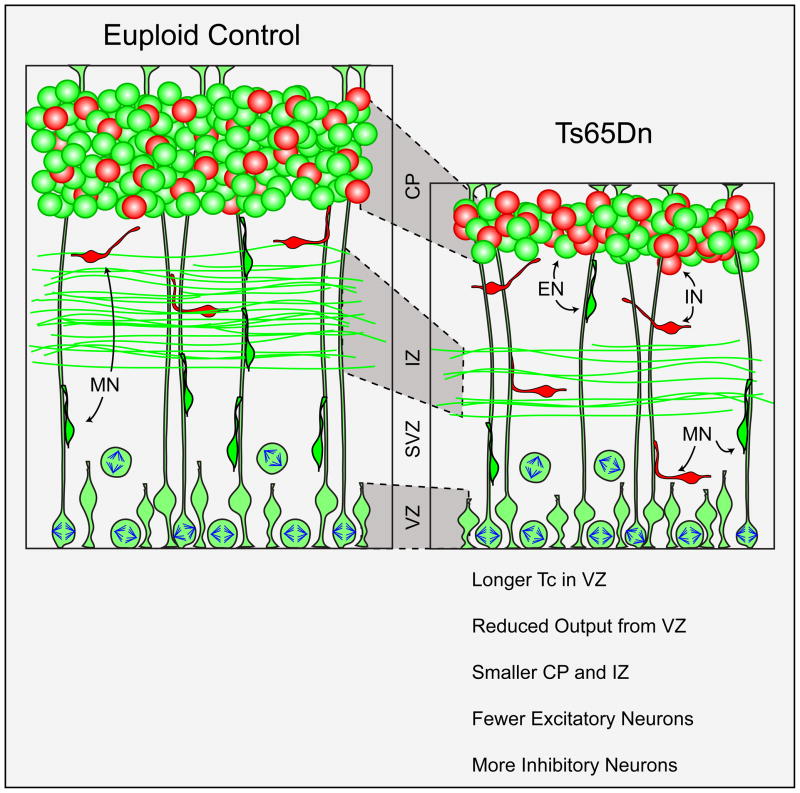
Figure 3. Abnormal cortical development in mouse models of Down syndrome (DS) results in an altered ratio of excitatory and inhibitory neurons.
Evidence from the Ts65Dn mouse model of DS indicates that longer cell cycle duration (Tc) in the ventricular and subventricular zones (VZ and SVZ, respectively) leads to reduced overall production of excitatory neurons (green cells, ENs) at the cortical plate (CP) [59]. This delayed arrival and differentiation of excitatory neurons results in a thinner intermediate zone (IZ) containing the axon tracts of descending and ascending cortical inputs. In addition, overexpression of the transcription factors Olig1 and Olig2 in the germinal zones of the ventral telencephalon leads to increased production of parvalbumin- and somatostatin-expressing interneurons (red cells, INs) in the cerebral cortex and hippocampus [99]. Altogether, these defects change the ratio of excitatory: inhibitory neurons during prenatal forebrain neurogenesis. Abbreviations: MN, migrating neuron.
One of the genes that is triplicated in DS and in several mouse models, and which is now strongly implicated in dorsal telencephalic VZ proliferation, is DYRK1A (formerly called minibrain) [62–66]. This gene encodes a dual specificity tyrosine phosphorylation-regulated kinase that is strongly expressed in neural structures, especially within the neural precursor populations during embryonic neurogenesis [67–69]. Haploinsufficiency or truncation of DYRK1A leads to brain abnormalities in mice and to microcephaly in humans [70, 71]. This kinase interacts with a wide variety of growth factors, transcription factors and cell cycle regulatory proteins known for their roles in neural cell proliferation and specification. These include Fibroblast growth factor beta (bFGF), Cyclin D1, the E2F family of transcription factor (E2F1), the mammalian homolog of the Drosophila transcription factor, Snf5-related 1 (SNR1), the type I transmembrane protein, Notch, neuron-restrictive silencer factor (NRSF, also known as REST) and the 53 kD tumor suppressor protein, p53 [72–78].
Mice that over-express Dyrk1a, either constitutively and promiscuously as a transgene with a leaky metallothionein promoter or under natural promoters in BAC and YAC transgenic mice, display subtle motor defects and spatial learning memory deficits [79]. This has been interpreted to suggest that Dyrk1a overexpression may perturb developmental pathways and lead to postnatal neurodevelopmental delays [80, 81]. DYRK1A is asymmetrically localized to the VZ precursors that divide to give rise to neuronal-committed progenitors or to neurons directly [68]. It has been hypothesized that the increased gene dosage and overexpression in DS may lead to precocious neuronal differentiation and depletion of the neural precursor pool during neurogenesis [82]. Indeed, Dyrk1a overexpression in embryonic VZ precursors, induced by in utero electroporation (IUE), inhibits proliferation and alters neuronal differentiation [77]. The latter study demonstrated that IUE-induced overexpression rapidly depletes the VZ proliferative pool by causing precocious neuronal differentiation. While the committed neurons were still able to migrate to the appropriate cortical layer, the overall production capacity of the germinal zones was decreased by this transient overexpression paradigm. The neurogenesis block induced by Dyrk1a overexpression is due, at least in part, to nuclear export and degradation of Cyclin D1 [77]. Further studies of the multiple roles of Dyrk1a in brain development are needed to explain how overexpression contributes to brain growth and function phenotypes of DS.
The ventral forebrain phenotype: over-production of inhibitory neurons
In addition to the under-production of cortical excitatory neurons caused by the dorsal phenotype, it is now clear that specific classes of forebrain inhibitory interneurons are overproduced during embryonic development in the Ts65Dn mouse model, further altering the excitatory: inhibitory neuron ratio in Ts65Dn [58]. In particular, a substantial increase in the numbers of parvalbumin- (PARV) and somatostatin- (SOM) expressing interneurons occurs in the dorsal neocortex and hippocampus at both embryonic and postnatal stages (Figure 3). These supernumerary interneurons result from elevated rates of cell production by precursors in the ganglionic eminence in the ventral telencephalon. Moreover, neurogenesis is specifically increased only in the medial ganglionic eminence (MGE)–the specific germinal zone for the PARV- and SOM- interneurons–and not in the progenitors of the lateral and caudal ganglionic eminence which generate other groups of unaffected interneurons. One physiological consequence of these over-abundant inhibitory neurons is increased inhibition of forebrain pyramidal neurons.
These results represent the first cellular evidence for the source of over-inhibition in the Ts65Dn forebrain. Excess inhibitory input in the hippocampus may lead to deficiencies in synaptic plasticity with associated cognitive deficits affecting learning and memory [83, 84]. These hippocampal circuits are now a target for therapies to improve cognition in DS (reviewed in [9, 11]). For example, blocking inhibitory GABAergic neurotransmission pharmacologically in adult Ts65Dn mice was shown to ameliorate the spatial learning deficits in these animals [85]. Thus, it is clear that increased inhibition may be a key mediator of cognitive dysfunction in DS.
During prenatal development, MGE precursors are bi-potential and can divide to generate either inhibitory interneurons or oligodendrocytes based on the expression of a combinatorial code of transcription factors [86–88]. Two of the major transcription factors expressed during MGE neurogenesis are Olig1 and Olig2. Both of these genes are triplicated and over-expressed in MGE of Ts65Dn and people with DS. By crossing Olig1+/−; Olig2+/− double heterozygotes to Ts65Dn females, the gene dosage of just Olig1 and Olig2 is returned to euploid levels (i.e., two copies) while maintaining the triplication of the other trisomic genes in the Ts65Dn background. The cortical interneurons in these Ts65DnOlig1/2+/+/− animals develops like that of their euploid littermates, showing normalized values of MGE neurogenesis and inhibitory neuron production [57]. Furthermore, the over-inhibition of hippocampal pyramidal neurons found in Ts65Dn mice is rescued in Ts65DnOlig1/2+/+/− mice. Thus, it appears that triplication of Olig1 and Olig2 is necessary for the ventral inhibitory neuron phenotype in Ts65Dn.
Interestingly, however, triplication of Olig1 and Olig2 is not sufficient for and may not directly affect synaptic plasticity, at least in the hippocampal DG, since synaptic deficits are observed in Ts1Rhr mice which are not trisomic for these two genes [89],[90](Figure 1). Unfortunately, the strength of these deficits has never been directly compared between models, a limitation that applies to behavioral tests as well (Box 2). However, whether triplication of Olig1 and Olig2 is involved in electrophysiological changes in the hippocampus and forebrain has not yet been tested. In future studies, it will be important to determine whether correcting the defects in the generation of excitatory and inhibitory neurons in Ts65Dn also rescues the spatial learning/memory and synaptic plasticity deficits. If this is the case, it will indicate that disturbances in neuronal production directly affect cognitive function in DS.
BOX 2. Outstanding questions.
How does trisomy affect cerebellar GCPs so as to attenuate the response to SHH? Does a trisomic gene directly affect the canonical pathway in an as yet unrecognized fashion? Does an undefined non-canonical pathway involved in GCP responses to the mitogenic effects of SHH include a trisomic gene? Is part of the effect due to a change in SHH production/release/diffusion?
It will be particularly important to understand how DYRK1A overexpression during the entire phase of embryonic neurogenesis affects cortical morphogenesis and function within the context of many other trisomic genes. One approach would be to assess the developmental consequences of normalizing gene dosage of DYRK1A specifically on the Ts65Dn trisomic background by crossing in a null allele of DYRK1A, thereby reducing the number of alleles from 3 to the normal 2 copies.
How does the paucity of excitatory neurons combine with the supernumerary inhibitory neurons to lead to the particular cognitive profile that is specific to DS? One approach is to engineer multiple individual gene rescues into trisomic mouse models as described for Olig1/2 and to assess cognition using electrophysiological and behavioral tasks. Up-regulation of Hsa21 genes in euploid cultured neurons or down-regulation to euploid levels in trisomic cells might provide a faster initial screen. Other models (eg. worms, flies, zebrafish) might provide additional insights into effects that disrupt normal cell-cell interactions in development and function.
The possible contributions of APP dosage to DS pathology continues to be a major focus of DS research. An overexpression of APP in Hsa21, and subsequent changes in the balance of its cleavage products, contribute to the AD pathology in DS, as well as to the retrograde transport of growth factors in basal forebrain cholinergic neurons [127] and to an impairment in the proliferation of neural precursors [105].
A major practical problem, not limited to DS research, is the absence of standardization for electrophysiological and behavioral testing. For DS models, comparisons are made between different mouse models tested in different ways in different labs, yet distinct names are not given to thetests. There has never been, for example, a head-to-head comparison of Ts65Dn, Ts1Cje and Ts1Rhr mice using electrophysiological or behavioral assays in the same experiment. This leads to considerable confusion in the literature.
As in many areas of biomedical research, translation from animal models to the human disorder is a significant challenge. Mice are the highest mammalian model for which there is currently a good genetic model of DS. While there are clear limitations in extrapolating findings from mice to humans, trisomic mouse models have proven useful in assaying for drugs that affect hippocampal function, including behavioral outcomes,. Indeed, results in Ts65Dn mice have led to multiple pilot studies and several clinical trials. A major question is whether the changes that have been observed in mice will produce effects in humans that result in improved cognitive functions and therapeutic outcomes to allow people with DS a broader range of opportunities, as well as an extended lifespan. This is, of course, the ultimate goal of experiments of the type discussed herein.
The cerebellar phenotype
Many features of DS show incomplete penetrance, that is, they are evident in some people but not in others. Where they do occur, they are likely to show variable expressivity/severity. One of the few DS features that occurs to a similar degree in all people with trisomy 21 is a markedly small cerebellum. This finding was recognized before the turn of the nineteenth century and expanded upon as the microscopic structure of the DS brain was characterized (see [91]). In a magnetic resonance imaging (MRI) study of adults with DS, the average cerebellar volume was less than 75% of euploid, which is proportionately smaller than the DS brain overall [14]. This volume was not further reduced with age.
Ts65Dn mice also have reduced cerebellar volume [15]. This reduction affects both the molecular layer and the internal granule layer (IGL) and is further characterized by significant reductions in the linear density of Purkinje cells and in the density of granule cell neurons (GC) in the IGL. Based on the mouse results, GC density was assessed for the first time in DS and shown to be reduced in people with trisomy 21 from 2–60 years old [15]. Both the volume of the IGL (where all cerebellar GC cell bodies reside) and the GC density were reduced by about 20% in Ts65Dn mice, suggesting that trisomic mice had about 65% as many GC as their euploid counterparts. Surprisingly, Ts65Dn mice do as well, or better, than euploid mice on the standard version of the accelerating RotaRod test, a behavioral task designed to test cerebellar functions of balance and motor coordination [15, 92], although a unique version requiring the investigator to place mice in the proper orientation on an already-moving rod found Ts65Dn mice to be impaired [93]. Another variant of this test, also called by the same name, has been used to test Tc1 mice which carry most of human chromosome 21 [35]. Different studies of this mouse show either presence or absence of impairments [36, 94]. The authors suggest that genetic background differences might contribute to the different results; variations in experimental technique might also play a role.
Mechanisms and abnormalities of cerebellar development
The cerebellum is immature at birth, and the IGL is absent early in its development. Instead of the IGL, a layer of granule cell precursors (GCPs) forms the external germinal layer (EGL) at the surface of the cerebellum. Beginning prenatally in human gestation and around the day of birth in mice, these GCPs begin dividing rapidly. Cells on the interior side of the EGL leave the cell cycle and migrate inward along Bergmann radial glia to take up a position in the forming IGL and complete their differentiation. This process occurs in the first two years of human postnatal life and during the first 3–4 postnatal weeks in mice. Proliferation of GCPs is dependent on the growth factor, SHH [95–97].
The cerebellar growth deficit in Ts65Dn mice has been documented morphologically from birth onward [98]. By one week after birth, the trisomic cerebellum was reduced in size in every parameter measured, and remained so during adulthood (Figure 4). On the day of birth, however, the overall size of the cerebellum and the thickness of the EGL were the same as in euploid. While the number of GCPs was the same as euploid, the number of dividing cells was reduced in trisomic pups [98]. When GCPs were isolated from trisomic or euploid cerebella and incubated with increasing amounts of SHH to measure the proliferative response, trisomic GCP cultures had fewer dividing cells at every SHH concentration [94]. Nonetheless, they did respond to SHH in a dosage sensitive manner. Based on these observations, trisomic mice were treated on the day of birth with a small molecule agonist of the SHH pathway (SAG 1.1) [99, 100]. Cerebellar morphology and growth parameters were restored to the same level as euploid at postnatal day 6, about a third of the way through cerebellar development. The mitotic index was also the same as euploid, suggesting that the normalized development of the cerebellum would continue as the animals age [98].
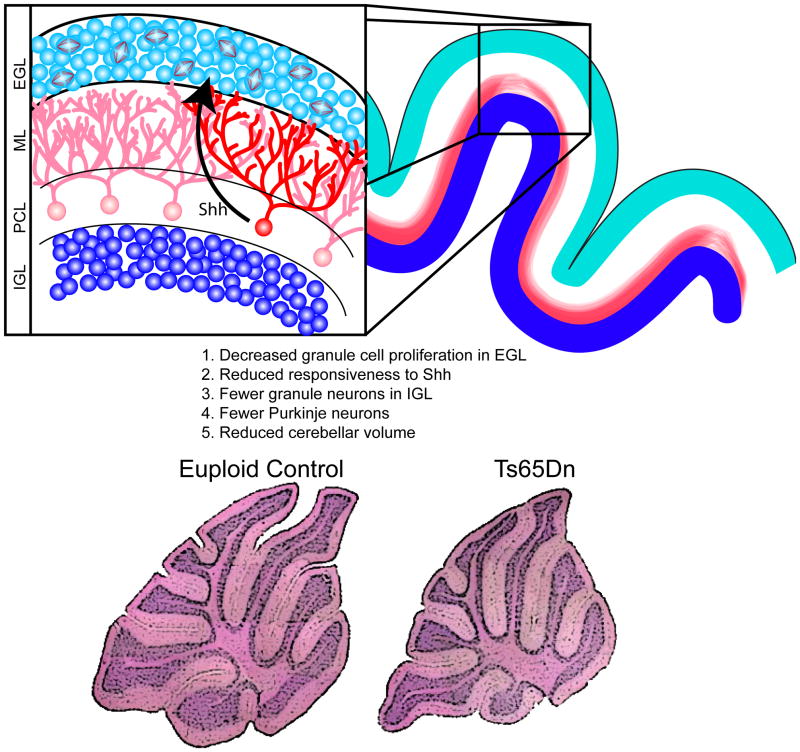
Figure 4. Proliferation deficits in the granule cell precursors (GCPs) lead to reduced cerebellar volume.
Fewer mitotic GPCs are found in the external granule cell layer (EGL) in mouse models of DS [15, 106] and this leads to a paucity of granule cells in the internal granule cell layer (IGL) following their migration inward, past the molecular layer (ML) and Purkinje cell layer (PCL). Reduced granule cell density is seen in the DS cerebellum, as well. The size of the Purkinje cell population is also reduced in trisomic mouse models [15] and, given the reduction in the overall volume of the DS cerebellum [14], this is likely also true in DS. These cell production abnormalities lead to reduced overall cerebellar volume in adult trisomic mice, as shown in the histological sections of euploid (bottom left) and Ts65Dn mouse model (bottom right) [98]. SHH, released by Purkinje cells, is known to influence granule cell production [96, 97], and granule cell precursors in the Ts65Dn cerebellum exhibit reduced responsiveness to SHH compared to euploid; application of a SHH agonist has been used to correct this proliferation defect in the Ts65Dn cerebellum [98]. Histological stainings adapted, with permission, from [98]. Copyright (2006) National Academy of Sciences, USA.
It is of obvious interest to determine the duration of these beneficial effects of SHH pathway stimulation. To date, there are no robust phenotypes of trisomy that can be attributed to the reduction in cerebellar GC and Purkinje cells. It was reported recently that individuals with DS display differences in some parameters of vestibulo-ocular reflex compared to age- and gender-matched people without DS [101]. It would be interesting to analyze this in trisomic mice, especially in combination with electrophysiological measurements as has been done for euploid mice [102]. Recent advances in our understanding of the role of the cerebellum in various learning and memory paradigms raises the question of whether restoration of cerebellar structure can correct not only functions that have historically been linked to the cerebellum, but also those attributed to other brain regions in Ts65Dn and DS [103, 104].
It has been reported that a C-terminal cleavage product of amyloid precursor protein (APP) can act as an accessory transcription factor of Protein patched homolog 1 (Ptch1), the SHH receptor [105]. This occurs in neurospheres derived from the SVZ and hippocampus. Since App is up-regulated in the cerebellum of Ts65Dn mice [35], and Ptch is a negative regulator of the pathway, this could contribute to the attenuated response of cerebellar GCPs to SHH. Several details of the response to SHH and the agonist SAG 1.1 differ between the transient cerebellar GCP population and the long term neuronal population from which these neurospheres are derived, including lower, not higher levels of Ptch transcript [Ref]. Nonetheless, in light of this finding, it is interesting that Ts1Cje mice, which are trisomic for about 80% of the genes triplicated in Ts65Dn (but not for App) (Figure 1) show a similar reduction in cerebellar volume but less of a reduction in GC density than does Ts65Dn [106]. Such a finding is consistent with App dosage effects on the SHH pathway. Finally, increasingly sophisticated bioinformatics analysis of protein network interactions should prove useful in identifying additional trisomic genes that might contribute to DS phenotypes [107].
Future Prospects
Since 1959 when DS was shown to result from trisomy 21 [108], a major focus of research has been to identify Hsa21 genes and correlate their overexpression with specific phenotypes of DS. However, the availability of genetically defined mouse models that reflect features of DS has led to a phenotype-based approach that has proven to be revolutionary for the prospects of improved cognition in DS. Behavioral tests have suggested specific brain regions that might be especially affected by trisomy 21. For example, most trisomic models show some degree of impairment in various aspects of spatial memory. Indeed, the poor performance of Ts65Dn in the Morris water maze, a test of visio-spatial integration that is dependent on the hippocampus, led behavioral neuropsychologists to study hippocampal function in children with DS [35, 109]. The Arizona Cognitive Test Battery is specifically designed to assess functions rooted in the hippocampus, forebrain and cerebellum in people with DS [110] and this battery is being proposed as a readout in several pending clinical trials.
Electrophysiological studies of mouse models have demonstrated impaired function in the forebrain. As discussed above, the hippocampus of Ts65Dn mice has an imbalance of inhibitory relative to excitatory input [83, 84]. These inhibitory neurons are GABAergic, and drugs such as pentylenetetrazole that antagonize activity of GABAA receptors can restore the electrophysiological imbalance and the ability of mice to perform hippocampal-based tasks, such as novel object recognition [85]. Indeed, efforts are underway to conduct clinical trials in DS with several drugs that are known to target GABAA receptors (clinical trial identifier NCT01436955; http://www.clinicaltrials.gov) and which have appeared promising in preclinical trials (eg. [111]).
While postnatal therapies targeting inhibitory neurotransmission may provide a measure of therapy for DS individuals, it is still unclear how efficacious these strategies will be for long-term improvements and whether they particularly act to offset the developmental changes described here. Several other genes triplicated in DS, including the ion channels subunits encoded by GRIK1 (glutamate receptor, ionotropic, kainate 1) and GIRK2 (G protein-coupled inwardly-rectifying potassium channel 2) and the synaptic vesicle trafficking regulator, synaptojanin 1 (SYNJ1), may also fundamentally impact neurotransmission and synapse maintenance after birth (Box 1). It will be important for future studies to distinguish between the relative roles of the prenatal versus postnatal defects with respect to the cognitive dysfunction in DS.
As discussed here, many of the phenotypes seen after birth are a result of gene dosage effects during early development. These early changes in brain formation are likely to substantially affect subsequent developmental events, such as cellular differentiation and synapse formation. Ameliorating or preventing these phenotypes will likely require early intervention. Methods for surgical and/or molecular interventions during late gestation are conceivable and in some cases have already been employed, as in the case of surgical repair of cardiac malformations. At the current time, however, many approaches for the treatment of human fetuses in early gestation are neither technically nor ethically possible, so gene-centric approaches in the prenatal period are limited.
While encouraging results demonstrate that the DYRK1A inhibitor, epigallocatechin gallate (a phenol found in green tea), may improve cognitive function in trisomic animals [112], it remains to be seen whether this therapeutic approach will correct any DYRK1A-associated developmental changes. Increased OLIG1/2 gene dosage and the role that these transcription factors play in the ventral forebrain phenotype presents another possible set of targets. If specific OLIG1/2 inhibitors can be found, the timing and localization of delivery will still present a challenge as these transcription factors are critically important in specifying proper inhibitory neuron, oligodendrocyte and neural tube development. The attenuated response to SHH by GCPs, if it represents a down-regulated response by all trisomic cells, may point to a common mechanism that may account for multiple DS phenotypes. The availability of trisomic mouse models in which development is affected in a similar manner to DS will continue to play a critical role in elucidating causes and identifying opportunities for therapeutic interventions in this complex disorder.
Acknowledgments
This work was supported in part by the Down Syndrome Research and Treatment Foundation (R.H.R), Research Down Syndrome (R.H.R), the Dana Foundation (T.F.H.) and the National Institute of Child Health & Human Development [ R01HD038384 (R.H.R.) and R01HD057580 (T.F.H.). The content of this manuscript is the responsibility of the authors and does not necessarily represent the views of the National Institutes of Health.
Footnotes
Disclosure statement
R.H.R. is a scientific advisor for Roche Pharmaceuticals, Inc. and a consultant to Elan Corp., plc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Canfield MA, et al. National estimates and race/ethnic-specific variation of selected birth defects in the United States, 1999–2001. Birth Defects Res A Clin Mol Teratol. 2006;76:747–756. doi: 10.1002/bdra.20294. [DOI] [PubMed] [Google Scholar]
- 2.Ferencz C, et al. Congenital cardiovascular malformations associated with chromosome abnormalities: an epidemiologic study. J Pediatr. 1989;114:79–86. doi: 10.1016/s0022-3476(89)80605-5. [DOI] [PubMed] [Google Scholar]
- 3.Levy J. The gastrointestinal tract in Down syndrome. Prog Clin Biol Res. 1991;373:245–256. [PubMed] [Google Scholar]
- 4.Fischer-Brandies H. Cephalometric comparison between children with and without Down’s syndrome. European Journal of Orthodontics. 1988;10:255–263. doi: 10.1093/ejo/10.3.255. [DOI] [PubMed] [Google Scholar]
- 5.Frostad W, et al. Craniofacial complex in the Trisomy 21 syndrome (Down’s syndrome) Archs oral BIol. 1971;16:707–722. doi: 10.1016/0003-9969(71)90116-6. [DOI] [PubMed] [Google Scholar]
- 6.Wechsler J, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 7.Yang Q, et al. Mortality associated with Down’s syndrome in the USA from 1983 to 1997: a population-based study. Lancet. 2002;359:1019–1025. doi: 10.1016/s0140-6736(02)08092-3. [DOI] [PubMed] [Google Scholar]
- 8.Chapman RS, Hesketh LJ. Behavioral phenotype of individuals with Down syndrome. Ment Retard Dev Disabil Res Rev. 2000;6:84–95. doi: 10.1002/1098-2779(2000)6:2<84::AID-MRDD2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 9.Reeves RH, Garner CC. A year of unprecedented progress in Down syndrome basic research. Ment Retard Dev Disabil Res Rev. 2007;13:215–220. doi: 10.1002/mrdd.20165. [DOI] [PubMed] [Google Scholar]
- 10.Wetmore DZ, Garner CC. Emerging pharmacotherapies for neurodevelopmental disorders. J Dev Behav Pediatr. 2010;31:564–581. doi: 10.1097/DBP.0b013e3181ee3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rissman RA, Mobley WC. Implications for treatment: GABA(A) receptors in aging, Down syndrome and Alzheimer’s disease. J Neurochem. 2011;117:613–622. doi: 10.1111/j.1471-4159.2011.07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golden JA, Hyman BT. Development of the superior temporal neocortex is anomalous in trisomy 21. J Neuropathol Exp Neurol. 1994;53:513–520. doi: 10.1097/00005072-199409000-00011. [DOI] [PubMed] [Google Scholar]
- 13.Sylvester PE. The hippocampus in Down’s syndrome. J Ment Defic Res. 1983;27 (Pt 3):227–236. doi: 10.1111/j.1365-2788.1983.tb00294.x. [DOI] [PubMed] [Google Scholar]
- 14.Aylward EH, et al. Cerebellar volume in adults with Down syndrome. Arch Neurol. 1997;54:209–212. doi: 10.1001/archneur.1997.00550140077016. [DOI] [PubMed] [Google Scholar]
- 15.Baxter LL, et al. Discovery and genetic localization of Down syndrome cerebellar phenotypes using the Ts65Dn mouse. Hum Mol Genet. 2000;9:195–202. doi: 10.1093/hmg/9.2.195. [DOI] [PubMed] [Google Scholar]
- 16.Crome R, et al. A statistical note on cerebellar and brain-stem weight in Down Syndrome. Journal of Mental Defect Research. 1966;10:69–72. [Google Scholar]
- 17.Jernigan TL, Bellugi U. Anomalous brain morphology on magnetic resonance images in Williams syndrome and Down syndrome. Arch Neurol. 1990;47:529–533. doi: 10.1001/archneur.1990.00530050049011. [DOI] [PubMed] [Google Scholar]
- 18.Raz N, et al. Selective neuroanatomic abnormalities in Down’s syndrome and their cognitive correlates: evidence from MRI morphometry. Neurology. 1995;45:356–366. doi: 10.1212/wnl.45.2.356. [DOI] [PubMed] [Google Scholar]
- 19.Becker LE, et al. Dendritic atrophy in children with Down’s syndrome. Ann Neurol. 1986;20:520–526. doi: 10.1002/ana.410200413. [DOI] [PubMed] [Google Scholar]
- 20.Benavides-Piccione R, et al. On dendrites in Down syndrome and DS murine models: a spiny way to learn. Prog Neurobiol. 2004;74:111–126. doi: 10.1016/j.pneurobio.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Takashima S, et al. Dendrites, dementia and the Down syndrome. Brain Dev. 1989;11:131–133. doi: 10.1016/s0387-7604(89)80082-8. [DOI] [PubMed] [Google Scholar]
- 22.Petit TL, et al. Synaptic development in the human fetus: a morphometric analysis of normal and Down’s syndrome neocortex. Exp Neurol. 1984;83:13–23. doi: 10.1016/0014-4886(84)90041-4. [DOI] [PubMed] [Google Scholar]
- 23.Weitzdoerfer R, et al. Fetal life in Down syndrome starts with normal neuronal density but impaired dendritic spines and synaptosomal structure. J Neural Transm Suppl. 2001:59–70. doi: 10.1007/978-3-7091-6262-0_5. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt-Sidor B, et al. Brain growth in Down syndrome subjects 15 to 22 weeks of gestational age and birth to 60 months. Clin Neuropathol. 1990;9:181–190. [PubMed] [Google Scholar]
- 25.Marin-Padilla M. Pyramidal cell abnormalities in the motor cortex of a child with Down’s syndrome. A Golgi study. J Comp Neurol. 1976;167:63–81. doi: 10.1002/cne.901670105. [DOI] [PubMed] [Google Scholar]
- 26.Zellweger H. Handbook of Clinical Neurology. North-Holland; 1977. Down Syndrome; pp. 367–469. [Google Scholar]
- 27.Kimura M, et al. Proliferation dynamics in cultured skin fibroblasts from Down syndrome subjects. Free Radic Biol Med. 2005;39:374–380. doi: 10.1016/j.freeradbiomed.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 28.Schneider EL, Epstein CJ. Replication rate and lifespan of cultured fibroblasts in Down’s syndrome. Proc Soc Exp Biol Med. 1972;141:1092–1094. doi: 10.3181/00379727-141-36940. [DOI] [PubMed] [Google Scholar]
- 29.Bernert G, et al. Decreased cyclin dependent kinase in brain of patients with Down syndrome. Neurosci Lett. 1996;216:68–70. doi: 10.1016/0304-3940(96)12988-8. [DOI] [PubMed] [Google Scholar]
- 30.Guidi S, et al. Neurogenesis impairment and increased cell death reduce total neuron number in the hippocampal region of fetuses with Down syndrome. Brain Pathol. 2008;18:180–197. doi: 10.1111/j.1750-3639.2007.00113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guidi S, et al. Widespread proliferation impairment and hypocellularity in the cerebellum of fetuses with down syndrome. Brain Pathol. 2011;21:361–373. doi: 10.1111/j.1750-3639.2010.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potier MC, et al. Transcriptional disruptions in Down syndrome: a case study in the Ts1Cje mouse cerebellum during post-natal development. J Neurochem. 2006;97(Suppl 1):104–109. doi: 10.1111/j.1471-4159.2005.03624.x. [DOI] [PubMed] [Google Scholar]
- 33.Roper RJ, Reeves RH. Understanding the basis for Down syndrome phenotypes. PLoS Genet. 2006;2:e50. doi: 10.1371/journal.pgen.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pletcher MT, et al. Use of comparative physical and sequence mapping to annotate mouse chromosome 16 and human chromosome 21. Genomics. 2001;74:45–54. doi: 10.1006/geno.2001.6533. [DOI] [PubMed] [Google Scholar]
- 35.Reeves R, et al. A mouse model for Down Syndrome exhibits learning and behaviour deficits. Nature Genetics. 1995;11:177–183. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 36.O’Doherty A, et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309:2033–2037. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu T, et al. A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum Mol Genet. 2010;19:2780–2791. doi: 10.1093/hmg/ddq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardiner KJ. Molecular basis of pharmacotherapies for cognition in Down syndrome. Trends Pharmacol Sci. 2010;31:66–73. doi: 10.1016/j.tips.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mo Z, et al. Human cortical neurons originate from radial glia and neuron-restricted progenitors. J Neurosci. 2007;27:4132–4145. doi: 10.1523/JNEUROSCI.0111-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kahlem P, et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res. 2004;14:1258–1267. doi: 10.1101/gr.1951304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das I, Reeves RH. Themes in therapeutics of Down syndrome converge in mouse models. Disease Models and Mechanisms. 2011 Aug 4; doi: 10.1242/dmm.007716. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradford HF. Glutamate, GABA and epilepsy. Prog Neurobiol. 1995;47:477–511. doi: 10.1016/0301-0082(95)00030-5. [DOI] [PubMed] [Google Scholar]
- 43.Haydar TF. Advanced microscopic imaging methods to investigate cortical development and the etiology of mental retardation. Ment Retard Dev Disabil Res Rev. 2005;11:303–316. doi: 10.1002/mrdd.20088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamain S, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson SM, et al. Mechanisms underlying the neuropathological consequences of epileptic activity in the rat hippocampus in vitro. J Comp Neurol. 1996;372:515–528. doi: 10.1002/(SICI)1096-9861(19960902)372:4<515::AID-CNE2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 46.Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 47.Corbin JG, et al. Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nat Neurosci. 2001;4(Suppl):1177–1182. doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- 48.de Carlos JA, et al. Dynamics of cell migration from the lateral ganglionic eminence in the rat. J Neurosci. 1996;16:6146–6156. doi: 10.1523/JNEUROSCI.16-19-06146.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson SA, et al. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 50.Tamamaki N, et al. Origin and route of tangentially migrating neurons in the developing neocortical intermediate zone. J Neurosci. 1997;17:8313–8323. doi: 10.1523/JNEUROSCI.17-21-08313.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ang ES, Jr, et al. Four-dimensional migratory coordinates of GABAergic interneurons in the developing mouse cortex. J Neurosci. 2003;23:5805–5815. doi: 10.1523/JNEUROSCI.23-13-05805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fairen A, et al. Times of generation of glutamic acid decarboxylase immunoreactive neurons in mouse somatosensory cortex. J Comp Neurol. 1986;251:67–83. doi: 10.1002/cne.902510105. [DOI] [PubMed] [Google Scholar]
- 53.Miller MW. Cogeneration of retrogradely labeled corticocortical projection and GABA-immunoreactive local circuit neurons in cerebral cortex. Brain Res. 1985;355:187–192. doi: 10.1016/0165-3806(85)90040-9. [DOI] [PubMed] [Google Scholar]
- 54.Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- 55.Valcanis H, Tan SS. Layer specification of transplanted interneurons in developing mouse neocortex. J Neurosci. 2003;23:5113–5122. doi: 10.1523/JNEUROSCI.23-12-05113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Q, et al. Origins of cortical interneuron subtypes. J Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Contestabile A, et al. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with down syndrome and in Ts65Dn mice. Hippocampus. 2007;17:665–678. doi: 10.1002/hipo.20308. [DOI] [PubMed] [Google Scholar]
- 58.Chakrabarti L, et al. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat Neurosci. 2010;13:927–934. doi: 10.1038/nn.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakrabarti L, et al. Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J Neurosci. 2007;27:11483–11495. doi: 10.1523/JNEUROSCI.3406-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haydar TF, et al. Consequences of trisomy 16 for mouse brain development: corticogenesis in a model of Down syndrome. J Neurosci. 1996;16:6175–6182. doi: 10.1523/JNEUROSCI.16-19-06175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng A, et al. Concurrent generation of subplate and cortical plate neurons in developing trisomy 16 mouse cortex. Dev Neurosci. 2004;26:255–265. doi: 10.1159/000082142. [DOI] [PubMed] [Google Scholar]
- 62.Dowjat WK, et al. Trisomy-driven overexpression of DYRK1A kinase in the brain of subjects with Down syndrome. Neurosci Lett. 2007;413:77–81. doi: 10.1016/j.neulet.2006.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fuentes JJ, et al. Genomic organization, alternative splicing, and expression patterns of the DSCR1 (Down syndrome candidate region 1) gene. Genomics. 1997;44:358–361. doi: 10.1006/geno.1997.4866. [DOI] [PubMed] [Google Scholar]
- 64.Guimera J, et al. A human homologue of Drosophila minibrain (MNB) is expressed in the neuronal regions affected in Down syndrome and maps to the critical region. Hum Mol Genet. 1996;5:1305–1310. doi: 10.1093/hmg/5.9.1305. [DOI] [PubMed] [Google Scholar]
- 65.Park J, et al. Function and regulation of Dyrk1A: towards understanding Down syndrome. Cell Mol Life Sci. 2009;66:3235–3240. doi: 10.1007/s00018-009-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song WJ, et al. Isolation of human and murine homologues of the Drosophila minibrain gene: human homologue maps to 21q22.2 in the Down syndrome “critical region”. Genomics. 1996;38:331–339. doi: 10.1006/geno.1996.0636. [DOI] [PubMed] [Google Scholar]
- 67.Hammerle B, et al. The spatio-temporal and subcellular expression of the candidate Down syndrome gene Mnb/Dyrk1A in the developing mouse brain suggests distinct sequential roles in neuronal development. Eur J Neurosci. 2008;27:1061–1074. doi: 10.1111/j.1460-9568.2008.06092.x. [DOI] [PubMed] [Google Scholar]
- 68.Hammerle B, et al. Mnb/Dyrk1A is transiently expressed and asymmetrically segregated in neural progenitor cells at the transition to neurogenic divisions. Dev Biol. 2002;246:259–273. doi: 10.1006/dbio.2002.0675. [DOI] [PubMed] [Google Scholar]
- 69.Okui M, et al. High-level expression of the Mnb/Dyrk1A gene in brain and heart during rat early development. Genomics. 1999;62:165–171. doi: 10.1006/geno.1999.5998. [DOI] [PubMed] [Google Scholar]
- 70.Fotaki V, et al. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol Cell Biol. 2002;22:6636–6647. doi: 10.1128/MCB.22.18.6636-6647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moller RS, et al. Truncation of the Down syndrome candidate gene DYRK1A in two unrelated patients with microcephaly. Am J Hum Genet. 2008;82:1165–1170. doi: 10.1016/j.ajhg.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fernandez-Martinez J, et al. Attenuation of Notch signalling by the Down-syndrome-associated kinase DYRK1A. J Cell Sci. 2009;122:1574–1583. doi: 10.1242/jcs.044354. [DOI] [PubMed] [Google Scholar]
- 73.Kinstrie R, et al. dDYRK2 and Minibrain interact with the chromatin remodelling factors SNR1 and TRX. Biochem J. 2006;398:45–54. doi: 10.1042/BJ20060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lepagnol-Bestel AM, et al. DYRK1A interacts with the REST/NRSF-SWI/SNF chromatin remodelling complex to deregulate gene clusters involved in the neuronal phenotypic traits of Down syndrome. Hum Mol Genet. 2009;18:1405–1414. doi: 10.1093/hmg/ddp047. [DOI] [PubMed] [Google Scholar]
- 75.Maenz B, et al. Characterization of the human DYRK1A promoter and its regulation by the transcription factor E2F1. BMC Mol Biol. 2008;9:30. doi: 10.1186/1471-2199-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raich WB, et al. Characterization of Caenorhabditis elegans homologs of the Down syndrome candidate gene DYRK1A. Genetics. 2003;163:571–580. doi: 10.1093/genetics/163.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yabut O, et al. Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells. J Neurosci. 2010;30:4004–4014. doi: 10.1523/JNEUROSCI.4711-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang EJ, et al. Protein kinase Dyrk1 activates cAMP response element-binding protein during neuronal differentiation in hippocampal progenitor cells. J Biol Chem. 2001;276:39819–39824. doi: 10.1074/jbc.M104091200. [DOI] [PubMed] [Google Scholar]
- 79.Dierssen M, de Lagran MM. DYRK1A (dual-specificity tyrosine-phosphorylated and -regulated kinase 1A): a gene with dosage effect during development and neurogenesis. ScientificWorldJournal. 2006;6:1911–1922. doi: 10.1100/tsw.2006.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Altafaj X, et al. Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down’s syndrome. Hum Mol Genet. 2001;10:1915–1923. doi: 10.1093/hmg/10.18.1915. [DOI] [PubMed] [Google Scholar]
- 81.Martinez de Lagran M, et al. Motor phenotypic alterations in TgDyrk1a transgenic mice implicate DYRK1A in Down syndrome motor dysfunction. Neurobiol Dis. 2004;15:132–142. doi: 10.1016/j.nbd.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 82.Tejedor FJ, Hammerle B. MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J. 2010;278:223–235. doi: 10.1111/j.1742-4658.2010.07954.x. [DOI] [PubMed] [Google Scholar]
- 83.Kleschevnikov AM, et al. Hippocampal long-term potentiation suppressed by increased inhibition in the Ts65Dn mouse, a genetic model of Down syndrome. J Neurosci. 2004;24:8153–8160. doi: 10.1523/JNEUROSCI.1766-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hanson JE, et al. The functional nature of synaptic circuitry is altered in area CA3 of the hippocampus in a mouse model of Down’s syndrome. J Physiol. 2007;579:53–67. doi: 10.1113/jphysiol.2006.114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fernandez F, et al. Pharmacotherapy for cognitive impairment in a mouse model of Down syndrome. Nat Neurosci. 2007;10:411–413. doi: 10.1038/nn1860. [DOI] [PubMed] [Google Scholar]
- 86.Butt SJ, et al. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 87.Flames N, et al. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nery S, et al. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- 89.Olson LE, et al. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science. 2004;306:687–690. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Belichenko NP, et al. The “Down syndrome critical region” is sufficient in the mouse model to confer behavioral, neurophysiological, and synaptic phenotypes characteristic of Down syndrome. J Neurosci. 2009;29:5938–5948. doi: 10.1523/JNEUROSCI.1547-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Davidoff LM. The brain in Mongolian idiocy. Arch Neurol Psychatry. 1928;20:1229–1268. [Google Scholar]
- 92.Hyde LA, et al. Motor learning in Ts65Dn mice, a model for Down syndrome. Dev Psychobiol. 2001;38:33–45. doi: 10.1002/1098-2302(2001)38:1<33::aid-dev3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 93.Costa AC, et al. Motor dysfunction in a mouse model for Down syndrome. Physiol Behav. 1999;68:211–220. doi: 10.1016/s0031-9384(99)00178-x. [DOI] [PubMed] [Google Scholar]
- 94.Galante M, et al. Impairments in motor coordination without major changes in cerebellar plasticity in the Tc1 mouse model of Down syndrome. Hum Mol Genet. 2009;18:1449–1463. doi: 10.1093/hmg/ddp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dahmane N, Ruiz-i-Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 96.Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- 97.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 98.Roper RJ, et al. Defective cerebellar response to mitogenic Hedgehog signaling in Down syndrome mice. Proc Natl Acad Sci U S A. 2006;103:1452–1456. doi: 10.1073/pnas.0510750103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen JK, et al. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Frank-Kamenetsky M, et al. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J Biol. 2002;1:10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Costa AC. An assessment of the vestibulo-ocular reflex (VOR) in persons with Down syndrome. Exp Brain Res. 2011;214:199–213. doi: 10.1007/s00221-011-2820-y. [DOI] [PubMed] [Google Scholar]
- 102.Migliaccio AA, et al. Characterization of the 3D angular vestibulo-ocular reflex in C57BL6 mice. Exp Brain Res. 2011;210:489–501. doi: 10.1007/s00221-010-2521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Burguiere E, et al. Role of the cerebellar cortex in conditioned goal-directed behavior. J Neurosci. 2010;30:13265–13271. doi: 10.1523/JNEUROSCI.2190-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rondi-Reig L, Burguiere E. Is the cerebellum ready for navigation? Prog Brain Res. 2005;148:199–212. doi: 10.1016/S0079-6123(04)48017-0. [DOI] [PubMed] [Google Scholar]
- 105.Trazzi S, et al. APP-dependent up-regulation of Ptch1 underlies proliferation impairment of neural precursors in Down syndrome. Hum Mol Genet. 2011;20:1560–1573. doi: 10.1093/hmg/ddr033. [DOI] [PubMed] [Google Scholar]
- 106.Olson LE, et al. Down syndrome mouse models Ts65Dn, Ts1Cje, and Ms1Cje/Ts65Dn exhibit variable severity of cerebellar phenotypes. Dev Dyn. 2004;230:581–589. doi: 10.1002/dvdy.20079. [DOI] [PubMed] [Google Scholar]
- 107.Ma’ayan A, et al. The cognitive phenotype of Down syndrome: insights from intracellular network analysis. NeuroRx. 2006;3:396–406. doi: 10.1016/j.nurx.2006.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lejeune J, et al. Etudes des chromosomes somatiques de neuf enfants mongoliens. CR Acad Sci (Paris) 1959;248:1721–1722. [PubMed] [Google Scholar]
- 109.Pennington BF, et al. The neuropsychology of Down syndrome: evidence for hippocampal dysfunction. Child Dev. 2003;74:75–93. doi: 10.1111/1467-8624.00522. [DOI] [PubMed] [Google Scholar]
- 110.Edgin JO, et al. Development and validation of the Arizona Cognitive Test Battery for Down syndrome. J Neurodev Disord. 2010;2:149–164. doi: 10.1007/s11689-010-9054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Braudeau J, et al. Specific targeting of the GABA-A receptor {alpha}5 subtype by a selective inverse agonist restores cognitive deficits in Down syndrome mice. J Psychopharmacol. 2011;25:1030–1042. doi: 10.1177/0269881111405366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guedj F, et al. Green tea polyphenols rescue of brain defects induced by overexpression of DYRK1A. PLoS One. 2009;4:e4606. doi: 10.1371/journal.pone.0004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Olson LE, et al. A chromosome 21 critical region does not cause specific down syndrome phenotypes. Science. 2004;306:687–690. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sago H, et al. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc Natl Acad Sci U S A. 1998;95:6256–6261. doi: 10.1073/pnas.95.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koo BK, et al. Magnetic resonance imaging evaluation of delayed myelination in Down syndrome: a case report and review of the literature. J Child Neurol. 1992;7:417–421. doi: 10.1177/088307389200700417. [DOI] [PubMed] [Google Scholar]
- 116.Wisniewski KE, Schmidt-Sidor B. Postnatal delay of myelin formation in brains from Down syndrome infants and children. Clin Neuropathol. 1989;8:55–62. [PubMed] [Google Scholar]
- 117.Sturgeon X, Gardiner KJ. Transcript catalogs of human chromosome 21 and orthologous chimpanzee and mouse regions. Mamm Genome. 2011;22:261–271. doi: 10.1007/s00335-011-9321-y. [DOI] [PubMed] [Google Scholar]
- 118.Vilardell M, et al. Meta-analysis of heterogeneous Down Syndrome data reveals consistent genome-wide dosage effects related to neurological processes. BMC Genomics. 2011;12:229. doi: 10.1186/1471-2164-12-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Salehi A, et al. Increased App expression in a mouse model of Down’s syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron. 2006;51:29–42. doi: 10.1016/j.neuron.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 120.Westmark CJ, Malter JS. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol. 2007;5:e52. doi: 10.1371/journal.pbio.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lipsky RH, Goldman D. Genomics and variation of ionotropic glutamate receptors. Ann N Y Acad Sci. 2003;1003:22–35. doi: 10.1196/annals.1300.003. [DOI] [PubMed] [Google Scholar]
- 122.Buccafusca R, et al. Characterization of the null murine sodium/myo-inositol cotransporter 1 (Smit1 or Slc5a3) phenotype: myo-inositol rescue is independent of expression of its cognate mitochondrial ribosomal protein subunit 6 (Mrps6) gene and of phosphatidylinositol levels in neonatal brain. Mol Genet Metab. 2008;95:81–95. doi: 10.1016/j.ymgme.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 123.Rezai Z, Yoon CH. Abnormal rate of granule cell migration in the cerebellum of “Weaver” mutant mice. Dev Biol. 1972;29:17–26. doi: 10.1016/0012-1606(72)90039-5. [DOI] [PubMed] [Google Scholar]
- 124.Cramer NP, et al. GABAB-GIRK2-mediated signaling in Down syndrome. Adv Pharmacol. 2010;58:397–426. doi: 10.1016/S1054-3589(10)58015-3. [DOI] [PubMed] [Google Scholar]
- 125.Arron JR, et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- 126.Martin LJ. Transgenic mice with human mutant genes causing Parkinson’s disease and amyotrophic lateral sclerosis provide common insight into mechanisms of motor neuron selective vulnerability to degeneration. Rev Neurosci. 2007;18:115–136. doi: 10.1515/revneuro.2007.18.2.115. [DOI] [PubMed] [Google Scholar]
- 127.Salehi A, et al. Restoration of norepinephrine-modulated contextual memory in a mouse model of Down syndrome. Sci Transl Med. 2009;1:7ra17. doi: 10.1126/scitranslmed.3000258. [DOI] [PubMed] [Google Scholar]