Abstract
The vast number of microbial sequences resulting from sequencing efforts using new technologies require us to re-assess currently available analysis methodologies and tools. Here we describe trends in the development and distribution of software for analyzing microbial sequence data. We then focus on one widely used set of methods, dimensionality reduction techniques, which allow users to summarize and compare these vast datasets. We conclude by emphasizing the utility of formal software engineering methods for development of computational biology tools, and the need for new algorithms for comparing microbial communities. Such large-scale comparisons will allow us to fulfill the dream of rapid integration and comparison of microbial sequence data sets, in a replicable analytical environment, in order to describe the microbial world we inhabit.
Introduction
Recent innovations in sequencing technologies allowed microbial ecologists to advance from analyzing a few hundred sequences per study to hundreds of millions (••1, ••2). These quantitative differences in the amount of sequence data produce qualitative differences in the types of studies that can be performed. For example, ten years ago, characterization of a single clone library from a single body site in one subject represented a substantial advance in knowledge about the human body. A few years ago, quantifying interpersonal differences in one body site, e.g. the gut, represented a major advance (3, 4). Three years ago, performing a multi-site microbial scan of the body, showing how the microbial communities that live on the same person’s body are clearly separated by body site, primarily skin, mouth and stool (5). Now, with higher-throughput sequencing technologies, we can observe the dynamics of the human microbiota across multiple sites and individuals through time, demonstrating that our microbial guests are highly volatile day-to-day even in healthy adults (••6). These examples also illustrate the daunting analytical challenges that microbial researchers face to handle datasets that are ever increasing in size. These challenges range from simply finding the right hypotheses to test, to finding the correct analytical tools and computational power to test them, to finding the methods for visualizing the key results. Here we review computational tools developed in the last three years and algorithms conceived over the last few decades, but only recently applied in microbial ecology; we conclude with suggestions for computational tool developers who wish to help the field continue its rapid pace of development over the next few years.
Microbial Diversity Analysis Tools
As 16S rRNA and shotgun metagenomic datasets grow dramatically, the need for easily accessible, well-documented and well-tested tools in the form of a pipeline becomes increasingly critical. In particular, the complexity of what is considered a “standard” analysis has increased rapidly, from small trees and pie chart to advanced analyses incorporating multivariate statistics, machine learning, and, increasingly, explicitly spatial and/or temporal analysis, Figure 1. These new challenges, and especially the need to integrate multiple tools, have forced researchers to move from ad hoc scripts developed in numerical computing environments like R (7) or MATLAB (8) to more general libraries that provide solutions to a specific research niche. Examples include vegan, which provides statistical functions for vegetation (and other) ecologists (9); ade4, which allows exploratory analyses for environmental sciences (10); and ape, which provides methods for phylogenetics and evolution (11); see Table 1. However, developing expertise in, appropriately formatting data, loading large datasets and transferring datasets among multiple packages can be time-consuming: for example, see the methods section and reference list of (12).
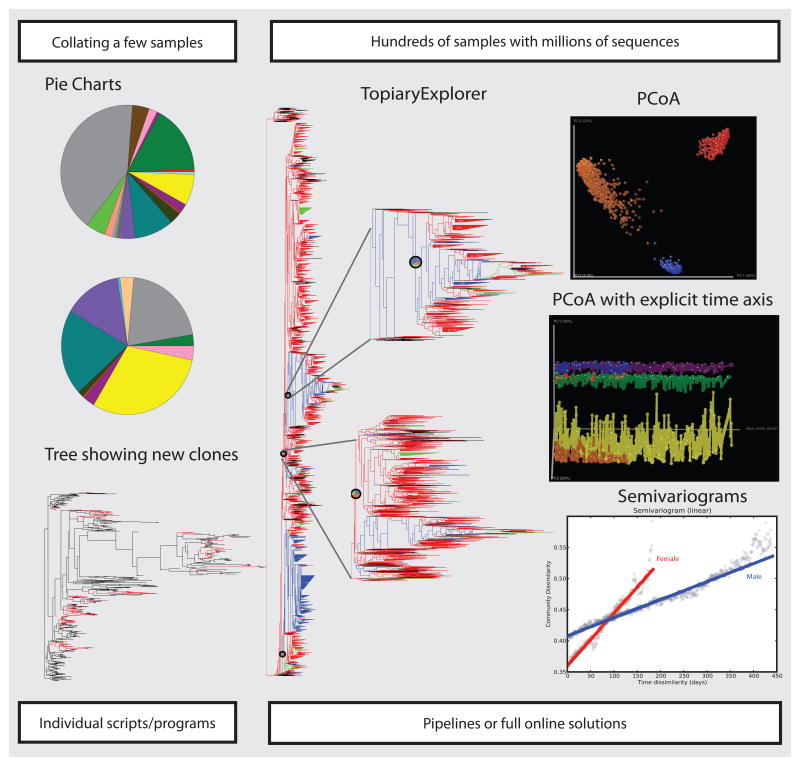
Figure 1. Moving away from pie charts and trees, current analytical methods.
With a few sequences from a small number of samples, pie charts and trees was sufficient for comparing microbial community samples. In contrast, with modern technologies that allow sequencing large number of samples with millions of reads, the new analysis “gold-standard” has moved towards deploying new tools. Here we show data from ref. (6) analyzed with several methods: TopiaryExplorer (41) allows visualization of large trees in the context of per-sample data, , in this example visualizing the GreenGenes reference tree colored by body site matches (red-stool, blue-oral, orange-skin), showing pie charts of most abundant sequences and zooming into the different clades; QIIME PCoA plot comparing all samples color by body site (same colors than in TopiaryExplorer), PCoA with explicit time axis and tracing to follow individuals over time in each body site, allowing visually inspection the changes over time (female: red-gut, blue-oral, orange-skin; male: green-gut, purple-oral, yellow-skin); and semivariograms to assess temporal correlation of observations in the stool samples separated by sex (red-female, blue-male).
Table 1. Different types of analytical tools for microbial diversity analysis.
This table highlights different options researchers have for performing microbial diversity analyses. Developers of new analytical tools should take advantage of existing options, and, especially, of pipeline integration to avoid duplication of effort.
| Name | Open Source | Analytical usage | Development method | Advantages | Disadvantages |
|---|---|---|---|---|---|
| vegan | Y | General purpose. | Single library | Several ordination methods, dissimilarity indices, alpha and beta diversity algorithms. | Incomplete analysis solution, the user has to be an R expert. |
| ade4 | Y | General purpose. | Single library | Multivariate analysis methods based on Euclidean distances. | Incomplete analysis solution, the user has to be an R expert. |
| ape | Y | General purpose. | Single library | Functions to manipulate and analyze phylogenetic trees . | Incomplete analysis solution, the user has to be an R expert. |
| mothur | Y | Data generated via high-throughput amplicon sequencing. | Standalone tool Workflow | Single program for complete analysis with basic visualizations. | Incomplete usage of software engineering techniques, custom command line interface, not designed for cluster. |
| QIIME | Y | Data generated via high-throughput amplicon sequencing. | Multiple scripts Workflow Virtualization | Multi-script pipeline for complete analysis with several advanced visualizations. Developed using formal software engineering methods. | Command line interface. Installation on local machine difficult for non-experts. |
| VAMPS | N | Data generated via high-throughput amplicon sequencing. | Web server Workflow | Complete online analytical solution. Provides compute resources for free. | Lack of access to source code, possible extended waiting times for results, limitations in data transfers and analysis. |
| MG-RAST | Y | Data generated via shotgun metagenomic sequencing. | Web server Workflow | Complete online analytical solution. Provides compute resources for free. | Possible extended waiting times for results, limitations in data transfer and analysis. |
| Smash Community | Y | Data generated via shotgun metagenomic sequencing. | Multi-scripts Workflow Virtualization | Complete analytical solution with visualizations. | Incomplete usage of software engineering techniques. Not fully parallelizable. |
| CLoVR | Y | Multipurpose pipeline creation. | Virtualization Workflow | Provides access to created analytical pipelines and the possibility of developing new ones in an easy interface. Fully integrated with academic and commercial clouds. | Potentially expensive for large analyses, difficult to change parameters once pipeline is running. |
| Galaxy | Y | Multipurpose pipeline creation. | Web server Workflows | Provides access to created analytical pipelines and the possibility of developing new ones. Fully integrated with academic and commercial clouds. | Potentially expensive for large analyses, difficult to change parameters once full pipeline running. Pipeline creation hard. |
A more recent approach has been to develop pipelines that provide complete analysis solutions, combining many steps. For example, if a researcher is interested in analyzing microbial community data generated via high-throughput amplicon sequencing data (such as SSU rRNA), starting with files containing a hundred million sequences to a set of meaningful statistics and visualizations, one tactic is to create a single workflow solution like mothur (13), which provides one program for analysis (for a use case see (•14)); an inherent downside of this approach is increased development time and support burden for a larger codebase, and errors arising from reimplementation of each specialized analysis step into a single tool. Another strategy is to wrap the original different applications in one single package; for example, Quantitative Insights Into Microbial Ecology (QIIME) (••15) provides workflows by splitting the steps into fully transparent scripts (for a use case see (6)); the cost is that the user must track down and install the individual tools, but the user has substantially more control over the analysis and knows they are using “name-brand” software. Another solution is to create analytical web servers, like Visualization and Analysis of Microbial Population Structures (VAMPS) (16), which allows researchers to upload their 16S rRNA data for analysis and visualization (for a use case see (••17)), or the Metagenomics RAST (MG-RAST) server (18) for studies based on shotgun metagenomic sequence. However, web servers usually limit the control users have over their analyses, some analysis steps and methods are hidden when source code is not available, and the user must fully commit to these tools rather than inserting data at later stages or retrieving partial results. A recent comparison of pipelines for metagenomic annotation and analysis pipelines, can be found in the supplementary material of SmashCommunity (•19), which is an open-source, local solution to some of these problems; see Table 1. Open source software, where the source code is available for download, is critical for research software in general as investigators can then check the correctness of the algorithms and make improvements.
The newest approach is to use virtual instances, either by virtualizing in a single computer (e.g. VirtualBox (https://www.virtualbox.org/) or VMWare (http://www.vmware.com/)), where resources are shared within a local machine (which can be a processing bottleneck), or virtualizing in the “cloud” (e.g. EC2 (http://aws.amazon.com/ec2) or Magellan (http://magellan.alcf.anl.gov)), where external resources are used, sometimes at cost. Both virtualization scenarios provide an environment to run virtual machines with preloaded operating systems and programs. For example, CLoVR (••20) can run several metagenomic analysis pipelines, and parallelizes some of these steps across virtual machines to speed up the analysis. Similarly, Galaxy (http://galaxy.psu.edu/) provides a web interface to create analysis pipelines, share them, and share data and results; see Table 1. Both resources are open source.
The QIIME pipeline in particular exemplifies several key software engineering methodologies. First, it is developed using agile software development techniques (21), which require constant interaction with end-users, rapid iterative development and updates, simplicity of implementations and interfaces, etc. QIIME also relies heavily on test-driven development (22), which is similar to the concept of positive and negative controls in lab research and reduces errors considerably. Furthermore, it is open source and distributes its software dependences for a range of computational options, such as direct personal computer installation, virtual machines images for single computer access via VirtualBox, and powerful cloud computing options such as EC2 and Magellan.
Summarizing and Understanding Microbial Diversity
The democratization of sequencing technology allows researchers to sequence large numbers of samples from diverse environments (1, 2). Large-scale collaborative projects have taken advantage of this possibility. For example, the Human Microbiome Project (23) sampled 250 individuals 2–3 times, in 5 main sites (the GI tract, the mouth, the vagina, the skin, and the nasal cavity), and the Earth Microbiome Project (24) will sequence up to 200,000 diverse environmental samples. A new challenge generated by these types of projects is to compare not only large numbers of sequences but also large numbers of samples, and to relate the variation in these samples to key clinical or environmental parameters. Although, as outlined above, many ways of examining the data can be valuable, we focus here on dimensionality reduction, an especially useful technique for examining these multidimensional matrices that have more variables than samples. Dimensionality reduction often yields easily interpretable results, while reducing computational costs, relative to trying to understand large taxon tables (25, 26).
Dimensionality reduction techniques help us simplify data represented by a large number of features compared to the number of samples (25, 26). There are two general strategies: feature transformation, which calculates a lower-dimension projection of the original features while retaining as much information as possible, and feature selection, which minimizes the number of variables by locating the “best” minimum subset of the original features (25). The two strategies can also be combined (27). In general, feature transformation has been more widely applied in microbial ecology, even though the transformed features may have no biological meaning (25, •28); feature selection has primarily been applied, often informally, in source tracking and biomarkers (29, 30). Feature transformation can be performed using unsupervised methods (that use only the data matrix itself), including metric and non-metric multidimensional scaling (MDS), or by supervised approaches (that use information about the samples, e.g. clinical or environmental categories) such as Linear Discriminant Analysis (LDA) (25, 31); see Table 2. Both supervised and unsupervised techniques are susceptible to noise in the category labels, e.g. due to mislabeling of samples or contamination. As these issues are a fact of life in projects covering thousands of samples, tools such as SourceTracker (30), which can detect contamination and mislabeling, are increasingly useful.
Table 2. Different unsupervised feature transformation techniques and their biological findings.
This table summarizes some of the unsupervised feature transformation techniques and some relevant biological findings achieved by the methods. It also shows other methods that need some future research to assess their performance while comparing microbial communities.
| Name | Advantages | Disadvantages | Example application |
|---|---|---|---|
| Principal Component Analysis (PCA) | Allows visualization of high dimensional data using lower dimensions. | Based on Euclidean distances for dissimilarity comparisons, which can hide biologically relevant patterns. Non-linear growth in processing time. Horseshoe effect. | Showed significant correlation between relative abundance of Bacteroidetes and metagenome functions associated with obesity (4). |
| Multidimensional Scaling (MDS) / Principal Coordinate Analysis (PCoA) | Allows visualization of high dimensional data using lower dimensions allowing the use of any dissimilarity metric. | Non-linear growth in processing time. Horseshoe effect. | Showed high variability in microbial community through time while preserving differences between body sites (6). |
| Non metric Multidimensional Scaling (NMDS) | In general, preserves the high-dimensional structure with fewer axes. Based on numerical optimization, relaxing linear assumptions. | Can be more time-consuming than MDS. Arch effect. | NMDS plots showed that short-term storage conditions for soil and human related samples do not affect community composition (42). |
| FastMap | MDS approximation that relies on a mapping technique that makes linear the processing time | As this is an approximation, it might miss interesting biological patterns | Has been used in other research areas but not for microbial community comparisons |
| MetricMap | MDS approximation that expands FastMap to work on many projections at once. | As this is an approximation it might miss interesting biological patterns | Has been used in other research areas but not for microbial community comparisons |
| Landmark MDS | MDS approximation that uses a small number of landmark points to derive new coordinates. | As this is an approximation, it might miss interesting biological patterns | Has been used in other research areas but not for microbial community comparisons |
One of the most commonly used dimensionality reduction techniques in microbial ecology is PCoA, also known as MDS. PCA, or principal coordinates analysis, is a special case of PCoA using Euclidean distance as a dissimilarity measure (32). PCoA takes as input an n × n matrix of distances, generally the results of beta diversity comparisons between n samples in p-dimensional space (traits) although phylogenetic distances such as UniFrac (33) can also be used. It produces a k-dimensional, k ≤ p, representation of the items such that the distances among the points in the new space preserve as closely as possible the distances in the original data (26). In other words, points that are close in the original space are also close in the new space. Results of MDS are indeterminate with respect to translation, rotation, and reflection; in other words, the direction of each axis is arbitrary, although typically the axes are chosen to maximize the variation in the data. PCoA can be used with any dissimilarity metric (beta diversity): for current best practices for non-phylogenetic metrics see (28), and for phylogenetic metrics see (34).
PCA and PCoA rely on solving the eigenvalue equation to find a linear representation of our samples by combining the original variables to generate the resulting k-dimensional representation of the data (32). Another approach that can reduce certain artifacts, such as the horseshoe effect (a pattern in which the two ends of an axis attract each other due to a shared lack of the taxa in the middle, thus obscuring the gradient pattern), is to use nonlinear methods (35). NMDS can better preserve the high-dimensional structure with few axes in some cases, although cannot fully avoid the arch effect in realistic microbial datasets (28). The main differences between PCoA and NMDS are that the former is based on distances, where the final configuration should match the original distances as close as possible, and the latter is based on ranks, which is robust to distribution effects, similar to the difference between Pearson and Spearman correlations (36). One drawback to MDS is that it is not based on an eigenvalue solution but on numerical optimization: for larger datasets, the calculations become time-consuming; see Table 2.
Because even PCoA is slow on large datasets, integrating new samples rapidly into large existing datasets poses a major algorithmic challenge. Such techniques are critical for integrating results from new studies, e.g. new environments or patient populations, into large-scale datasets such as those provided by the Human Microbiome Project (23) or Earth Microbiome Project (24). There has been substantial recent improvement in the performance of some of these approximate algorithms for PCoA. For example, Nystrom techniques such as FastMap, which uses a mapping technique to derive the k-dimension representation, are linear-time algorithms rather than quadratic like PCoA (i.e. the time increases in proportion to the number of samples rather than to the square of the number of samples) (37). MetricMap expands FastMap to assess many projections at once, whereas FastMap calculates one dimension at the time (38). Landmark MDS (LMDS) uses a small number landmark points, either manually or randomly selected, to derive new coordinates (39); see Table 2. For a performance comparison of these methods see (40). The accuracy of these techniques have been assessed by methods that determine how much of the variance is explained by the new set of axes (R2) or how much the distances change in the low-dimensional projection (Kruskal stress). The inherent problem of these methods for determining accuracy, however, is that they do not relate well to clustering quality or ability to interpret the patterns in the data (as has been previously observed for different distance metrics, where the metric that explains most of the variance may produce results that have no biological meaning (28)). Thus improved, and biologically informed, evaluations of these methods are a key area of current interest.
Conclusions
We are currently faced with daunting bioinformatics and computational challenges because of the large numbers of sequences and samples now examined in microbial ecology studies, which require the use of defined software engineering methods to create pipelines that are user-driven and well-tested. Although these pipelines integrate many different techniques for visualizing and understanding data, dimensionality reduction techniques such as PCoA have proven especially valuable for understanding patterns in the data. However, these techniques are reaching their limits as very large numbers of samples are analyzed in large-scale, and ongoing studies could potentially reach a processing bottleneck as these methods do not scale linearly to the number of samples; approximate algorithms, which can be much faster, provide a way out of this conundrum, but could also create a complication if research do not focus in exact approximations. Thus, substantial additional work will be required in order to realize the dream of rapid integration of new samples into large existing frameworks that cover our bodies or our planet.
Highlights.
New technologies permit dramatic increases in number of sequences/sites per study.
Multisite spatial/temporal studies with hundreds of millions of sequences possible.
New software pipelines are required to analyze these vast datasets.
Field is increasingly moving from ad hoc scripts towards integrated pipelines.
Multivariate techniques often introduce computational bottlenecks.
Cloud computing and improved approximation methods needed to avoid bottlenecks.
Acknowledgments
We thank Greg Caporaso, Jesse Stombaugh and Meg Pirrung for assistance in creating Figure 1, and Jessica Metcalf for helpful comments and edits on the manuscript. The work described in this review was supported by the National Institutes of Health, the Bill and Melinda Gates Foundation, the Crohns and Colitis Foundation of America, the Colorado Center for Biofuels and Biorefining and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- ••1.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011 Mar 15;108(Suppl 1):4516–22. doi: 10.1073/pnas.1000080107. Demonstrates that the Illumina platform's short reads can be used successfully for 16S rRNA profiling in a wide range of environments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••2.Bartram AK, Lynch MD, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl Environ Microbiol. 2011 Jun;77(11):3846–52. doi: 10.1128/AEM.02772-10. Demonstrates the advances in sampling depth and quality by using Illumina technologies; also shows the possibility of reducing erroneous sequences by using paired-end reads. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007 Aug 21;104(34):13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009 Jan 22;457(7228):480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009 Dec 18;326(5960):1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••6.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol. 2011 May 30;12(5):R50. doi: 10.1186/gb-2011-12-5-r50. Largest published timeseries of human associated microbial communities; the authors show the tremendous variability on those communities through time even within a subject. Also provides a recent example of a QIIME analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2009. [Google Scholar]
- 8.MATLAB. version 7.10.0 (R2010a) Natick, Massachusetts: The MathWorks Inc; 2010. [Google Scholar]
- 9.Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos P, et al. vegan: Community Ecology Package. 2009. [Google Scholar]
- 10.Dray S, Dufour A, Chessel D. The ade4 package-II: Two-table and K-table Methods. R News. 2007;7(2):47–52. [Google Scholar]
- 11.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20(2):289–90. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 12.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005 Mar 25;307(5717):1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 13.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Applied and Environmental Microbiology. 2009;75(23):7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •14.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010 Aug 17;107(33):14691–6. doi: 10.1073/pnas.1005963107. A gut microbial community comparison between European and Burkina Faso children that suggested that rural African children have a higher alpha diversity. The authors suggest that this difference is due to diet and low calorie intake. Provides a recent example of a mothur analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••15.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010 May;7(5):335–6. doi: 10.1038/nmeth.f.303. Describes a microbial ecology analysis pipeline built using a set of well-defined software engineering techniques for design, development, testing, documentation, and distribution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sogin M, Welch DM. VAMPS: Visualization and Analysis of Microbial Population Structure. doi: 10.1186/1471-2105-15-41. [cited]; Available from: http://vamps.mbl.edu/ [DOI] [PMC free article] [PubMed]
- 17.Gobet A, Boer SI, Huse SM, van Beusekom JE, Quince C, Sogin ML, et al. Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J. 2011 Oct 6; doi: 10.1038/ismej.2011.132. The results of this paper show the enormous variability of coastal sand microbial communities over time and their relationship with seasonal fluctuations. Provides a recent example of analysis with VAMPS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9(1):386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •19.Arumugam M, Harrington ED, Foerstner KU, Raes J, Bork P. SmashCommunity: a metagenomic annotation and analysis tool. Bioinformatics. 2010 Dec 1;26(23):2977–8. doi: 10.1093/bioinformatics/btq536. This tool collates and expands existing metagenomic tools, and provides an example of an open-source pipeline that runs locally. [DOI] [PubMed] [Google Scholar]
- ••20.Angiuoli S, Matalka M, Gussman A, Galens K, Vangala M, Riley D, et al. CloVR: A virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC bioinformatics. 2011;12(1):356. doi: 10.1186/1471-2105-12-356. This tool collates and expands existing metagenomic tools, and provides an example of an open-source pipeline that runs locally. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abrahamsson P, Salo O, Ronkainen J, Warsta J. Agile software development methods. Vtt Publications. 2002;478(3):167–8. [Google Scholar]
- 22.Janzen DS, Saiedian H, editors. Software Engineering Education and Training, 2006 Proceedings 19th Conference on. 2006. On the Influence of Test-Driven Development on Software Design. [Google Scholar]
- 23.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. The NIH Human Microbiome Project. Genome Res. 2009 Dec;19(12):2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert JA, Meyer F, Antonopoulos D, Balaji P, Brown CT, Desai N, et al. Meeting report: the terabase metagenomics workshop and the vision of an Earth microbiome project. Stand Genomic Sci. 2010;3(3):243–8. doi: 10.4056/sigs.1433550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunningham Pd. Dimension Reduction. 2007. Contract No.: Document Number|. [Google Scholar]
- 26.Fodor I. A Survey of Dimension Reduction Techniques. 2002 [updated 2002. cited]; Available from: citeulike-article-id:5467879 http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.8.5098.
- 27.Lal T, Chapelle O, Weston J, Elisseeff A, Guyon I, Nikravesh M, et al. Embedded Methods Feature Extraction. Springer; Berlin / Heidelberg: 2006. pp. 137–65. [Google Scholar]
- •28.Kuczynski J, Liu Z, Lozupone C, McDonald D, Fierer N, Knight R. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods. 2010 Oct;7(10):813–9. doi: 10.1038/nmeth.1499. Demonstrates importance of using the correct distance metric to compare microbial communities: critically, they show that often the distance metric that explains the most variance is oftens not actually the one that best recovers biological patterns either previously discovered in, or explicitly simulated in, the data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenny L, Dunn W, Ellis D, Myers J, Baker P, Consortium G, et al. Novel biomarkers for pre-eclampsia detected using metabolomics and machine learning. Metabolomics. 2005;1(3):227–34. [Google Scholar]
- 30.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8(9):761–3. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Celussi M, Bussani A, Cataletto B, Del Negro P. Assemblages' structure and activity of bacterioplankton in northern Adriatic Sea surface waters: a 3-year case study. FEMS Microbiol Ecol. 2011 Jan;75(1):77–88. doi: 10.1111/j.1574-6941.2010.00997.x. [DOI] [PubMed] [Google Scholar]
- 32.Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. ZAMM - Journal of Applied Mathematics and Mechanics / Zeitschrift für Angewandte Mathematik und Mechanik. 1981;61(3–5):206. [Google Scholar]
- 33.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005 Dec;71(12):8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010 Jan;4(1):17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shepard RN, Carroll JD. Parametric representation of nonlinear data structures. In: Krishnaiah PR, editor. Multivariate analysis. Academic Press; New York: 1966. pp. 561–92. [Google Scholar]
- 36.O'Brien CM. Analysing Ecological Data by Alain F. Zuur, Elena N. Ieno, Graham M. Smith. International Statistical Review. 2007;75(3):426–7. [Google Scholar]
- 37.Faloutsos C, Lin K-ID. FastMap: A Fast Algorithm for Indexing, Data-Mining and Visualization of Traditional and Multimedia Datasets. 1995:163–74. [Google Scholar]
- 38.Wang JTL, Wang X, Shasha D, Zhang K. MetricMap: An embedding technique for processing distance-based queries in metric spaces. IEEE Transactions on Systems, Man, and Cybernetics, Part B. 2005;35 doi: 10.1109/tsmcb.2005.848489. [DOI] [PubMed] [Google Scholar]
- 39.De Silva V, Tenenbaum JB. Sparse multidimensional scaling using landmark points. Technology. 2004:1–41. [Google Scholar]
- 40.Platt JC. Fastmap, metricmap, and landmark MDS are all nystrom algorithms. Proceedings of 10th International Workshop on Artificial Intelligence and Statistics; 2005. [Google Scholar]
- 41.Pirrung M, Kennedy R, Caporaso JG, Stombaugh J, Wendel D, Knight R. TopiaryExplorer: Visualizing large phylogenetic trees with environmental metadata. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lauber CL, Zhou N, Gordon JI, Knight R, Fierer N. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol Lett. 2010 Jun;307(1):80–6. doi: 10.1111/j.1574-6968.2010.01965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]