Abstract
Recent solved structures of G protein-coupled receptors (GPCRs) provide insights into variation of the structure and molecular mechanisms of GPCR activation. In this review we provide evidence for the emerging paradigm of domain coupling facilitated by intrinsic disorder of the ligand-free state in GPCRs. The structure-function and dynamic studies suggest that ligand-bound GPCRs exhibit multiple active conformations in initiating cellular signals. Long-range intra-molecular and inter-molecular interactions at distant sites on the same receptor are crucial factors that modulate signaling function of GPCRs. Positive or negative coupling between the extracellular, the transmembrane and the intracellular domains facilitates cooperativity of activating “switches” as requirements for the functional plasticity of GPCRs. Awareness that allosteric ligands robustly affect domain coupling provides a novel mechanistic basis for rational drug development, small molecule antagonism and GPCR regulation by classical, as well as non-classical modes.
G protein-coupled receptors (GPCRs)
With more than 5000 members unique to eukaryotes, the superfamily of GPCRs is the largest that senses changes in the extracellular milieu and conveys this information to the interior of the cell [1-6]. In humans, more than 1200 GPCRs mediate responses to hormones, neurotransmitters, metabolites, ions, fatty acids, pathogens, and physical stimuli, such as light, smell, taste, and mechanical stretch [7-9]. GPCRs are integral membrane glycoproteins, containing a seven-transmembrane (7TM) helical protein-fold. These receptors can signal through heterotrimeric G-protein-dependent or -independent pathways to control various physiological functions. Abnormal GPCR signaling which could result from genetic variation, acquired or inherited receptor mutations, changes in ligand binding specificity, improper regulation of receptor functions and antibodies directed against the receptors causes disorders in most tissues and organs [10,11]. More than 60% of current therapeutic drugs for human diseases are surrogate modulators of only a small fraction of known GPCRs [12]. Thus, a better understanding of the mechanism of GPCRs is crucial to novel drug discovery.
Canonical GPCR architecture consists of three functional domains (Box 1), a TM domain (TMD) that links the intracellular domain (ICD) to the extracellular domain (ECD) [13]. The ICD recruits G-proteins and other signaling molecules to exert a specific intracellular effect. Core of the TMD harbors the native ligand-binding site in the GPCR prototypes, rhodopsin and β-adrenergic receptor (βAR), as well as other GPCRs (Figure 1). Conserved structural features such as micro-switches, hydrogen-bond network extending from the orthosteric ligand-binding site to the ICD are thought to coordinate global movements essential for the remarkable ability of GPCRs to elicit a ligand-specific signaling response [14]. Recent studies suggest a greater role of ECD in ligand specificity; however, the canonical function of the ECD in operation of GPCRs is not firmly established. The ECD and ICD are remarkably diverse in GPCRs, which suggests potentially different specialization for ligand or G protein sensing and receptor activation (Figure 2) [15].
Box1. Domains of GPCRs.
The intracellular, the transmembrane and the extracellular domains of GPCRs are conventionally referred to as units of function. Each of these domains consists of discontinuous part of the polypeptide chain of a GPCR. However, Gaussian Network Model (GNM) and the Floppy Inclusions in Rigid Substructure Topography (FIRST) (http://firstweb.asu.edu) analysis of solved GPCR structures suggest that there are three clusters of structural stability. Three stable parts of the structure corresponds to the cytoplasmic and the extracellular domains separated by the transmembrane bundle [10]. As detailed below, these regions of GPCRs also harbor the ability to fold into stable functional domains in experimental systems.
Trans-membrane domain (TMD): The 25-35 amino acid long hydrophobic segments, thermodynamically driven to form independently stable alpha-helices in the lipid environment and then to make helix-helix contacts resulting in the transmembrane domain structure. The TMD of bacteriorhodopsin can be refolded in vitro from fragments. However, in contrast to bacteriorhodopsin, only a subset helix fragments of mammalian GPCRs associate in vivo [52]. Mutations in the TM helices severely affect alignment of helices and disrupt formation of the TM helical bundle.
Extra-cellular domain (ECD): The largest cluster of stability in GPCRs corresponds to interactions of the extracellular loops including the cysteine residues that form the crucial disulfide bond. Point mutations and deletions not only in the transmembrane helices but also in the extracellular loops of GPCRs result in misfolding via disruption of a disulfide bond that is highly conserved throughout the GPCR family. The three dimensional structure of all GPCRs solved until now confirm this disulfide bond located at the interface between the TM and the ECD. These findings suggest that the ECD and TMD are tightly coupled structurally [10,13].
Intra-cellular domain (ICD): Two additional clusters of structural stability in GPCRs are present at the cytoplasmic end of the transmembrane domain. One of these clusters contains the D/E R Y motif, which is conserved and is important for the interaction with the G protein. The cytoplasmic domain of rhodopsin has been reconstructed in proof of concept studies [53,54] which demonstrated that reconstructed cytoplasmic domain of bovine opsin is functional. Biochemical studies show that the reconstructed domain effectively mimics the activated rhodopsin in stimulating G-protein, in the phosphorylation by rhodopsin kinase, and arrestin binding. These results suggest that inter helical segments of the cytoplasmic surface of rhodopsin can adopt functionally discrete conformation in the absence of the transmembrane helices.
Long C-terminal domain (LCD): The functional autonomy of LCD is less clear. However, the LCD when present in a GPCR is now recognized as the main domain for the regulation of interactions with cytosolic proteins involved in a wide range of functions including targeting, trafficking and signaling of GPCRs in specialized cells. Splice variants of GPCRs generally differ in their intracellular domains in particular forming LCD that provide additional regulation in 5-Hydroxytryptamine 4 (5-HT4) receptor in mouse, rat, and human brain tissue [55]. With new unique C-terminal sequence and only expressed in brain tissue, these receptors are less prone to constitutive activation and recycling than the splice variants with shorter C-terminal tails. Similarly, the formation of a whole functional glutamate “receptosome” and its association with the dendritic cell cytoskeleton is a G-protein independent function of the LCD [55].
Long N-terminal domain (LND): A large number of non-rhodopsin family of GPCRs is defined by their relatively large and unique >100 residue long N-terminal domain (LND) that contain multiple disulfide bonds. These GPCRs utilize their ECD to recognize polypeptide hormones of about 30–40 amino acids, which include parathyroid hormone, glucagon-like peptide, calcitonin, and the calcitonin gene-related peptide. Recombinant functional LNDs have been produced and high-resolution structures have been obtained with bound ligand [56]. The canonical LND has two central antiparallel beta sheets and an N-terminal alpha helix interconnected by several loops and stabilized by disulfide bonds. The C-terminal portion of the peptide-ligand is recognized by the LND, while the N-terminal portion of the peptide interacts distinctly with the ECD and TMD of the receptor. Thus, LND is structurally independent of the ECD.
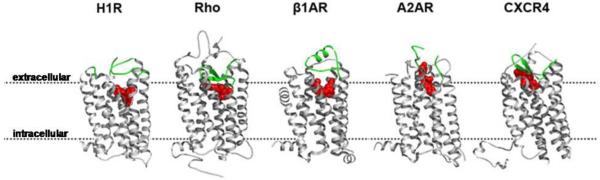
Figure 1. Variable position of the ligand binding pocket as seen in crystal structures of GPCRs.
Structures of histamine H1 receptor (PDB: 3RZE), bovine rhodopsin (PDB: 1U19), β1AR (PDB: 2VT4), adenosine A2A receptor, (PDB: 3EML), and CXCR4 (PDB: 3ODU) are depicted. The ligand is shown in red and the ECL2 is shown in green. Overall molecular architecture of these GPCRs is similar. The intracellular half of TMD structure is more conserved than the extracellular half. The extracellular loops can adopt very different structural forms. The ligand pocket is located ~5Å deeper in the histamine H1 receptor than in rhodopsin and β1AR structures. The antagonist orientation is in the plane of the membrane in H1R, rhodopsin and β1AR, but perpendicular to the plane of the membrane and collinear with TM7 in the A2A adenosine receptor while interacting with ECL2 and ECL3. The binding cavity for the small-molecules and the peptide ligands is located closer to ECD in the CXCR4.
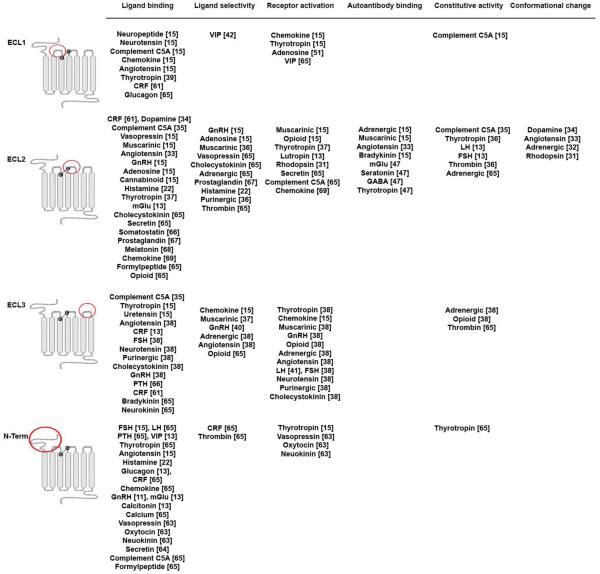
Figure 2. Experimentally determined roles of extracellular loops in different GPCRs.
The GPCR secondary structures with the highly conserved disulfide bond between TM3 and ECL2 are indicated. The ECL regions with indicated functions are circled in red. Abbreviations are: CRF; corticotropin-releasing factor, mGluR; metabotropic glutamate receptor, GABA; gamma-aminobutyric acid, GnRH; gonadotropin-releasing hormone, FSH; follicle stimulating hormone, PTH; parathyroid hormone, LH; luteinizing hormone, VIP; vasoactive intestinal peptide.
In this review, we will describe insights from pharmacology, molecular genetics, crystallography, and biophysical studies for the emerging paradigm for coupling between the domains of a GPCR. Long-range interactions and cooperativity are crucial factors for signaling in GPCRs, which undergo ligand-specific global conformational changes as requirements for structural coupling between extra-membranous and transmembrane regions. If domains are independent, perturbation of a domain should have no effect on the function of the other domains, whereas coupling would allow neighboring domains to “sense” the perturbation and respond appropriately. Domain coupling is important for explaining the in vivo scenario of tweaking the GPCR functions, not only by classical agonists and antagonists, but also by a variety of non-classical factors including receptor-interacting proteins, dimerization and activation by autoantibody. Binding of structurally different ligands could produce different active receptor conformations and downstream signaling. However, this review points out that the common principle applicable to all types of ligands is change in the intrinsic disorder state of the receptor affecting domain coupling. Awareness of domain coupling will help solve key challenges in pharmacological research concerning GPCRs with extended ligand binding domains and highlights the mechanism of small molecule antagonism. A number of new experiments will follow based on this idea and specifically advance our understanding of GPCR structure and function. The review provides new insight into the dynamic behavior of GPCR domains.
Diversity of GPCR architecture
The GPCR repertoire has expanded through evolution, during which the structural and functional properties have been preserved in a way that suggests a common ancestry (Box 2). Most mammalian GPCRs belong to one of five GPCR families: glutamate (G), rhodopsin (R), adhesion (A), frizzled/taste2 (F) and secretin (S) [1,8]. Mammals lack certain GPCRs (e.g. cAMP receptors, present in other classes) but contain some that are unique to mammals (e.g. the ocular albinism GPCRs). Although the 7TM architecture is the unifying feature of this superfamily, the N-terminal and C-terminal tails vary greatly among different GPCR families and within a single GPCR family as well (Table 1). The receptors in all of the main families, except the rhodopsin family, have long N-termini; whereas the rhodopsin family has only a few members with this characteristic (e.g. luteinizing hormone, follicle stimulating hormone, and thyrotropin receptors) [13]. Long N-termini are especially evident among receptors in the adhesion family; however, secretin, glutamate, and frizzled receptors also have long N-termini that are fairly rich in cysteine residues. The C-terminal tail shows similar features. It is reasonable, then, to consider these GPCRs with a long N-terminal domain (LND) or long C-terminal domain (LCD) as having four domains. There is less variation in the length of extracellular loops in the GPCR superfamily and also a disulfide bond linking TM3 and ECL2 is highly conserved [13].
Box 2. GPCR repertoire through evolution.
A bona fide GPCR is not found in prokaryotes. However, prokaryote genomes encode 7TMD proteins, such as the light-sensitive proteo-, halo-, and bacterio-rhodopsins that are involved in non-photosynthetic energy harvesting [6,57). Their abundance is less than 5 per species, and the 7TM protein-fold did not evolve into GPCRs in prokaryotes. All eukaryotes contain GPCRs, indicating that these proteins are of ancient origin [7,8,58]. The malaria parasite P. falciparum without GPCRs seems to be the exception [1]. Viruses such as human cytomegalovirus, herpes viruses, Epstein-Barr virus, and poxvirus encode GPCRs that are homologous to human chemokine receptors. This suggests that virus-host adaptation involved a GPCR repertoire to either “counteract” or “exploit” the host defense mechanisms [59]. In plants, the mildew resistance O (MLO) family of GPCRs provides resistance to leaf cell death and broad-spectrum disease caused by pathogenic, powdery mildew fungus [58,60].
A striking feature of metazoan evolution is the transformation of the 7TM protein-fold into GPCRs, and the origin of ≈15 GPCR families [1]. Primordial GPCRs essential for basic functions such as sensing cAMP, ions, amino acids, nucleotides, pheromones, and cell-adhesion are common among invertebrates. A strong expansion in the diversity and abundance of GPCRs accompanied vertebrate evolution. For example, more than 500 GPCRs per mammalian species form five distinct high-functioning GPCR families [1]. The number of GPCRs in these families differs in classes of species. There is a strong correlation between the repertoire of GPCRs and the complexity of the organism. It can therefore be inferred that a larger GPCR repertoire enables the organism to extract more information from its environment, allowing for a more complex homeostatic regulation.
Table 1. The length of intrahelical loops , N-terminal and C-terminal tails in GRAFS family receptors1.
| Family | Glutamate | Rhodopsin | Adhesion | Frizzled | Secretin |
|---|---|---|---|---|---|
| # of receptors | 15 | 701 | 24 | 24 | 15 |
| N-terminus | 518±76 | 54±42 | 1300±1048 | 101±100 | 139±41 |
| ECL1 | 12±6 | 15±10 | 12±11 | 20±11 | 25±8 |
| ECL2 | 30±7 | 24±19 | 28±12 | 30±10 | 17±2 |
| ECL3 | 11±5 | 14±11 | 6±4 | 8±4 | 12±4 |
| ICL1 | 16±4 | 16±12 | 17±9 | 23±11 | 23±16 |
| ICL2 | 21±7 | 16±12 | 18±12 | 17±12 | 15±3 |
| ICL3 | 14±2 | 126±114 | 34±22 | 33±8 | 22±4 |
| C-terminus | 205±180 | 63±61 | 205±192 | 25±12 | 41±33 |
|
Characteristics and conserved motifs |
Venus Flytrap | S-S bond DRY inTM3 NPXXY in TM7 |
EGF-like repeats Mucin-like repeats GPS domain |
IFL in TM2 SFLL in TM5 SXKTL in TM7 |
CWP, CP, GXW in N-term |
The numbers were derived from the GPCR database (http://www.gpcr.org) and represent the number of residues in each region (mean ± S.D.) of human GPCRs only. The length of the TM domain is between 25-35 residues for GPCR superfamily
Relative positioning of TM-helices and ligand binding pocket
Although the 7TM architecture is preserved in all vertebrate and invertebrate GPCRs, the highest sequence identity within the TMD is 20% to 50%. The sequence variability in the rhodopsin family is thought to be important for accommodating diverse ligands by the TMD [2,5,7], leading to the notion that direct contact between TMD and the ligand regulates signal transduction.
This view is not universal. Many ligands binding to the ECD or LND also modulate the function in GPCRs. The location of the ligand pocket and the orientation of ligands vary in the 3D-structures available now. The overall molecular architecture of these GPCRs is similar, but, unsurprisingly, the relative positions of the various helices differ in different structures, which have structural and biochemical implications (Figure 1). The relative position of helices and the ligand-binding pocket are similar in rhodopsin and βAR [14,16-18]. However, the structures of A2A adenosine, D2 dopamine, CXCR4 chemokine and histamine H1 receptors suggest that there is no general, family conserved ligand-pocket position or inter-helical interaction network in the TMD [19-22]. Flexible positioning of the helices in the GPCRs indicates that, under native conditions, intrinsic disorder in the TMD of GPCRs may be more common than previously anticipated. The ligand interaction at many different positions of TMD or outside TMD could facilitate transition to an ordered state.
TM helical dynamics coupled to ligand binding
The rigid body movement of TM helices, associated with agonist-activation of a GPCR allows the TMD to act as a mechanical lever that transduces the signal across the membrane [13]. Regardless of its location, the ligand-binding pocket is the epicenter for conformational changes. The interaction of agonists changes the rotamer conformation of side chains that are directly involved [14,23]. A comparison of active and inactive structures of rhodopsin, β2AR, and A2A adenosine receptors reveals structural rearrangements in the helices TM3, TM5, TM6, and TM7 [4,24-27]. Conformation of TM helices 1, 2, and 4 do not change. However, the extent and direction of the helical motion differ. For instance, TM6 motion is more pronounced in the A2A adenosine receptor, and the activation of β2AR is mainly facilitated by TM5. The versatile structural and conformational flexibility of the helices tolerates an intrinsic disordered state of TMD which facilitates binding of a specific ligand to the receptor. Ligand-induced TM helical movement leads to an ordered state, but the order of helical movement may differ significantly in different GPCRs. Intrinsic disorder may play an important functional role in regulating the specificity of the interactions between receptors and ligands by allowing receptor conformation to change into an ordered state that structurally accommodates the ligand with high specificity and affinity by establishing the interactions between different residues and influencing the kinetics. The formation of the agonist-receptor complex with lower intrinsic disorder is the preliminary step, and full activation occurs more readily when molecules within cells, such as G-proteins, bind to the ICD of a GPCR [23,27,28]. The GTP effect on agonist-affinity in classical pharmacological experiments also suggested the same. The crystal structure of the active state ternary complex composed of agonist-β2AR-Gs heterotrimer indeed shows the G-protein induced conformational changes propagating to the TMD [24]. This evidence clearly shows the reciprocal coupling between the G-protein site in ICD and the agonist site in TMD. In all GPCRs, ICL2, and ICL3 are important for selective binding and activation of G proteins [23,24,29]. The ICL1 conformation is similar in the solved structures of GPCRs, but the ICL2 conformation significantly varies, although the specific function of this region is highly conserved. The ICL3 conformational change is only known for opsins, and the observed structure is non-physiological due to mutations introduced to crystallize other GPCRs [16-22].
The mechanisms of structural coupling between the ICD and TMD vary among GPCRs and actually do not involve a conserved network. An “ionic lock” is formed between the conserved D/ERY sequence motif and TM4, which accounts for coupling ICD and TMD in the inactive conformation of rhodopsin and dopamine receptor [14]. The “ionic lock” breaks upon light-activation of rhodopsin, allowing G protein binding to the ICD. Other GPCR structures lack an “ionic-lock”; instead the DRY motif restrains the conformation of ICL2. In the histamine H1 receptor, the Arg residue in the DRY motif makes a hydrogen bond instead of an ion-pair with TM6 [22]. As seen in the β2AR-Gs complex [24], initial interaction of G-protein with the cytoplasmic loops initiates conformational changes in the ICD, which subsequently influences conformation of the TMD so that the agonist pocket is apt to nestle the agonists. Thus, reciprocal coupling between TMD and ICD conformation is clear. By contrast, the coupling of agonist-induced activation and the conformation of ECD is not systematically analyzed and the structural determinants for possible coupling are not delineated.
Extracellular domain conformation coupled to activation of GPCRs
Overall, the conformation of ECD in GPCRs is markedly different (Figure 1), largely dictated by ECL2 and its interactions with the bound ligands in each receptor [4,14,16-22]. The ECL2 in bovine rhodopsin adopts a β-hairpin structure that folds into the ligand binding pocket and extensively interacts with retinal as a “lid” [16]. Activation-induced displacement of this “lid” from the retinal-binding pocket is coupled to TMD through the Cys110-Cys187 bond linking ECL2 to TM3 [30]. An equivalent of this S-S bond is a highly conserved feature in GPCR superfamily, which ensures structural coupling between the ECD and TMD [13]. In GPCRs lacking this S-S bond, other bonding interactions may mediate domain coupling. The order of TM helical motion coupled to ECL2 may differ during the activation of different GPCRs [31].
Does the ECD conformation change in GPCRs that respond to diffusible ligands? A “lid” similar to that formed by ECL2 of rhodopsin is absent in other GPCR structures, but the highly variable conformations of ECL2 appear to contribute to ligand specificity in all GPCR structures except the histamine H1 receptor [22]. The ECD in these GPCRs often harbor disulfide bonds in addition to the highly conserved S-S bond, which may stabilize an ECL, or link the N-terminal tail (or LND) and ECLs. Disulfide links between TM-helices and cytoplasmic loops are generally absent in GPCRs. The frequent occurrence of disulfides in the ECD, perhaps, confers a high degree of coupling between the ECD and TMD as well as the ECD and LND.
Drugs displaying different efficacies that bind within the TMD of β2AR stabilize distinct conformations of the ECD in nuclear magnetic resonance (NMR) studies, which illustrates conformational coupling between the ECD and the orthosteric ligand binding site [32]. The ECL2 in the angiotensin II type 1 receptor (AT1R) adopts a ligand-specific “lid” conformation and that also slows down the dissociation of bound ligands [33]. Bound agonists and antagonists induce distinct conformations around the conserved disulfide bond, suggesting that the S-S bond coupling the ECD to the TMD plays a role in producing different functional states of the AT1R [13,33]. Ligand-induced conformational changes in the ECL2 have been documented in several GPCRs, including the D2 dopamine receptor and C5A complement receptor [34,35]. The ligand-induced conformational changes observed in these studies suggest that ECL2 is a crucial regulator and that the motion of ECL2 is generally coupled to GPCR activation (Table 1). New and unanticipated roles of ECL regions, including (i) forming the ECD, (ii) binding ligands or allosteric modulators, (iii) switching agonist/antagonist properties and (iv) receptor activation have been documented (Figure 2) [13,15,36-42).
In GPCRs with LNDs, the peptide ligand mediates interaction between the LND and ECD, which is crucial for activation of receptor functions (Box 1). The LND regions alone can bind the hormone in these receptors; however, the signal transduction depends on different ECLs, especially ECL2 and ECL3 [13]. Several intermediate conformational changes are essential to activate the TMD and ICD for biological function upon binding of the ligand to ECD in the metabotropic glutamate receptor (mGluR). These steps may be common for GPCRs with LNDs (Figure 2). In the secretin family of GPCRs for hormones, such as glucagon, parathyroid hormone, and vasoactive intestinal peptide, residues in ECLs are essential for receptor activation, although the ligands bind primarily to their LND [13]. In the protease-activated receptors, the attached ligand is activated by proteolysis and binds to the ECLs to initiate the physiological signal [36]. The interaction of ligand-bound ECLs with the TMD is crucial for the activation of the glutamate receptor family [13]. The ligand-induced activation of the other two GPCR families, adhesion and frizzled receptors, has been shown to be impaired by the mutations in ECLs. These observations suggest a conserved role for the ECD throughout the GPCR superfamily [13,15].
Non-classical GPCR activation
There is increasing evidence for modulation of GPCR functions through interaction with non-classical binding partners [44,45], an important mode of in vivo regulation beyond the classical action mediated by pharmacological modulators. Non-classical factors include extrinsic perturbations (the interaction with agonist, antagonist, autoantibodies, cholesterol, lipids, other GPCRs or transmembrane receptors and G proteins, etc.) and factors intrinsic to the receptor (naturally occurring and experimental modifications such as gain or loss of function mutations, disruption of disulfide bonds or salt-bridges, chemical modification of a residue e.g. phosphorylation).
As portrayed in Figure 3, the binding of non-classical factors to any domain can modulate agonist-mediated activation and sometimes cause agonist-independent activation of a GPCR. Autoantibodies targeting specific GPCRs that harbor agonist-like activity are found in patients with chronic disorders of various etiologies. Agonistic antibodies directed against the thyrotropin (TSH) receptor in Graves’ disease; β1AR and β2AR in Chagas disease, idiopathic-, ischemic-, and dilated-cardiomyopathies; α1-AR and B2 bradykinin receptor in malignant hypertension; and the muscarinic M1 and M2 receptors in schizophrenia cause hyperactivation of the receptors [15,46,47]. Activating antibodies against ECL2 of the AT1R cause preeclampsia, malignant hypertension, rejection of kidney transplantation, and vascular injury [47,48]. An agonistic antibody alone is sufficient to cause disease. Antibodies purified from plasma of pregnant women directly bind to exposed epitopes in the ECL2 of AT1R [33]. When bound to AT1R, these antibodies activate the signaling cascade in vitro and cause preeclampsia in pregnant mice. Additionally, the antibodies increase angiotensin II sensitivity observed in preeclampsia. These effects of the antibodies can be inhibited by losartan, a AT1R-selective antagonist. Likewise, the anti-β1-ECL2 antibody causes signaling in vitro and heart-failure, ventricular arrhythmias, and sudden cardiac death in vivo. The antibody-induced activation represents an intriguing example of a common form of activation seen in many GPCRs initiated at the ECL2 [47].
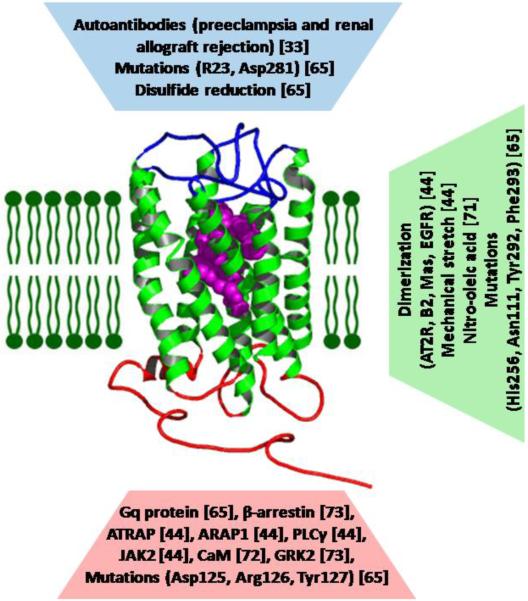
Figure 3. Non-classical interactions that modulate the function of AT1R.
Extent of functional coupling between domains can change from perturbation of any domain of the AT1R. In AT1R, extrinsic and intrinsic perturbations are observed at three distinct domains: (i) at the ECD where autoantibodies can directly activate AT1R or allosterically facilitate or inhibit activation by AngII; (ii) at the TMD where intrinsic disorder of TM helices can be reduced to facilitate activation by AngII; (iii) at the ICD where canonical GPCR-associated protein complexes can allosterically modulate activation by AngII AngII. The factors affecting specific domains; extracellular domain (blue), TM domain (green), or intracellular domain (red) and modulate AT1R activity are shown. Bound angiotensin II is shown in magenta. The abbreviations are: AT2R; angiotensin II type 2 receptor, B2; bradykinin receptor, EGFR; epidermal growth factor receptor, ATRAP; angiotensin II type 1 receptor-associated protein, ARAP1; angiotensin II type 1 receptor-associated protein 1, PLCγ; phospholipase γ, JAK2; Janus kinase, CaM; calmodulin.
Activating mutations found in the ECD of several GPCRs emphasize its participation in GPCR activation [36,46]. The βAR chimera containing the ECL2 of α1AR is constitutively active [38]. Mutations of ECL2 in thyrotropin (TSH) [37] complement C5A [35], and thrombin receptors [36] cause constitutive activity. In the mGluR, the frizzled, and the secretin receptor families, mutations of the LND result in constitutive activity. Gain-of-function mutations in any domain of GPCRs can cause human diseases. The activating mutations in ECD of the glycoprotein hormone, luteinizing hormone, follicle-stimulating hormone, and thyrotropin result in endocrine disorders in humans [15,37,46,49]. These mutated residues in ECD alter the local structure and overall dynamics of the receptors in the absence of the ligand.
Treatment with dithiothreitol () alters the affinity of GPCRs for their ligands and activates some GPCRs while it inactivates others, presumably by reducing the conserved disulfide bond. DTT causes functional activation of purified GPCRs, such as β2AR, β1AR, α1AR, 5HTR, and muscarinic receptor, even in the absence of agonists. The DTT-induced activation and high agonist-affinity state of β2AR is conferred by the intra-loop Cys184-Cys190 disulfide bond, whereas reduction of the conserved Cys106-Cys191 bond by DTT inactivates β2AR [13,17,18]. Reduction of an equivalent disulfide bond has a similar effect in most GPCRs. Bovine rhodopsin mutants, which lack this conserved disulfide bond, are functionally unstable when activated [13]. The disulfide bond is crucial for folding and stability of the native structure in most GPCRs which are inactivated by DTT [13]. However, the disulfide bond in other GPCRs could be protected from inactivation or could be inactivated at a higher concentration of DTT. On the other hand, DTT treatment could mimic agonist-mediated disulfide bond reduction in some GPCRs, therefore, could be involved in receptor activation.
Other examples of non-classical activation include a high G protein to receptor ratio in cells, as well as changes in the fluid membrane environment. Both promote transitional changes in the receptor structure. Recently, nanobody-mediated activation of β2-AR initiated in the ICD was reported [26]. Many GPCRs have been previously shown to homo- and hetero-dimerize, constitutively and/or upon ligand binding. Although the functional importance of dimerization is unclear, considerable data suggest that it has important in vivo pharmacological effects [50,51]. The specific nature of the interactions within the cell membrane may facilitate the ability of a GPCR to heterodimerize with other subtypes, as well as GPCRs outside of the family (Figure 3). Dimerization-induced conformational change could explain the cooperative binding of ligands and G proteins, as well as the effects of allosteric modulators. Specifically, binding of a ligand to one receptor could induce a structural change in the TMD of the second receptor, which could modify the G protein-binding affinity to the second receptor. Negative or positive cooperativity could result from dimerization, an observation that may have implications for drug efficacy. Therefore, non-classical activation may be quite common in nature, supporting the conclusion that the initial trigger for activating conformational change in GPCRs can occur in any domain [44].
Coupling between GPCR domains
In multi-domain allosteric proteins, individual domains are usually found in thermodynamically driven intrinsically disordered states. Accordingly, the conformational fluctuations of the canonical structure form the basis of the intrinsic disorder states of domains in a GPCR. The intrinsic disorder of a domain decreases when the domain engages a classical or non-classical ligand (e.g. agonist or autoantibody), leading to a decrease of intrinsic disorder in a coupled domain and allowing subsequent binding of the ligand that is specific for the coupled domain (e.g. G protein). Thus, the inherent intrinsic disorder of the GPCR domains allows function only when a ligand binding to one domain can influence further the binding of cognate ligands to the coupled domains. Inherent intrinsic disorder of individual domains in these transmembrane signaling proteins will allow mutual communication between domains, which is crucial for coordinated operation of the domains and the regulatory capacity of a GPCR. This contrasts with the classical view that efficient coupling between the binding sites for the orthosteric ligand and the G-protein would require a family-wide conserved structural arrangement linking the two sites. The observation that functionally analogous domains in different GPCRs bind G-protein or agonist with very little sequence conservation supports this idea. The regulatory concept based in the intrinsic disorder-to-order transition would be robust and effective in vivo.
GPCRs can be represented as a group of 3 or 4 interacting domains (Figure 4). The simplest GPCR would be one with ECD-TMD-ICD domain organization, as seen in the rhodopsin family. This arrangement can change in other families, as LND-ECD-TMD-ICD and ECD-TMD-ICD-LCD. The state of intrinsic disorder in each domain is governed independently, resulting in possible virtual states (shown as, R, R’1, R’2, and R*), representing combinations of disordered/ordered states of domains (Figure 4). Agonist or autoantibody binding to a domain initiates ID-to-order transition. Coupling allows each domain to “sense” the transition in the neighboring domain directly because of the physical interaction between them. For instance, with a covalent S-S bond and complementary hydrophobic surfaces, it would be energetically more favorable for ECD to interact with the TMD upon ligand binding. In principle, the extent of coupling between domains would change after a perturbation applied to any domain (Figure 3).
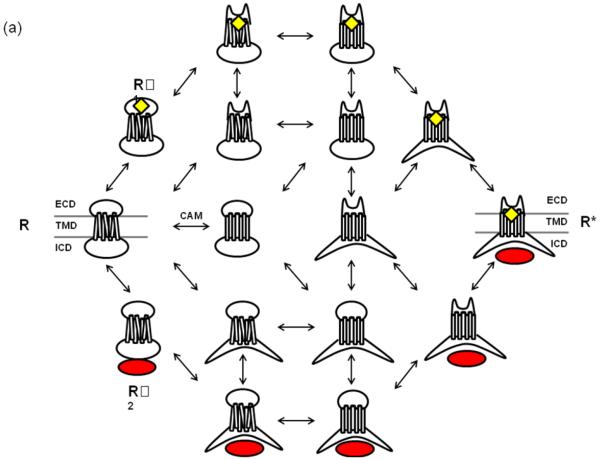
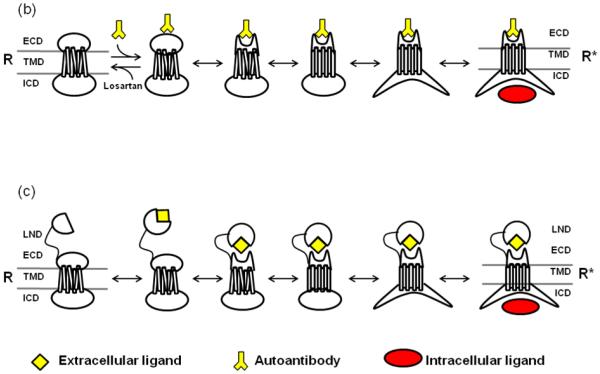
Figure 4. Dynamic coupling of GPCR domain conformation.
(a) The schematic transition intermediates between inactive and active states of a three domain GPCR. Independent conformational changes in the ECD, TMD and ICD are shown, in which each state differs by only one conformational change in a specific domain. Each step indicates a reversible transition of the intrinsic disorder within a specific domain. Each transition triggers a similar change of intrinsic disorder in the coupled domain, ultimately leading to the fully activated conformation of the receptor. Native or non classical ligands including agonists and antagonists (yellow) and other interacting proteins including G-protein (red) are indicated. When the ligand (L) encounters the ECD (or TMD in some GPCRs), a binding site apt for that L is induced. Similarly when a G-protein (G) encounters ICD, a binding site for that G is induced in the ICD. Initial encounter of the L or G, is followed by induced change of intrinsic disorder, such that the affinity of the L and the G respectively to the ordered conformation of ECD and the ICD is greater than the affinity of the L or the G for the disordered conformation. The kinetic scheme is such that each domain can productively bind its cognate ligand utilizing intrinsic disorder of the domain. Thus, state R’1 can only bind L, state R’2 can only bind G, state R* can bind both L and G, and state R can bind neither the L nor the G. Intrinsic disorder is higher in constitutively active mutant GPCRs, which increases the probability of the R* state in these mutant GPCRs. The model suggests that two neighboring domains can be either positively (left to right) or negatively coupled (right to left). In the case of positive coupling, upon adding the L, the ordered state of the ECD (or TMD in some receptors) will be preferentially stabilized. The ECD and TMD are positively coupled (S-S bond and ECL2), therefore the energy of breaking the interaction between them will be unfavorable. As a result, stabilizing the binding site within the ECD will have the effect of also stabilizing the TMD (or binding an allosteric modulator in TMD) and so on. The degree of response to a given amount of L represents extent of coupling, which is a measure of the sensitivity of coupled domains and a quantifiable metric of allosteric coupling potential. (b) Autoantibody mediated transition to active state and mechanism by which antagonist, such as losartan, reverse activation. (c) Ligand induced activation in receptors with LND (Long N-terminal domain). In this representation, a portion of the ligand is recognized by the LND, while the other portion of the ligand is shown to interact with the ECD of the receptor.
The schematic in Figure 4 illustrates that each domain can productively bind a selective ligand utilizing the intrinsic disorder of the domain. R’1 binds extracellular ligand (L), R’2 binds intracellular ligand (G), R* binds both L and G and R binds neither L nor G. Domains can be coupled either positively (left to right, activation) or negatively (right to left, inhibition). The assumption in this model is that the ECD and ICD are intrinsically most disordered, and the TMD is partially disordered at any time. ECD-TMD (S-S bond and ECL2) and ICD-TMD (ionic lock and other interactions) are positively coupled; therefore the energy of breaking the interaction between them will be unfavorable. As a result, stabilizing the ECD will have the effect of also stabilizing the TMD (or binding in TMD) and so on.
The classic view is that two sites are coupled through a conserved network of interactions that directly connect the sites. Paradoxically, in GPCRs, an inverse relationship between G-protein coupling and the stability of the native receptor is observed. Inverse agonists increase stability at the expense of coupling to G proteins. This relationship provides insight into the rules governing domain coupling in GPCRs: (i) coupling is maximized when the domains are intrinsically disordered for a significant fraction of the time, in the absence of a ligand; and (ii) introducing additional domains (e.g. LND) is not expected to affect the responsiveness of a GPCR.
The observation that a disproportionately higher amount of diversity is present in the ECD indicates that in the evolution of coupling between GPCR domains, nature uses an ensemble-mediated mechanism for their regulatory role. The interactions between the different domains in the GPCR produce an ensemble of states that is “optimally poised” to respond to classical as well as non-classical ligands. Upon binding a ligand, the ensemble is redistributed to elicit a specific function. The proposed mechanism depends only on the ensemble interactions of the domains and not on the specific structural details. The current model provides significant insight into how a GPCR can transmit signals through interaction with many different types of ligands (Figure 3).
Concluding remarks
Recently there has been remarkable progress in GPCR structural biology with elucidation of three dimensional structures of several GPCRs in both active and inactive states. These studies not only provide information about the ligand-dependent receptor activation mechanisms, but they also allow us develop ideas about the dynamic nature of these proteins. Here we reviewed recent studies that provide evidence for a variable mode of ligand binding accompanied with variable position of TM helices and hydrogen bonding networks even within a receptor family. The intrinsically flexible ECD and ICD are functionally coupled; the coupling is mediated by the TMD. This arrangement facilitates coordinated domain functions. When a domain engages a ligand, a decrease in intrinsic disorder of the domain cooperatively influences the conformation of the neighboring domain. Negative and positive cooperativity between domains, respectively can account for antagonism and agonism. This model may explain the full complexity of both ligand-dependent and ligand-independent GPCR activation. Information provided by future structures from different families of GPCRs will provide a more complete picture that should be valuable in drug discovery.
Acknowledgements
We thank members of the Karnik laboratory for fruitful discussions and advice and NIH for RO1 grant (HL57470) to Karnik, S. and an NRSA award (HL007914) to Unal, H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fredriksson R, Schioth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol. Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbaum DM, et al. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofmann KP, et al. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem. Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Costanzi S, et al. Rhodopsin and the others: a historical perspective on structural studies of G protein-coupled receptors. Curr. Pharm. Des. 2009;15:3994–4002. doi: 10.2174/138161209789824795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millar RP, Newton CL. The year in G protein-coupled receptor research. Mol. Endocrinol. 2010;24:261–274. doi: 10.1210/me.2009-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strotmann R, et al. Evolution of GPCR: Change and continuity. Mol. Cell. Endocrinol. 2011;331:170–178. doi: 10.1016/j.mce.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Rompler H, et al. G-protein coupled time travel. Evolutionary aspects of GPCR research. Mol. Interv. 2007;7:17–25. doi: 10.1124/mi.7.1.5. [DOI] [PubMed] [Google Scholar]
- 8.Schoneberg T, et al. Learning from the past: evolution of GPCR functions. Trends Pharmacol. Sci. 2007;28:17–121. doi: 10.1016/j.tips.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 10.Klein-Seetharaman J. Dual role of interactions between membranous and soluble portions of helical membrane receptors for folding and signaling. Trends Pharmacol. Sci. 2005;26:183–9. doi: 10.1016/j.tips.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Insel PA. Impact of GPCRs in clinical medicine: monogenic diseases, genetic variants and drug targets. Biochim. Biophys. Acta. 2007;1768:994–1005. doi: 10.1016/j.bbamem.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundstrom K. An overview on GPCRs and drug discovery: structure-based drug design and structural biology on GPCRs. Methods Mol. Biol. 2009;552:51–66. doi: 10.1007/978-1-60327-317-6_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karnik SS, et al. Activation of GPCRs: A common molecular mechanism. Trends Endocrinol. Metab. 2003;14:431–437. doi: 10.1016/j.tem.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Nygaard R, et al. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 2009;30:249–59. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Peeters MC, et al. Importance of the extracellular loops in G protein-coupled receptors for ligand recognition and receptor activation. Trends Pharmacol. Sci. 2011;32:35–42. doi: 10.1016/j.tips.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Palczewski K, et al. Crystal structure of rhodopsin: a G-protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 17.Rasmussen SG, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 18.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G-protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaakola VP, et al. The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimamura T, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Rasmussen SG, et al. Crystal structure of the b2 adrenergic receptor-Gs protein complex. Nature. 2011 doi: 10.1038/nature10361. doi: 10.1038/nature10361. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu F, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobilka BK. Structural insights into adrenergic receptor function and pharmacology. Trends Pharmacol. Sci. 2011;32:213–218. doi: 10.1016/j.tips.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunyady L, et al. Agonist induction and conformational selection during activation of a G-protein-coupled receptor. Trends Pharmacol. Sci. 2003;24:81–86. doi: 10.1016/S0165-6147(02)00050-0. [DOI] [PubMed] [Google Scholar]
- 29.Rovati GE, et al. The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Mol. Pharmacol. 2007;71:959–964. doi: 10.1124/mol.106.029470. [DOI] [PubMed] [Google Scholar]
- 30.Yan EC, et al. Retinal counterion switch in the photoactivation of the G protein-coupled receptor rhodopsin. Proc. Natl. Acad. Sci. USA. 2003;100:9262–9267. doi: 10.1073/pnas.1531970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahuja S, et al. Helix movement is coupled to displacement of the second extracellular loop in rhodopsin activation. Nat. Struct. Mol. Biol. 2009;16:168–175. doi: 10.1038/nsmb.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bokoch MP, et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463:108–112. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unal H, et al. Ligand-specific conformation of extracellular loop-2 in the angiotensin II type 1 receptor. J. Biol. Chem. 2010;285:16341–16350. doi: 10.1074/jbc.M109.094870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi L, Javitch JA. The second extracellular loop of the dopamine D2 receptor lines the binding-site crevice. Proc. Natl. Acad. Sci. 2004;101:440–445. doi: 10.1073/pnas.2237265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klco JM, et al. Essential role for the second extracellular loop in C5a receptor activation. Nat. Struct. Mol. Biol. 2005;12:320–326. doi: 10.1038/nsmb913. [DOI] [PubMed] [Google Scholar]
- 36.Massotte D, Kieffer BL. The second extracellular loop: a damper for G protein-coupled receptors? Nat. Struct. Mol. Biol. 2005;12:287–288. doi: 10.1038/nsmb0405-287. [DOI] [PubMed] [Google Scholar]
- 37.Kleinau G, Krause G. Thyrotropin and homologous glycoprotein hormone receptors: structural and functional aspects of extracellular signaling mechanisms. Endocr. Rev. 2009;30:133–151. doi: 10.1210/er.2008-0044. [DOI] [PubMed] [Google Scholar]
- 38.Lawson Z, Wheatley M. The third extracellular loop of G-protein-coupled receptors: more than just a linker between two important transmembrane helices. Biochem. Soc. Trans. 2004;32:1048–1050. doi: 10.1042/BST0321048. [DOI] [PubMed] [Google Scholar]
- 39.Sura-Trueba S, et al. An inactivating mutation within the first extracellular loop of the thyrotropin receptor impedes normal posttranslational maturation of the extracellular domain. Endocrinology. 2009;150:1043–1050. doi: 10.1210/en.2008-1145. [DOI] [PubMed] [Google Scholar]
- 40.Mizutori Y, et al. The thyrotropin receptor hinge region is not simply a scaffold for the leucine-rich domain but contributes to ligand binding and signal transduction. Mol. Endocrinol. 2008;22:1171–1182. doi: 10.1210/me.2007-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li JH, et al. Extracellular loop 3 (EL3) and EL3-proximal transmembrane helix 7 of the mammalian type I and type II gonadotropin-releasing hormone (GnRH) receptors determine differential ligand selectivity to GnRH-I and GnRH-II. Mol. Pharmacol. 2005;67:1099–1110. doi: 10.1124/mol.104.004887. [DOI] [PubMed] [Google Scholar]
- 42.Hashimoto H, et al. Vasoactive intestinal polypeptide and pituitary adenylate cyclase activating polypeptide receptor chimeras reveal domains that determine specificity of vasoactive intestinal polypeptide binding and activation. Mol. Pharmacol. 52:128–135. doi: 10.1124/mol.52.1.128. [DOI] [PubMed] [Google Scholar]
- 43.Siu FY, Stevens RC. RAMP-ing up class-B GPCR ECD structural coverage. Structure. 2010;18:1067–8. doi: 10.1016/j.str.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Mogi M, et al. New insights into the regulation of angiotensin receptors. Curr. Opin. Nephrol. Hypertens. 2009;18:138–43. doi: 10.1097/MNH.0b013e328324f5fa. [DOI] [PubMed] [Google Scholar]
- 45.Pierce KL, et al. Seven-transmembrane receptors. Nat. Rev. Mol .Cell Biol. 2002;3:639–50. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 46.Tao YX. Constitutive activation of G protein-coupled receptors and diseases: insights into mechanisms of activation and therapeutics. Pharmacol. Ther. 2008;120:129–148. doi: 10.1016/j.pharmthera.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unal H, et al. Mechanism of GPCR-directed Autoantibody in Diseases. The proceedings of the IXth ISCSM (2011) Advances in Exp Biol. and Med. 2011 in press. [Google Scholar]
- 48.Zhou CC, et al. Angiotensin receptor agonistic autoantibodies induce preeclampsia in pregnant mice. Nat. Med. 2008;14:855–862. doi: 10.1038/nm.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nurwakagari PJ, et al. A conformational contribution of the luteinizing hormone-receptor ectodomain to receptor activation. Mol. Endocrinol. 2007;38:259–275. doi: 10.1677/jme.1.02160. [DOI] [PubMed] [Google Scholar]
- 50.Terrillon S, Bouvier M. Roles of G-protein-coupled receptor dimerization. From ontogeny to signalling regulation. EMBO rep. 2004;5:30–34. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gurevich VV, Gurevich EV. How and why do GPCRs dimerize? Trends Pharmacol. Sci. 2008;29:234–40. doi: 10.1016/j.tips.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wess J. Considerations in the design and use of chimeric G protein-coupled receptors. Methods Enzymol. 2002;343:295–312. doi: 10.1016/s0076-6879(02)43143-6. [DOI] [PubMed] [Google Scholar]
- 53.Karnik S. Analysis of structure-function from expression of G protein-coupled receptor fragments. Methods Enzymol. 2002;343:248–59. doi: 10.1016/s0076-6879(02)43140-0. [DOI] [PubMed] [Google Scholar]
- 54.Abdulaev NG, et al. Functionally discrete mimics of light-activated rhodopsin identified through expression of soluble cytoplasmic domains. J. Biol. Chem. 2000;275:39354–63. doi: 10.1074/jbc.M005642200. [DOI] [PubMed] [Google Scholar]
- 55.Bockaert J, et al. GPCR-interacting proteins (GIPs): nature and functions. Biochem. Soc. Trans. 2004;32:851–5. doi: 10.1042/BST0320851. [DOI] [PubMed] [Google Scholar]
- 56.Siu FY, Stevens RC. RAMP-ing up class-B GPCR ECD structural coverage. Structure. 2010;18:1067–8. doi: 10.1016/j.str.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 57.Fuhrman JA, et al. Proteorhodopsins: an array of physiological roles? Nat. Rev. Microbiol. 2008;6:488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- 58.Nordstrom KJV, et al. The secretin GPCRs descended from the family of adhesion GPCRs. Mol. Biol. Evol. 2009;26:71–84. doi: 10.1093/molbev/msn228. [DOI] [PubMed] [Google Scholar]
- 59.Vischer HF, et al. A viral conspiracy: hijacking the chemokine system through virally encoded pirated chemokine receptors. Curr. Top. Microbiol. Immunol. 2006;303:121–154. doi: 10.1007/978-3-540-33397-5_6. [DOI] [PubMed] [Google Scholar]
- 60.Hok S, et al. Getting the most from the host: how pathogens force plants to cooperate in disease. Mol. Plant Microbe Interact. 2010;23:1253–1259. doi: 10.1094/MPMI-04-10-0103. [DOI] [PubMed] [Google Scholar]
- 61.Peeters MC, et al. GPCR structure and activation: an essential role for the first extracellular loop in activating the adenosine A2B receptor. FASEB J. 2011;25:632–643. doi: 10.1096/fj.10-164319. [DOI] [PubMed] [Google Scholar]
- 62.Gkountelias K, et al. Alanine scanning mutagenesis of the second extracellular loop of type 1 corticotropin-releasing factor receptor revealed residues critical for peptide binding. Mol. Pharmacol. 2009;75:793–800. doi: 10.1124/mol.108.052423. [DOI] [PubMed] [Google Scholar]
- 63.Wheatley M, et al. Extracellular loops and ligand binding to a subfamily of Family A G-protein-coupled receptors. Biochem. Soc. Trans. 2007;35:717–720. doi: 10.1042/BST0350717. [DOI] [PubMed] [Google Scholar]
- 64.Dong M, et al. Importance of each residue within secretin for receptor binding and biological activity. Biochemistry. 2011;50:2983–93. doi: 10.1021/bi200133u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunyady, Catt Pleiotropic AT1 receptor signaling pathways mediating physiological and pathogenic actions of angiotensin II. Mol. Endocrinol. 2006;20:953–70. doi: 10.1210/me.2004-0536. [DOI] [PubMed] [Google Scholar]
- 66.Greenwood MT, et al. Ligand binding pocket of the human somatostatin receptor 5: mutational analysis of the extracellular domains. Mol. Pharmacol. 1997;52:807–14. doi: 10.1124/mol.52.5.807. [DOI] [PubMed] [Google Scholar]
- 67.Stillman BA, et al. Importance of the extracellular domain for prostaglandin EP(2) receptor function. Mol. Pharmacol. 1999;56:545–51. doi: 10.1124/mol.56.3.545. [DOI] [PubMed] [Google Scholar]
- 68.Mazna P, et al. Ligand binding to the human MT2 melatonin receptor: the role of residues in transmembrane domains 3, 6, and 7. Biochem. Biophys. Res. Commun. 2005;332:726–34. doi: 10.1016/j.bbrc.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 69.Brelot A, et al. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J. Biol. Chem. 2000;275:23736–44. doi: 10.1074/jbc.M000776200. [DOI] [PubMed] [Google Scholar]
- 70.Oro C, et al. Type 1 angiotensin receptor pharmacology: signaling beyond G proteins. Pharmacol. Ther. 2007;113:210–26. doi: 10.1016/j.pharmthera.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J, et al. Nitro-oleic acid inhibits angiotensin II-induced hypertension. Circ. Res. 2010;107:540–548. doi: 10.1161/CIRCRESAHA.110.218404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas WG, et al. Identification of a Ca2+/calmodulin-binding domain within the carboxyl-terminus of the angiotensin II (AT1A) receptor. FEBS Lett. 1999;455:367–71. doi: 10.1016/s0014-5793(99)00904-7. [DOI] [PubMed] [Google Scholar]
- 73.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–7. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]