Abstract
Previous studies have identified multiple conserved noncoding sequences (CNS) at the mouse Ifng locus sufficient for enhancer activity in cell-based assays. These studies do not directly address biology of the human IFNG locus in a genomic setting. IFNG enhancers may be functionally redundant or each may be functionally unique. We test the hypothesis that each IFNG enhancer has a unique necessary function using a bacterial artificial chromosome transgenic model. We find that CNS-30, CNS-4 and CNS+20 are required at distinct stages of Th1 differentiation while CNS-16 has a repressive role in Th1 and Th2 cells. CNS+20 is required for IFN-γ expression by memory Th1 cells and by NKT cells. CNS-4 is required for IFN-γ expression by effector Th1 cells. In contrast, CNS-16, CNS-4 and CNS+20 are each partially required for human IFN-γ expression by NK cells. Thus, IFNG CNS enhancers have redundant necessary functions in NK cells, but unique necessary functions in T helper cells. These results also demonstrate that distinct CNSs are required to transcribe IFNG at each stage of the Th1 differentiation pathway.
Introduction
A primary signal in the defense against intracellular infections is the cytokine interferon gamma (IFN-γ) (1). IFN-γ is expressed by NK and NKT cells, CD8+ cytotoxic T cells and CD4+ T helper (Th) 1 cell subsets. During the initial CD4+ T cell maturation stage, a naïve CD4+ T cell can polarize into various T helper cell subsets including Th1 and Th2 subsets (2). Th2 cells must repress IFNG. This IFNG repression in Th2 cells is dependent upon the noncoding segment of the genome. Mice carrying an 8.6 kb transgene of the human IFNG gene fail to repress IFNG in Th2 cells (3). In contrast, mice carrying a 190 kb bacterial artificial chromosome (BAC) transgene with IFNG and the surrounding noncoding region both correctly express human IFN-γ in Th1 cells and repress human IFN-γ production in Th2 cells (3). As such, cell-type selective expression of human IFN-γ depends upon the noncoding segment of the genome.
The majority of the conserved portion of the human genome is noncoding. Further, the majority of the human common single nucleotide polymorphisms associated with disease traits are noncoding (4). As such, understanding the noncoding segments of the genome will be important to understanding human health. The noncoding portion of the genome includes various types of functional elements, including enhancers. Enhancers are thought to be necessary for driving tissue-specific, as well as species-specific gene expression (5, 6). Some genes are regulated by multiple, redundant enhancers, which some models propose are necessary to allow for expression under sub-optimal signaling conditions (7). As enhancers drive tissue-specific expression, and tissue-specific expression of IFNG is critical for protection from intracellular infections in humans (8), a relevant question is the mechanism of how IFNG enhancers drive tissue-specific expression of IFNG.
Transcription factor binding to mouse Ifng distal regulatory elements is cell type and stimulus type-specific. The transcription factors T-bet (9, 10), STAT4 (10), STAT5 (11, 12), NF-κB family members (13), and Runx3 (14) positively regulate IFNG expression and directly bind to distinct conserved noncoding sequences (CNS) of the mouse Ifng locus in a Th1 and stimulus dependent manner. Transcription factor binding is accompanied by Th1-specific covalent histone modifications at conserved noncoding sequences (13, 15, 16). These observations have led to the hypothesis that proper regulation of interferon gamma is conferred by transcription factor interactions with CNSs. In transgenic model systems, a mouse CNS −16 kb from the Ifng start site (mCNS-16) is needed for Thy1.1 reporter expression from a mouse Ifng BAC (9). In addition to mCNS-16, additional mCNSs display enhancer activity in reporter assays (15, 17, 18) and also function with other Ifng CNS to synergistically stimulate transcriptional activity (18).
Our understanding of human IFNG distal regulation in the setting of an intact genome is incomplete. We considered two non-exclusive hypotheses. First, CNSs may have redundant function where each CNS is necessary for a fraction of IFNG expression in all responder cell types in response to diverse stimuli. Second, CNSs may possess unique functions such that each individual CNS provides a unique contribution to developmental decisions and stimulus-specificity to achieve proper IFNG transcriptional regulation. To test these hypotheses, we employed an IFNG-BAC transgenic system (3, 14). In this model, mice are created with a transgene that contains IFNG and surrounding regulatory regions with or without specific CNSs. Normal production of mouse IFN-γ is not affected and serves as an internal control. We have previously characterized a conserved noncoding sequence −30 kb from the IFNG start site (CNS-30) necessary for transgenic IFNG expression in T cells but not NK cells (14). Here, we extend those studies to comprehensively characterize IFNG CNS, and detail the necessary function of CNS-16, CNS-4, CNS+20 and CNS+120. We find an essential role for CNS-4 in effector/memory Th1 cells. CNS+20 is required for human IFN-γ production by in vivo generated memory Th1 cells, as well as NKT cells. Lastly, we find that CNS-16 has a repressive role in both Th1 and Th2 cells, opposite to what was found in the mouse BAC transgenic system (9). In contrast, each of these CNSs, CNS-16, CNS-4, and CNS+20 contribute a fraction to IFNG transcription in NK cells. These results demonstrate that the necessary functions of distal regulatory elements are dependent upon developmental context, as well as species, cell type and stimuli.
Materials and Methods
Mice and preparation of transgenic reporter lines
C57BL/6 mice were obtained from Jackson Laboratory (Bar Harbor, ME), housed in the Vanderbilt University animal facilities, and used between 4-5 weeks of age. Preparation of human IFNG-BAC transgenic lines was preformed as described previously (14). Briefly, human IFNG-BAC CTD-3002C24 was used to make 210 kb IFNG-BAC mice. Alternatively, CTD-3002C24 was moved into EL250 E. coli and CNS deletion was achieved using homologous recombination followed by FRT-mediated removal of the selection marker. Targeting primers are described in supplemental table 1. We used human-specific PCR primers to verify insertion integrity of the different IFNG-BAC transgenes. Lines without full-length insertions were excluded from analysis. All animal studies were approved by the Institutional Animal Care and Use Committee of Vanderbilt University.
Cell purification and cultures
CD4+ and CD8+ T cells were purified from splenocytes by positive selection per manufactures instructions (Miltenyi Biotec). T cells were cultured with plate bound 2 μg/ml anti-CD3 (hybridoma 2C11, American Type Culture Collection, Manassas, VA) and anti-CD28 (BD Biosciences). For Th1/Tc1 cultures, cells were cultured with 10 ng/ml IL-12 and 10 μg/ml anti-IL4 (11B11 hybridoma, ATCC). For Th2 cultures, CD4+ T cells were cultured with 20 ng/ml IL-4 and 10 μg/ml anti-IFN-γ (hybridoma, ATCC). In addition, cultures were harvested after three days and re-cultured with IL-2 for two additional days. Effector Th1/Tc1 cells were re-stimulated with either plate bound anti-CD3, 10 ng/ml IL-12 and 10 ng/ml IL-18, 10 ng/ml IL-12 and 10 ng/ml IL-2, or 50 ng/ml PMA and 1 μM ionomycin. NK cells were purified from spleen by negative selection (Miltenyi Biotech).
Immunization
Five-week old transgenic mice were immunized with 50 μg soluble ovalbumin in complete Freund’s adjuvant (Sigma Chemical Co.) by intraperitoneal injection. At day ten, mice received an additional boost of Ova in incomplete Freund’s adjuvant (IFA). Splenocytes were harvested at day 35 for analysis. Splenocytes were either re-stimulated with 0.1 mg/ml Ova, 0.1 mg/ml hen egg lysozyme, or 10 μg/ml Ova 257-264 peptide. After two days, human and mouse IFN-γ was determined in culture supernatants by ELISA. Alternatively, splenocytes were restimulated overnight with 10 ng/ml IL-12 and 10 ng/ml IL-18. The next day, BD GolgiPlug was added, and cells were cultured for an additional six hours and analyzed by flow cytometry.
Results
Creation of 210 kb BAC ΔCNS-16, ΔCNS-4 and ΔCNS+20 transgenic mice
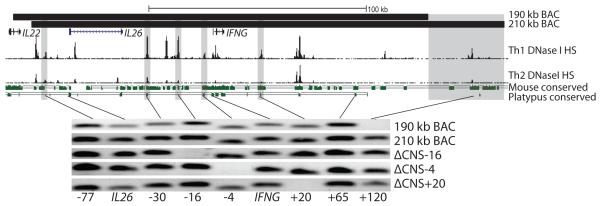
Previous reports have detailed that mouse CNS-22, mCNS-6, mCNS+22 and mCNS+67 contribute to Ifng transcriptional regulation (19). We used UCSC genome alignments to identify respective orthologs at human CNS-16, CNS-4, CNS+20 and CNS+120. CNS-4, CNS+20 and CNS+120 showed strong conservation in placental and non-placental mammals, but we were unable to find a non-placental ortholog to CNS-16 (Fig. 1). We verified correct enhancer ortholog location by referencing Th1 and Th2 DNase I hypersensitivity tracks. We next designed new IFNG-BAC transgenes with or without these conserved noncoding sequences. While a 190 kb IFNG-BAC transgene correctly recapitulates developmental and signaling-dependent expression of IFNG including dependence upon the transcription factors T-bet and STAT4 (14), the 190 kb IFNG-BAC transgene does not contain CNS+120. We identified a new 210 kb IFNG-BAC transgene which differed mainly by the inclusion of CNS+120 (Fig. 1). To assay functional roles of CNS-16, CNS-4 and CNS+20 we created new 210 kb IFNG-BACs with 1 kb deletions of CNS-16 and CNS-4 or a 3 kb deletion of CNS+20. A 3 kb region was chosen for CNS+20 to include all Th1 specific DNase I hypersensitivity sites as well as all regions homologous to mouse CNS+22. The exact location of deletions, transgene locations, and homologous mouse locations are found in supplemental table 1. We next created new transgenic mice with the full 210 kb IFNG-BAC transgene, or with the ΔCNS-16, ΔCNS-4 or ΔCNS+20 transgenes.
FIGURE 1.
BAC transgenes used in this study. Top: Locations of 190 kb and 210 kb BAC transgenes, Th1 and Th2 DNase I hypersensitivity and conserved sequences between humans and mice or between humans and platypus. Grey bars represent deletion locations. For conservation analysis, green bars are conserved sequences and solid lines represent a fully aligned genome. Bottom: Integration check of transgenes. Shown are human-specific PCR integration checks for BAC transgenic mice.
Transgenic mice displayed no overt abnormalities, were born at Mendelian ratios and gained weight at appropriate rates. Mouse IFN-γ production was consistent among different transgenic lines and was not affected by the presence of the transgene. The large size of BAC transgenes is thought to help protect against position integration effects, but lead to potential partial integration effects (20). We verified full integration of all transgenes by human-specific PCR of the BAC transgene from transgenic mouse genomic DNA (Fig. 1). Transgenic mice without full integration were not analyzed further. We have previously observed that IFN-γ expression from the 190 kb IFNG-BAC is dependent upon copy number (14). We next analyzed relative copy number by quantitative PCR. Copy number of all new lines of BAC transgenic mice was consistent, equivalent to two copies in a 190 kb IFNG-BAC control. We next assayed for human IFN-γ expression in the 210 kb IFNG-BAC, ΔCNS-16, ΔCNS-4 and ΔCNS+20 BAC transgenic mice.
CNS-4 is necessary for IFN-γ production by effector Th1 cells
We began our assays by using an in vitro tissue culture system. To test for expression in Th1 cells and repression in Th2 cells, CD4+ T cells were isolated and cultured under Th1 polarizing conditions (IL-12 and anti-IL4) or Th2 conditions (IL-4) with plate bound anti-CD3 and soluble anti-CD28 for three days. To test for expression in CD8 cells, CD8+ T cells were isolated and cultured under Th1 polarizing conditions (IL-12). At day three, human and mouse IFN-γ levels in cultures were assayed by ELISA. Alternatively, Th1 cells were cultured for two additional days in IL-2. On day five, Th1 cells were re-stimulated with either anti-CD3, IL-12 and IL-18, or IL-2 and IL-12 to test for defects specific to either T cell receptor stimulation or coordinated cytokine stimulation. IFN-γ was measured in cultures two days later.
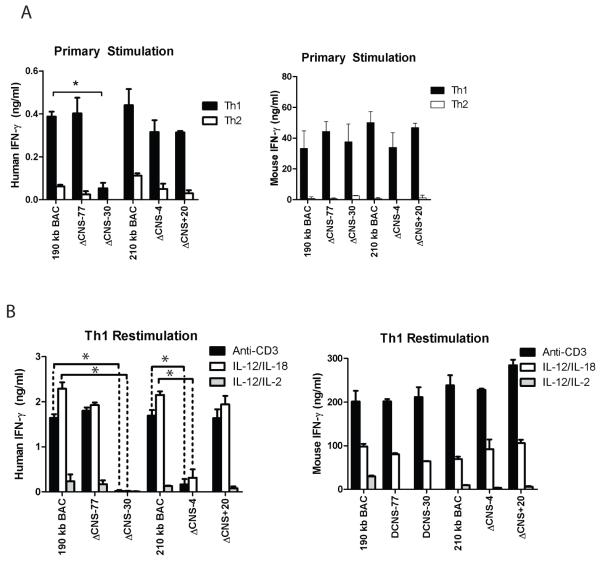
We first assayed for human IFN-γ expression from T cells with the 210 kb IFNG-BAC, the ΔCNS-4 and the ΔCNS+20 IFNG-BAC transgenes. For comparison, we assayed for human IFN-γ expression from previously created mice with the 190 kb IFNG-BAC transgene, or 190 kb IFNG-BAC transgenes lacking CNS-30 or CNS-77, which have been previously described (14). In day three Th1 cultures, human IFN-γ culture supernatant concentrations did not significantly differ between 190 kb and 210 kb IFNG-BAC cultures. Further, presence or absence of CNS-77, CNS-4 or CNS+20 did not significantly change human IFN-γ concentrations in Th1 or Th2 cultures (Fig. 2A: Left). Control mouse IFN-γ concentrations did not vary among transgenes (Fig. 2A: Right). As previously described (14), removal of CNS-30 led to a loss of detectable human IFN-γ in cultures.
FIGURE 2.
CNS-4 is required for human IFN-γ production. A, Transgenic CD4+ cells were cultured under Th1 and Th2 polarizing conditions for three days. Human IFN-γ (left) or mouse IFN-γ (right) concentrations were determined from cultures. B, Day five CD4+ Th1 cultures were restimulated with anti-CD3, or IL-12 and IL-18, or IL-12 and IL-2. Human and mouse IFN-γ concentrations were determined by ELISA. Results are representative of independent replicates. Error bars are standard deviations. * P < 0.05.
We continued our in vitro protocol and rested day three Th1 cultures in IL-2 for two days. CNS-4 has been proposed as an IL-2 responsive element (11). Effector Th1 cells were re-stimulated with plate-bound anti-CD3, IL-12 and IL-18, or IL-12 and IL-2. Human IFN-γ was measured in cultures two days later by ELISA. Human IFN-γ levels did not significantly differ between Th1 cells with the 190 kb IFNG-BAC transgene or the 210 IFNG-BAC transgene (Fig. 2B). Presence or absence of CNS-77 or CNS+20 also did not significantly alter concentrations of human IFN-γ in cultures. As previously reported, and as seen in primary cultures, removal of CNS-30 resulted in loss of detectable human IFN-γ in re-stimulated Th1 cell cultures. However, unlike in primary cultures, removal of CNS-4 resulted in a significant decrease in the concentration human IFN-γ in culture supernatants of re-stimulated effector Th1 cells. The CNS-4 defect was observed in both T cell receptor re-stimulated and IL-12 and IL-18 re-stimulated Th1 cells. As such, removal of CNS-4 resulted in a loss of human IFN-γ production by differentiated effector Th1 cells.
Lack of CNS-16 leads to increased human IFN-γ expression
We next investigated the role of CNS-16 in the regulation of IFN-γ in the human IFNG-BAC transgenic system. Using UCSC genome alignments, we identified CNS-16 as the ortholog of mouse CNS-22. mCNS-22 is necessary for Thy1.1 reporter expression from a murine BAC (9). mCNS-22 is also a T-bet responsive enhancer in two independent cell-based reporter assays (9, 15). As such, we hypothesized that CNS-16 would be required for human IFN-γ production in the IFNG-BAC transgenic system. We began by assaying human IFN-γ expression from cultures of 210 kb IFNG-BAC or ΔCNS-16 IFNG-BAC transgenic Th1 and Th2 cells. In direct contrast to our hypothesis, removal of CNS-16 led to relatively higher human IFN-γ in Th1 cultures, compared to controls (Fig. 3A). In addition, removal of CNS-16 led to increased human IFN-γ in Th2 cells. Control mouse IFN-γ was appropriately repressed in Th2 cultures (Fig. 3A: Right). In this study, and in previous studies of IFNG-BAC transgenic mice (14), measurable human IFN-γ above background levels was a phenotype unique to ΔCNS-16 Th2 cultures. These results indicate that ΔCNS-16 plays a key role repressing IFN-γ expression in both Th1 and Th2 cell cultures.
FIGURE 3.
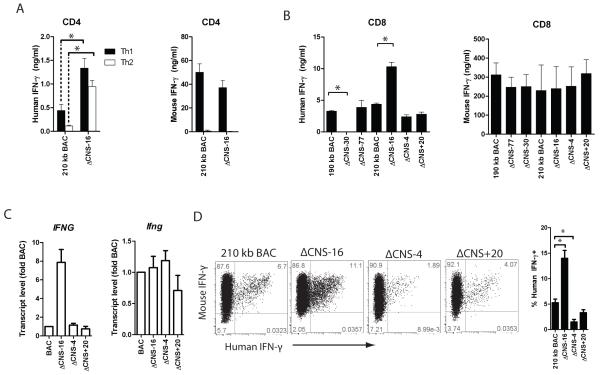
CNS-16 represses human IFN-γ production. A, Transgenic CD4+ cells were cultured under Th1 and Th2 polarizing culture conditions for three days. Human IFN-γ (left) or mouse IFN-γ (right) concentrations were determined in cultures. B, Transgenic CD8+ cells were cultured under Th1 polarizing conditions for three days. IFN-γ was measured in culture by ELSIA. Results are representative of independent replicates. C, Transgenic CD4+ cells were cultured under Th1 conditions for three days and mRNA levels were determined by quantitative PCR. Error bars are standard deviation. D, Transgenic CD8+ T cells were cultured under Th1 polarizing conditions for seven days and restimulated with PMA/Ionomycin. Interferon gamma was determined by intracellular cytokine staining. Quantification of the percentages of human IFN-γ+ cells from three independent replicates is shown. * P < 0.05.
We next assayed for human IFN-γ in cultures of CD8+ T cells. We cultured 190 kb IFNG-BAC, ΔCNS-77, ΔCNS-30, 210 kb IFNG-BAC, ΔCNS-16, ΔCNS-4 and ΔCNS+20 transgenic CD8+ T cells for three days and measured human IFN-γ by ELISA. As previously reported (14), removal of CNS-30 resulted in concentrations of human IFN-γ below levels of detection (Fig. 3B). Similar to Th1 cell cultures, removal of CNS-16 resulted in relatively high concentrations of human IFN-γ in CD8+ T cell cultures (Fig. 3B). Choice of 190 kb IFNG- or 210 kb IFNG-BAC or presence or absence of CNS-4 or CNS+20 did not significantly affect human IFN-γ production by CD8+ T cells under these culture conditions. To verify our results at the transcript level we cultured transgenic CD4+ T cells for three days under Th1 polarizing conditions. After cDNA synthesis, we determined human IFNG and mouse Ifng transcript levels by quantitative PCR. We observed relatively high human IFNG transcripts in ΔCNS-16 Th1 cultures compared to 210 kb IFNG-BAC cultures. Mouse Ifng transcripts were not different among cultures with the different transgenes (Fig. 3C). To verify that results were not due to contaminating NK cells, we assayed human and mouse IFN-γ expression by intracellular cytokine staining. 210 kb IFNG-BAC, ΔCNS-16, ΔCNS-4 and ΔCNS+20 transgenic CD4+ T cells were cultured under Th1 conditions for 5 days, rested for 2 days in IL-2 containing media, re-stimulated with PMA/ionomycin, and analyzed for IFN-γ by flow cytometry. Similar to measures of levels of human IFN-γ in cultures of effector cells, we observed a high percentage of human IFN-γ positive ΔCNS-16 Th1 cells and a low percentage of human IFN-γ positive ΔCNS-4 Th1 cells, relative to controls. As such, in our in vitro cultures of CD4+ and CD8+ T cells, production of human IFN-γ was dependent upon CNS-4 and CNS-30 with CNS-16 having a repressive role.
STAT4 and T-bet bind to the transgenic IFNG promoter
We next determined if any deletion was associated with altered T-bet and STAT4 binding to the IFNG-BAC promoter. We cultured 210 kb IFNG-BAC, ΔCNS-16, ΔCNS-4 and ΔCNS+20 transgenic CD4+ T cells for three days under Th1 polarizing conditions and processed cells for chromatin immunoprecipitation assays using antibodies against T-bet, STAT4 or an IgG isotype control. We observed specific T-bet and STAT4 binding to the human IFNG and mouse Ifng promoters in BAC transgenic Th1 cells (Supplemental Fig. 1). Levels of STAT4 binding to the transgenic IFNG and mouse Ifng promoters were essentially equivalent. Similarly, levels of T-bet binding to the transgenic IFNG and mouse Ifng promoters were essentially equivalent Further, deletion of CNS-16, CNS-4, or CNS+20 did affect binding of STAT4 or T-bet to the IFNG promoter. As such, these deletions were not associated with changes T-bet or STAT4 binding to the IFNG promoter.
The CNS-16 ortholog, mCNS-22, is required for Thy1.1 reporter expression from a mouse Ifng BAC transgene (9). As mCNS-22 and human CNS-16 have opposing functions, we analyzed the mouse and human interferon gamma loci for obvious differences. We first compared conservation of the human IFNG locus (Supplemental Fig. 2A). A small region of the human CNS-16 was not conserved in the mouse and rat genomes, but was conserved in other rodents and mammals outside Muridae. We also observed a poorly conserved site within this region marked by Th1-specific DNase I hypersensitivity. As the ΔCNS-16 deletion covered the murine-conserved portion of CNS-16, we reanalyzed conserved transcription factor binding sites across CNS-16. Analysis of transcription factor binding sites in the segment of CNS-16 not conserved in mice identified a cluster of T-bet and Runx3 binding sites (Supplemental Fig. 2B). To confirm the ability of the non-conserved segment of CNS-16 and the poorly conserved CNS-21 to bind transcription factors, we analyzed publically available chromatin immunoprecipitation followed by sequencing (ChIP-seq) data for binding of STAT4, STAT5A/B and YY1 (21, 22) in Th1 cultures because of their known ability to regulate IFNG transcription. This analysis showed that STAT4 and STAT5 bound to both CNS-21 and the portion of CNS-16 not conserved in mice. These data identify CNS-21 and the portion of CNS-16 not conserved in mice as likely regulatory elements that may contribute to their observed functional differences.
CNS+20 is necessary for IFNG expression by memory Th1 cells
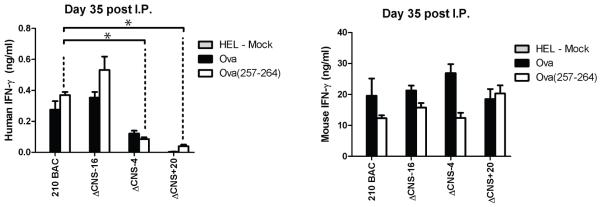
Our previous experiments relied upon in vitro maturation and polarization of T helper cells using experimental procedures optimized for murine IFN-γ production. Because previous reports have described differences in long-range chromatin modifications of the mouse Ifng locus during in vivo and in vitro Th1 polarization (15), we addressed the necessary functional requirements of human IFNG distal regulatory elements, in vivo. Mice were immunized by i.p. injection with OVA in CFA and boosted at day ten with OVA in IFA. At day thirty-five splenocytes were isolated and re-stimulated with irrelevant protein, HEL, OVA protein, or OVA 257-264 SIINFEKL peptide, which is recognized by CD8+ T cells on the C57BL/6 background (23). After two days, human and mouse IFN-γ concentrations in cultures were determined by ELISA (Fig. 4). Human and murine IFN-γ production were antigen-specific and not observed in HEL stimulated cultures. Concentrations of human IFN-γ were higher in ΔCNS-16 cultures when re-stimulated with either OVA or OVA (257-264) peptide relative to controls. Consistent with results from in vitro generated Th1 cells, removal of CNS-4 resulted in a significant decrease in human IFN-γ production in the antigen-specific memory responses. Further, while human IFN-γ production by in vitro generated ΔCNS+20 Th1 cells did not significantly differ from controls, human IFN-γ concentrations in the cultures of ΔCNS+20 T cells were significantly lower in response to antigen-specific re-stimulation of in vivo generated memory cells. These results demonstrate a necessary role for CNS+20 in memory responses in vivo.
FIGURE 4.
CNS+20 is required for antigen specific recall responses. Transgenic mice were immunized with OVA in CFA, and boosted at day 10 with OVA in IFA. At day 35 mice splenocytes were restimulated with HEL protein, OVA, or OVA 257-264 peptide. IFN-γ concentrations were determined by ELISA. Results are means of three independent replicates. Error bars are standard error of the mean. * P < 0.05.
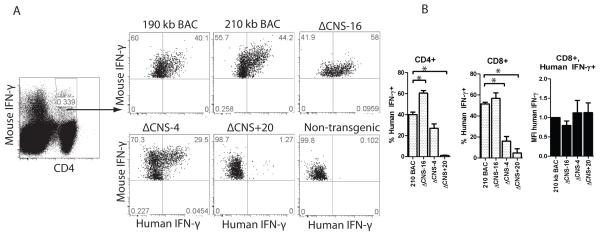
Among T cells, IFN-γ expression in response to IL-12 and IL-18 stimulation is restricted to effector/memory Th1/Tc1 lineages. Therefore, we analyzed human IFN-γ expression by freshly isolated CD4+ and CD8+ T cells after stimulation with IL-12 and IL-18 overnight. Eighteen hours later single cells producing human and mouse IFN-γ were quantified by flow cytometry (Fig. 5A & B). Cells were gated on murine IFN-γ positive populations to restrict analysis to effector/memory populations. Under these stimulation conditions, up to 45% of mouse IFN-γ positive BAC-transgenic CD4+ T cells were also human IFN-γ+. Percentages of human IFN-γ+ positive cells did not differ between 190 kb IFNG-BAC and 210 kb IFNG-BAC transgenic mice. Consistent with antigen-specific re-stimulation of in vivo generated antigen-specific Th1 cell responses, removal of CNS-4 or removal of CNS+20 resulted in a significant decrease in the percentages of human IFN-γ+ cells compared to 210 kb IFNG-BAC T cells. Further, stimulation of ΔCNS-16 T cells resulted in a relatively higher percentage of human IFN-γ+ cells compared to controls. Results were similar for CD8+ T cells. Unlike changes in the percentages of human IFN-γ producing cells, mean fluorescence intensities of human IFN-γ staining did not differ among transgenes. As such, CNS-4 and CNS+20 determine numbers of IFN-γ+ cells rather the amount of human IFN-γ produced per cell.
FIGURE 5.
CNS+20 is required for in vivo generated Th1 cells. A, Splenocytes from day 35 immunized transgenic mice were restimulated with IL-12 and IL-18 and IFN-γ was assayed by intracellular cytokine staining. B, Quantification of the percentages of human IFN-γ positive cells in mouse IFN-γ+, CD4+ or mouse IFN-γ+, CD8+ populations or MFI of human IFN-γ positive cells. Results are means of three independent replicates and error bars are standard error of the mean. * P < 0.05.
Natural killer T cells require CNS+20 for human IFN-γ production
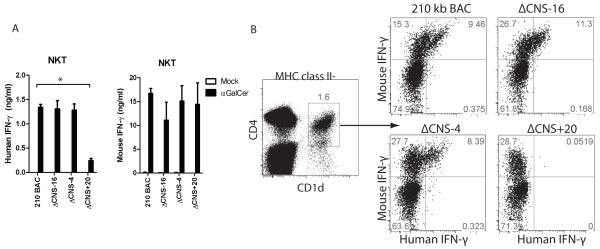
Natural killer T (NKT) cells represent an innate-like T cell subset arising from the T cell lineage but which express NK cell markers. Upon recognition of glycolipid antigens, NKT cells rapidly express IFN-γ and other cytokines characteristic of multiple T cell lineages (24). Therefore, we determined which distal regulatory elements were required for IFNG expression by NKT cells. We first assayed NKT cell expression after antigen-specific stimulation. Splenocytes from 210 kb IFNG-BAC, ΔCNS-16, ΔCNS-4 and ΔCNS+20 transgenic mice were isolated and stimulated with the glycolipid α-galactosylceramide (α-GalCer), or a mock stimulus. After two days, human and mouse IFN-γ concentrations in cultures were determined by ELISA (Fig. 6A). Concentrations of human IFN-γ did not significantly differ among 210 kb IFNG-BAC, ΔCNS-16, or ΔCNS-4 cultures. In contrast, removal of CNS+20 resulted in a marked decrease in production of human IFN-γ.
FIGURE 6.
CNS+20 is required for IFN-γ production by antigen-stimulated NKT cells. A, Splenocytes were isolated from transgenic mice and stimulated with a mock treatment or with alpha GalCer. Human and mouse IFN-γ concentrations in culture supernatants were determined by ELISA. Results are representative of three independent experiments. Error bars are standard error of the mean. B, Splenocytes from transgenic mice were stimulated with IL-12 and IL-18 and IFN-γ was assayed by intracellular cytokine staining. NKT cells were gated on MHC class II−, CD1d+ populations. * P < 0.05.
To directly compare NKT cell regulation with T and NK cell regulation of the IFNG-BAC transgene, we examined NKT cell expression of human and mouse IFN-γ after stimulation with IL-12 and IL-18. Isolated splenocytes were stimulated with IL-12 and IL-18 for 16 hours and analyzed by intracellular cytokine staining (Fig. 6B). To identify NKT cell populations, we used a tetramer specific for the NKT cell receptor CD1d. Cells were gated on MHC class II-negative populations to remove non-specific tetramer staining. Percentages of human IFN-γ positive cells did not vary among 210 kb IFNG-BAC, ΔCNS-16 and ΔCNS-4 transgenic NKT cells. By contrast, we did not detect ΔCNS+20 transgenic NKT cells positive for human IFN-γ. These results are consistent with a necessary role for CNS+20, but not other CNS, for NKT cells to express human IFN-γ from the IFNG-BAC transgene.
Natural killer cells partially require CNS-16, CNS-4 and CNS+20
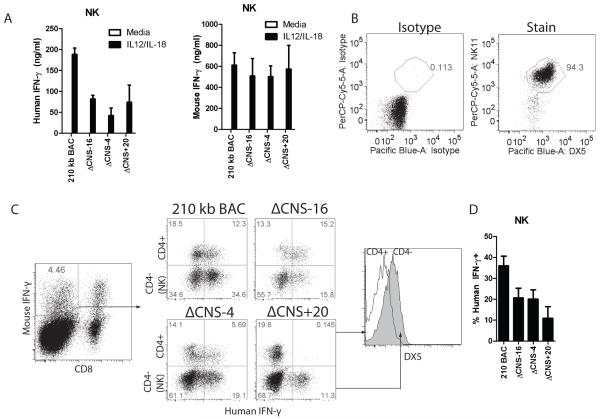
In previous work, we demonstrated that CNS-30 is not required for human IFN-γ expression by NK cells (14). Human CNS-4 is acetylated in natural killer cells, and is an enhancer in reporter assays using human peripheral blood mononuclear cells (11). Further, NK cells immigrate to the periphery fully able to rapidly produce IFN-γ, much like effector Th1 cells, which required CNS-4 for human IFN-γ expression. As such, we hypothesized that CNS-4 was required for human IFN-γ expression by NK cells. To determine distal regulatory usage by NK cells, purified 210 kb IFNG-BAC, ΔCNS-16, ΔCNS-4, and ΔCNS+20 transgenic NK cells were stimulated with IL-12 and IL-18. After two days, we measured human and mouse IFN-γ in cultures (Fig. 7A). In contrast to T cells, removal of CNS-16 resulted in a significant decrease in human IFN-γ concentrations in NK cell cultures. In addition, removal of CNS-4 or CNS+20 resulted in a significant decrease in human IFN-γ concentrations in culture supernatants, compared to controls. Concentrations of human IFN-γ in NK cell culture supernatants were noticeably higher than concentrations of human IFN-γ in T cell cultures under identical stimulation conditions (Fig. 2B). We verified that cultures were 95% pure DX5+, NK1.1+ (Fig. 7B). The high concentrations of human IFN-γ in the cultures and functional requirements for CNS-16 suggests that NK cell regulation of the BAC transgene differed from T cell regulation.
FIGURE 7.
CNS-16, CNS-4 and CNS+20 are required for IFN-γ expression by NK cells. A, NK cells were purified from transgenic splenocytes and stimulated with IL-12 and IL-18, or given a mock stimulus. IFN-γ concentrations were determined by ELISA. Results are representative of four independent experiments and error bars represent standard deviation. B, Representative cell purity of purified NK cells. C, Splenocytes from day 35 immunized transgenic mice were restimulated with IL-12 and IL-18 and IFN-γ was determined by intracellular cytokine staining. NK cells were determined as CD8−, CD4−, DX5+. D, Quantification of the percentages of human IFN-γ positive cells in mouse IFN-γ+ CD4−, CD8-populations. Results are means of three independent replicates and error bars represent standard error of the mean.
We directly compared distal regulatory element usage in T cells and NK cells by intracellular cytokine staining of IL-12 and IL-18 stimulated splenocytes from immunized transgenic mice. After gating on mouse IFN-γ+, CD8− populations, we directly compared T cells and NK cells by comparing CD4+ T cells to CD4−, DX5+ cells (Fig. 7C). In 210 kb IFNG BAC NK cells, 40% of cells were human IFN-γ+ (Fig. 7D), similar to expression by T cells. Consistent with measurements of protein concentrations, each of the CNS-16, CNS-4 and CNS+20 deletions individually resulted in a slight decrease in percentages of human IFN-γ+ NK cells (Fig. 7D). Taken together, CNS-16, CNS-4 and CNS+20 all contribute to a fraction of the total human IFN-γ+ expression by NK cells but have markedly different functions in CD4+ and CD8+ T cells, and NKT cells.
Discussion
Here we determined the necessary functional roles of IFNG distal regulatory elements CNS-16, CNS-4, CNS+20 and CNS+120, using a BAC transgenic system. In T cells, these distal regulatory elements all have discrete functions. CNS-16 is a repressor of IFN-γ expression in both Th1 and Th2 cells. CNS-4 is necessary for IFN-γ expression by effector Th1 cells in response to secondary TCR stimulation. CNS+20 is necessary for memory responses in vivo. In contrast, only CNS+20 is required by NKT cells to produce human IFN-γ in response to antigen stimulation. Unlike T cells and NKT cells, CNS-16, CNS-4 and CNS+20 each are partially required for IFNG expression in NK cells, and removal of no individual distal regulatory element completely abolishes human IFN-γ expression. As such, NK, NKT and Th1 cells employ very different usage of distal regulatory elements to achieve lineage-specific IFNG transcription.
Conserved noncoding sequences at −30 kb, −4 kb and +20 kb from the IFNG start site are necessary for expression of human IFN-γ. In an earlier report, we used 40 and 80 kb deletions in the IFNG-BAC transgenic model to identify the −30 to +20 kb region as necessary for IFNG expression while regions outside this core regulatory element, −80kb to −40 kb and +20 kb to +100 kb, were dispensable (14). CNS-30, CNS-4, and CNS+20 show clear non-placental orthologs in our conservation analysis. In addition, CNS-30, CNS-4, and CNS+20 possess Th1-specific DNase I HS. Distal regulatory regions which are either not conserved in non-placental mammals or do not show Th1-specific DNase I hypersensitivity were not required for IFNG expression. Thus, in our model system the combination of Th1-specific DNase I hypersensitivity and non-placental mammalian conservation appears to separate necessary distal regulatory elements from distal regulatory elements that are not necessary for IFNG expression.
Consistent with our results, other reports have demonstrated functional roles for CNS-4 in responding to Stat5a, Stat5b, NFAT and T-bet transcription factors (11-13, 18, 19). IL-2 signaling through Jak3 induces Stat5 binding at CNS-4 and is important for remodeling of the Ifng locus after 72 hours of culture (12), consistent with a role for CNS-4 in effector cultures. CNS+20 has little enhancer activity on its own, but acts synergistically with CNS-4 in reporter assays (17) and loops into CNS-4 upon Th1 differentiation (25). In the IFNG-BAC transgenic system, CNS+20 is required for IFN-γ expression from in vivo generated memory cells. One model to explain these observations would be that IFNG distal regulation has successive requirements during T cell maturation. As Th1 cells develop further away from an initial polarization signal more distal regulatory elements would become necessary. In this model CNS-30 is required for human IFN-γ expression in primary cultures and later stages of differentiation, CNS-4 is required in effector Th1 cells, and CNS+20 is required in memory Th1 cells. An alternative hypothesis would be that there are yet-undefined in vivo factors which regulate IFN-γ expression and distal regulatory element utilization. In this model, an in vivo factor allows CNS-4 to be dispensable in primary cultures and makes CNS+20 absolutely required during in vivo differentiation. Future work will differentiate between these hypothesizes.
Counter to our initial hypothesis, removing CNS-16 leads to relatively high human IFN-γ expression by both Th1 and Th2 cells and is necessary for IFN-γ expression only in NK cells. These results are surprising because in a similar BAC transgenic system, the murine CNS-16 ortholog is absolutely necessary for transcription of an Ifng-BAC transgene Thy1.1 reporter by both T cells and NK cells (9). However, CNS-16 is not conserved beyond non-placental mammals (Fig. 1), is not required for IFN-γ expression in our human BAC system, and is therefore not universally required for IFN-γ expression by T cells. Numerous reports have demonstrated species-specific roles of distal regulatory elements (5, 26, 27). Thus, it is likely that the CNS-16 ortholog, mCNS-22, is required for murine IFN-γ expression in T cells while CNS-16 plays an opposing role in human IFNG regulation repressing IFN-γ expression. Indeed, the mouse IFN-γ locus has undergone substantial rearrangements relative to non-rodent species (28). Future comparison of the mouse and human interferon gamma loci will provide additional insights into the species-specific functions of distal regulatory elements.
Although regulation of human IFN-γ expression from the 190 kb IFNG-BAC transgene correctly depends upon the transcription factors T-bet and STAT4 (14) and is Th1/Th2 selective, it is likely that regulation of IFNG-BAC transgenes does not perfectly mirror regulation in humans. Analysis is based upon the assumption that regulation of transgenic human IFN-γ in a mouse fully recapitulates regulation in the endogenous human genome, which may not be true. As such, these findings need to be confirmed in humans.
Transgenic T cells with a 190 kb or 210 kb IFNG-BAC transgene express human IFN-γ at equivalent levels. The two transgenes differ mainly by the inclusion of CNS+120 in the 210 kb IFNG-BAC. CNS+120 is thought to facilitate three-dimensional organization of the IFNG locus via CCCTC-binding factor (CTCF) binding at CNS+120, CNS-63 and the IFNG first intron (29, 30). CTCF has been described as a transcriptional activator (31), insulator (32) and repressor (33). However, most of these functional studies come from reporter assay systems which do not take genomic context into account and a relevant question is the exact function of CTCF binding sites in the genome. Arguing against an insulating role of the +120 CTCF site in the IFNG locus are previous experiments reporting copy-number dependence for 190 kb IFNG-BAC transgenes (14), and equivalent expression between 190 kb and 210 kb IFNG BAC transgenes. Another hypothesis would be that the +120 kb and −63 kb CTCF sites serve to bring the IFNG locus into close physical location to other genes in the surrounding regions. IFNG is adjacent to IL22 and IL26, both of which are expressed in Th17 cell subsets upon T cell receptor signaling. IFNG locus CTCF sites may play a role in co-regulation of IFNG, IL22 and IL26. Alternatively, the CTCF sites may govern intrachromosomal looping interactions, such as those observed between the Ifng locus and the Il4 locus (25).
Transgene deletion effects are dissimilar between NK cells and T cells. In NK cell assays, each distal regulatory element is required for a fraction of human IFN-γ expression. These results support models proposing that distal regulation serves to allow expression under sub-optimal signaling conditions (7) and functions in evolution to modify the level of gene expression in individual species (27). A second model is that distal regulatory elements are both cell-type and stimulus selective and function in evolution by allowing changes in the timing and location of gene expression (5, 27). This model is consistent with distal regulatory element usage in T cells. In T cell assays, CNS-30 is needed in primary cultures, CNS-4 in effector cultures and CNS+20 in memory experiments. Thus, our results support both models of distal regulation. More importantly, choice of cell type determines which model is supported. These results illustrate the importance of analyzing multiple cell types and phenotypes when analyzing the key contributions distal regulatory elements make to achieve proper gene regulation.
Supplementary Material
Acknowledgements
We would like to acknowledge the Vanderbilt Transgenic Mouse/Embryonic Stem Cell Shared Resource for assistance in creating transgenic mice and the Vanderbilt Flow Cytometry shared resource core. We acknowledge Rachel Henry for her careful review of this manuscript.
This work was supported by the National Institute of Health grant AI44924 and training grant HL069765. The VUMC Flow Cytometry Shared Resource is supported in part by Vanderbilt CTSA grant 5UL1 RR024975-03 from NCRR/NIH, the Vanderbilt Digestive Disease Research Center (DK058404). The Vanderbilt Transgenic/Embryonic Stem Cell Shared Resource is supported in part by NIH grant CA68485.
Abbreviations
- BAC
Bacterial artificial chromosome
- CNS
Conserved noncoding sequence
Footnotes
Disclosures The authors have no financial conflicts of interest.
References
- 1.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 2.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 3.Soutto M, Zhou W, Aune TM. Cutting edge: distal regulatory elements are required to achieve selective expression of IFN-gamma in Th1/Tc1 effector cells. J Immunol. 2002;169:6664–6667. doi: 10.4049/jimmunol.169.12.6664. [DOI] [PubMed] [Google Scholar]
- 4.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 5.Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, Pennacchio LA, Rubin EM, Noonan JP. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler KJ, Chandler RL, Mortlock DP. Identification of an ancient Bmp4 mesoderm enhancer located 46 kb from the promoter. Dev Biol. 2009;327:590–602. doi: 10.1016/j.ydbio.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, Weaver CT. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Chang S, Aune TM. Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells. Proc Natl Acad Sci U S A. 2005;102:17095–17100. doi: 10.1073/pnas.0502129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bream JH, Hodge DL, Gonsky R, Spolski R, Leonard WJ, Krebs S, Targan S, Morinobu A, O’Shea JJ, Young HA. A distal region in the interferon-gamma gene is a site of epigenetic remodeling and transcriptional regulation by interleukin-2. J Biol Chem. 2004;279:41249–41257. doi: 10.1074/jbc.M401168200. [DOI] [PubMed] [Google Scholar]
- 12.Shi M, Lin TH, Appell KC, Berg LJ. Janus-kinase-3-dependent signals induce chromatin remodeling at the Ifng locus during T helper 1 cell differentiation. Immunity. 2008;28:763–773. doi: 10.1016/j.immuni.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balasubramani A, Shibata Y, Crawford GE, Baldwin AS, Hatton RD, Weaver CT. Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity. 2010;33:35–47. doi: 10.1016/j.immuni.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins PL, Chang S, Henderson M, Soutto M, Davis GM, McLoed AG, Townsend MJ, Glimcher LH, Mortlock DP, Aune TM. Distal regions of the human IFNG locus direct cell type-specific expression. J Immunol. 2010;185:1492–1501. doi: 10.4049/jimmunol.1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W, Chang S, Aune TM. Long-range histone acetylation of the Ifng gene is an essential feature of T cell differentiation. Proc Natl Acad Sci U S A. 2004;101:2440–2445. doi: 10.1073/pnas.0306002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferongamma (IFN-gamma) locus revealed by genome sequence comparison. J Biol Chem. 2004;279:4802–4810. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- 18.Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, Wilson CB. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci U S A. 2004;101:12622–12627. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balasubramani A, Mukasa R, Hatton RD, Weaver CT. Regulation of the Ifng locus in the context of T-lineage specification and plasticity. Immunol Rev. 2010;238:216–232. doi: 10.1111/j.1600-065X.2010.00961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandler KJ, Chandler RL, Broeckelmann EM, Hou Y, Southard-Smith EM, Mortlock DP. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm Genome. 2007;18:693–708. doi: 10.1007/s00335-007-9056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011;12:551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuddapah S, Schones DE, Cui K, Roh TY, Barski A, Wei G, Rochman M, Bustin M, Zhao K. Genomic profiling of HMGN1 reveals an association with chromatin at regulatory regions. Mol Cell Biol. 2011;31:700–709. doi: 10.1128/MCB.00740-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc Natl Acad Sci U S A. 2003;100:15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godfrey DI, Kronenberg M. Going both ways: immune regulation via CD1d-dependent NKT cells. J Clin Invest. 2004;114:1379–1388. doi: 10.1172/JCI23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 26.Menke DB, Guenther C, Kingsley DM. Dual hindlimb control elements in the Tbx4 gene and region-specific control of bone size in vertebrate limbs. Development. 2008;135:2543–2553. doi: 10.1242/dev.017384. [DOI] [PubMed] [Google Scholar]
- 27.Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474:598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levi-Acobas F, Mars LT, Orth A, Bureau JF, Bonhomme F. Adaptive evolution of interferon-gamma in Glire lineage and evidence for a recent selective sweep in Mus. m. domesticus. Genes Immun. 2009;10:297–308. doi: 10.1038/gene.2009.22. [DOI] [PubMed] [Google Scholar]
- 29.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, Wilson CB. CCCTC-Binding Factor and the Transcription Factor T-bet Orchestrate T Helper 1 Cell-Specific Structure and Function at the Interferon-gamma Locus. Immunity. 2009 doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vostrov AA, Quitschke WW. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem. 1997;272:33353–33359. doi: 10.1074/jbc.272.52.33353. [DOI] [PubMed] [Google Scholar]
- 32.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 33.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dumoutier L, Van Roost E, Ameye G, Michaux L, Renauld JC. ILTIF/IL-22: genomic organization and mapping of the human and mouse genes. Genes Immun. 2000;1:488–494. doi: 10.1038/sj.gene.6363716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.