Abstract
Standard Illumina mate-paired libraries are constructed from 3- to 5-kb DNA fragments by a blunt-end circularization. Sequencing reads that pass through the junction of the two joined ends of a 3–5-kb DNA fragment are not easy to identify and pose problems during mapping and de novo assembly. Longer read lengths increase the possibility that a read will cross the junction. To solve this problem, we developed a mate-paired protocol for use with Illumina sequencing technology that uses Cre-Lox recombination instead of blunt end circularization. In this method, a LoxP sequence is incorporated at the junction site. This sequence allows screening reads for junctions without using a reference genome. Junction reads can be trimmed or split at the junction. Moreover, the location of the LoxP sequence in the reads distinguishes mate-paired reads from spurious paired-end reads. We tested this new method by preparing and sequencing a mate-paired library with an insert size of 3 kb from Saccharomyces cerevisiae. We present an analysis of the library quality statistics and a new bio-informatics tool called DeLoxer that can be used to analyze an IlluminaCre-Lox mate-paired data set. We also demonstrate how the resulting data significantly improves a de novo assembly of the S. cerevisiae genome.
INTRODUCTION
Paired-end and mate-paired sequencing libraries both are methodologies that, in addition to sequence information, give information about the physical distance between the two reads in the reference genome. The ability to map reads to a reference using distance information is useful to resolve larger structural rearrangements (insertions, deletions, inversions). Distance information also has a major impact on the overall success of de novo assembly with short reads, helping to assemble across repetitive regions: if one read cannot be mapped because it falls in a highly repetitive region, but the paired read is unique, the distance information can be used to map both reads. When the two reads of a pair can be mapped to two different contiguous sequences from an assembly (contigs), they specify the contigs’ order, orientation and approximate distance in the genome. This ability greatly facilitates de novo genome assembly of complex organisms. The difference between paired-end and mate-paired is typically that mate-paired is used to indicate a longer insert size compared to paired-end, with insert sizes measuring between 2 and 20 kb.
Illumina mate-paired libraries
Illumina mate-paired libraries are constructed from 3- to 5-kb DNA fragments by a blunt-end circularization and a secondary fragmentation step (1). A biotin molecule on the circularization junction is used to enrich for fragments containing the junction. Still, a typical Illumina mate-paired library will have fragments that lack the junction and map as paired-end reads with short inserts. When sequencing a mate-paired library, Illumina recommends a read length no longer than 36 bases. Although short reads are not ideal in de novo assembly of genomes with a high repeat content or when looking for structural variations, the 36-bp limit aims to decrease the possibility that a sequence read will pass through the junction of the two joined ends of a 3- to 5-kb DNA fragment. When using standard mapping software like the Illumina pipeline, such junction reads are discarded, since they would not align to the reference sequence. To map junction reads, specifically adapted software like the Novoalign mate-paired algorithm can be used to detect junction reads and split the read at the junction. Junction reads are problematic for de novo assembly software, where they can reduce the performance of the assembly. To further reduce the number of junction reads, Illumina recommends a final library size range selection of 400–600 bp, which is larger than a typical paired-end library of 200–300 bp. Increasing the size range of the library in the mate-paired protocol minimizes the number of sequence reads that will pass through a junction.
Roche GS-FLX paired-end libraries
In Roche GS-FLX library preparation, the 3- to 20-kb DNA fragments are circularized by a Crerecombinase-mediated recombination event between LoxP sites, which are added to both ends of the fragment by ligating circularization adapters (2). The resulting circularized DNA molecules bear one recombined biotinylated circularization adapter sequence at the junction site. This LoxP sequence makes it possible to detect the junction computationally in paired read and split them at the junction without mapping to a reference sequence.
Illumina mate-paired libraries using Cre-Lox
To sequence Illumina mate-paired libraries with a read length >36 bp without running into the problem of a high percentage of unusable junction reads, we adapted the Illumina mate-paired protocol to use Cre-Lox recombination instead of blunt end circularization. In this way, a Cre-Lox sequence is incorporated between both joined ends at the junction site. This sequence allows screening for junction reads and makes it possible to trim or split those reads at the junction. We tested this new method by preparing and sequencing a mate-paired library with an insert size of 3 kb from Saccharomyces cerevisiae DNA. We present an analysis of the library quality statistics: ratio of mate-paired reads versus paired-end reads, number of junction reads, fragment size statistics, yield of usable mate-paired bases and library diversity. We show that all of the read pairs identified as mate-pairs map to the reference genome with a mean distance of ∼3 kb. We also present a bioinformatics tool that can be used to analyze an IlluminaCre-Lox mate-paired data set and to produce FASTQ files containing categorized mate-paired, paired-end and LoxP negative reads which are split or trimmed at the junction site to eliminate LoxP adapter sequences. Finally, we show how the sequencing data resulting from the library improves a de novo assembly of the S. cerevisiae genome.
METHODS
The IlluminaCre-Lox mate-paired library preparation protocol presented here is similar to the Illumina Mate Pair Library v2 Sample Preparation Guide for 2–5 kb libraries (1). The first part of this protocol was modified to allow for Cre-Lox recombination instead of blunt end circularization. The protocol was also changed to achieve a higher yield of DNA that can be used in the PCR library amplification step. Doing so allows for using fewer PCR cycles, increasing library diversity and reducing PCR bias. Instead of nebulization or hydroshearing the DNA in combination with a 15-h gel size selection step, the DNA is fragmented using a Covaris S2 Adaptive Focused Acoustic (AFA) instrument. This method yields fragmented DNA with a size distribution small enough to eliminate the need for gel size selection. It not only saves time, but also encompasses a major yield gain. Supplementary Figure S1 shows a typical Agilent High Sensitivity DNA trace of genomic DNA fragmented to 3 kb. The fragmented DNA undergoes an additional circularization adapter ligation step to ligate the LoxP adapter sequences to both ends of the fragments, followed by a Bst DNA polymerase fill in reaction. The ligation product is then used in a 1-h Cre recombination reaction instead of an overnight blunt end circularization. Note that only the 50% of LoxP adapter containing fragments with both a forward and a reverse adapter can circularize by CreLox recombination. The library preparation steps following the Cre recombination are an adaptation of the Illumina Mate Pair Library v2 Sample Preparation Guide for 2–5 kb libraries. The complete, detailed protocol is available in Supplementary Methods. Supplementary Figure S2 shows a typical Invitrogen 4% E-gel after library PCR amplification. Figure 1 shows the used LoxP adapter oligos while Figure 2 shows a schematic of the Cre recombination related library preparation steps.
Figure 1.
LoxP adapter oligos. Both double-stranded oligos have a 3-bp overhang to allow for directional ligation.
Figure 2.
Schematic of the Cre recombination-related library preparation steps. NNNNN denotes 2- to 5-kb DNA fragments taken into the LoxP adaptor ligation. LoxP sequences are in red and the 8-bp spacer between the two palindromic elements are in green. Orientation of the spacer region determines direction of recombination. Marked in yellow are the biotinylatedthymidines.
IlluminaHiSeq 2000 sequencing and data analysis
A mate-paired library was created using 5 µg of S. cerevisiae S288C genomic DNA (ATCC 204508) obtained from American Type Culture Collection (Manassas, VA, USA). Sequencing was performed in one lane of a v1.5 flowcell on an IlluminaHiSeq 2000 sequencer, using the TruSeq Paired-End Cluster Kit v2.5 (Illumina PE-401-2510) and the TruSeq SBS HS Kit v1 200 cycles (Illumina FC-401-1001), generating 2 × 100 bp reads. Image analysis and basecalling was done using the HiSeq Control Software version 1.1.37.19 and the Off-Line Base Caller v1.9.
LoxP detection, trimming and classification of paired reads using DeLoxer
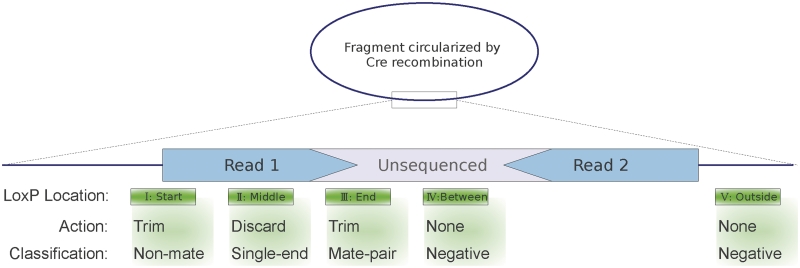
A software tool named DeLoxer was written in R (www.r-project.org) to classify read pairs based on the presence and position of the LoxP sequence in the paired reads. The tool, including the open-source R script, a manual and installation instructions, is available at http://genomes.ucsd.edu/downloads. DeLoxer aligns the LoxP adapter sequence to each pair of reads in an input data set and uses the alignment to infer the position of the LoxP sequence within the DNA fragment from which the read pair was generated (see Figure 3, Cases I–V). With Illumina sequencing, each read in a pair starts from an outside end of the sequenced DNA fragment and reads toward the center. Thus, a LoxP sequence at the tail end of either read is actually between the two reads, and such a read pair is classified as ‘mate-paired’ (Case III). Similarly, a LoxP sequence at the beginning of either read is at the end of the fragment and not between the reads, and yields a classification of non-mate, or ‘paired-end’ (Case I). If the LoxP sequence is not detected in either read, the pair is classified as ‘LoxP-negative’, indicating one of the following cases: The LoxP sequence lies within the unsequenced portion of the fragment (Case IV), the fragment does not contain the LoxP sequence (Case V) or the reads were not of sufficient quality to be aligned. Finally, the LoxP aligned regions are trimmed from the reads, and any reads <36 bp are discarded, leaving the other read as unpaired (case II). A perfect overlap of at least 10 bp (or equivalent-scoring imperfect overlap) is required to make a LoxP-positive call, but an overlap of any size is always trimmed.
Figure 3.
Schematic of the classification scheme used by DeLoxer. The dark blue line represents the original circularized DNA fragment obtained by Cre recombination and polymerase fill-in, while the block represents a single linear piece of that fragment obtained during the mate-pair sequencing prep. The ends are sequenced (light blue), yielding a read pair that must be classified, while the center of the fragment may be unsequenced (light gray). The possible positions of the LoxP adapter relative to the sequenced fragment are shown as green boxes. Case I: the LoxP site aligns to the start of one read, and the overlap is trimmed off. This LoxP site is outside the two reads, so the read pair is not a mate pair and is labeled as ‘paired-end’. Case II: The LoxP site aligns near the center of one read. The read is discarded as it does not contain at least 36 contiguous bp of genomic DNA, and the other read in the pair is retained as an unpaired read. Case III: The LoxP site aligns to the end of one (or both) reads, and the overlap is trimmed off. This site lies between the two reads, making this pair a mate-pair. Case IV: The LoxP site lies entirely in the unsequenced center portion of the DNA fragment, and does not overlap either read, so the pair is LoxP-negative. Case V: The LoxP site does not occur anywhere within the sequenced fragment, so the pair is LoxP-negative. Note that although Case IV represents a mate-pair, it is indistinguishable from Case V.
DeLoxer can trim and classify ∼80 million read pairs in under 7 h using 48 cores and ∼128 GB of memory. Time and memory requirements are proportional to the input size. The memory requirement can be reduced by splitting the input into small chunks.
Mapping of the DeLoxer output to the S. cerevisiae reference genome
The S. cerevisiae S288C genome (Assembly EF 2 February 2010; Database version 62.2d; Base pairs: 12 162 995; Gene build by SGD, last updated/patched, March 2010) was downloaded from the Ensembl ftp site. NovoAlign 2.07.10 (www.novocraft.com) was used to map reads from the Deloxer output files to the indexed reference S288C genome, storing the unique alignments in a SAM file. To map mate-paired reads, reverse–forward orientation with insert sizes between 10 and 10 000 were considered. To map paired-end reads, forward–reverse insert sizes between 10 and 5000 were considered. LoxP-negative read pairs were mapped both using reverse–forward (insert size 10–10 000) and forward–reverse (insert size 10–5000) orientation.
Quality correction of paired reads
Read pairs categorized as LoxP-negative by Deloxer were filtered based on quality to eliminate low quality reads. A perl script published online by Nik Joshi, The Bioinformatics Core at UC Davis Genome Center (last revised in September 2010), was used with parameters to trim reads at locations where the average quality in the next window of 10 bases is <20. Reads <30 after trimming were discarded. If only one member of a pair was discarded, the remaining read was stored in a single read FASTQ file.
RESULTS
Library quality
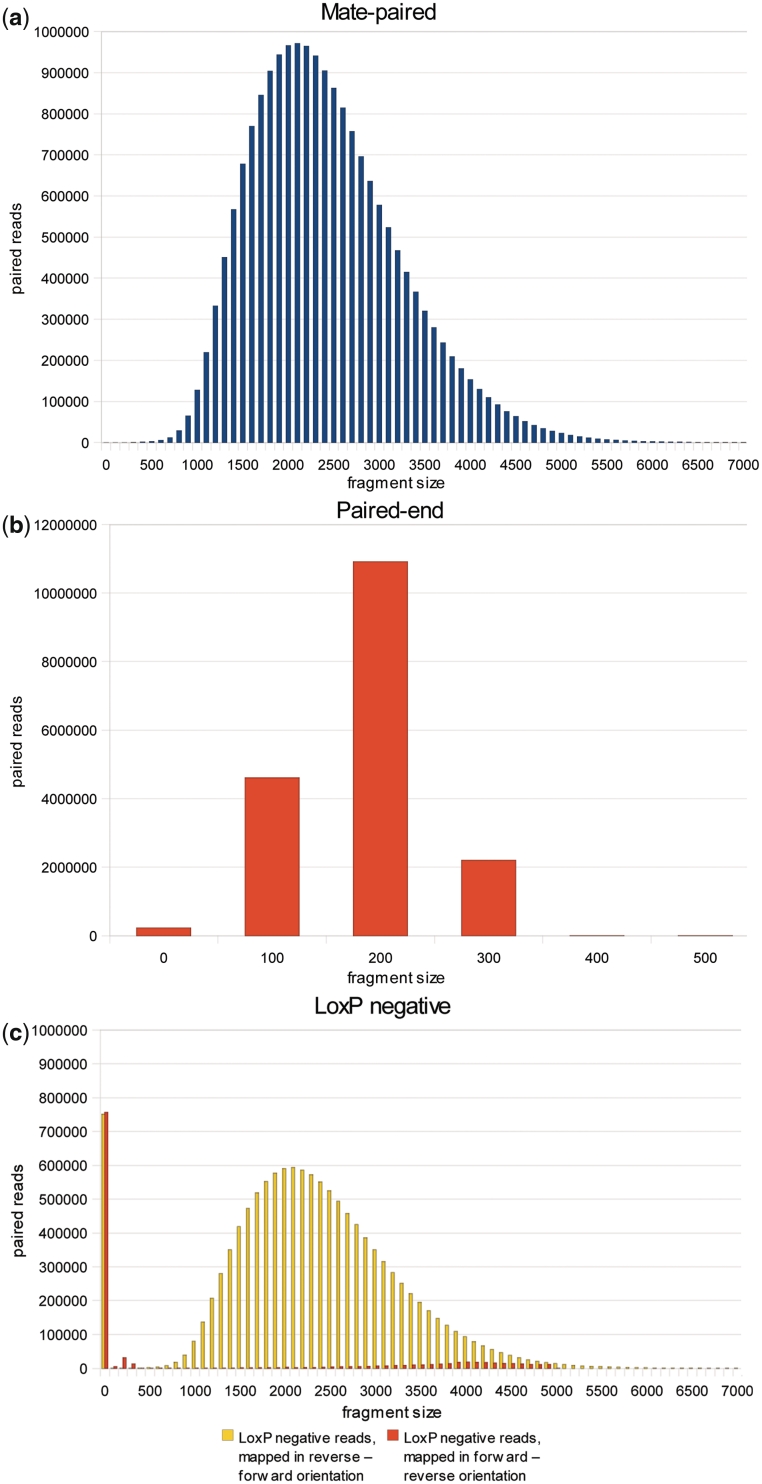
The ideal mate-paired library would contain full-length reads only, all with an insert of 3 kb between the paired reads. Because biotin enrichment is not perfect, in practice, some fragments present in the library do not contain biotin and hence do not overlap the circularization junction. Paired reads resulting from these fragments will align to a reference genome with a much smaller insert size. With longer read lengths, a read is more likely to cross the circularization junction. All junction sites should contain the LoxP sequence. This makes it possible to detect the junction. However, reads that cross the junction are not full length: The LoxP sequence must be trimmed from the reads before they can be used. An ideal library would also have a small variation in insert length. Because no gel size selection was performed after the initial 3 kb fragmentation, insert sizes vary. Ligation errors at the circularization adapter ligation step and inter-molecular recombination during the circularization step can introduce some chimeras (1). Figure 4 and Table 1 show statistics on the frequency with which these issues arise derived from a 3-kb mate-paired S. cerevisiae DNA library (prepared using 14 cycles of PCR), sequenced in one IlluminaHiSeqflowcell lane. The processed reads produced by DeLoxer (mate-paired, paired-end and the LoxP-negative read pairs) were mapped against the S. cerevisiae genome using Novoalign. The LoxP-negative reads were quality filtered (see Methods) before mapping because this data set contains a subset of low quality reads with no useful sequence. Since these reads inherently do not have the LoxP sequence, DeLoxer categorizes them together with the high quality LoxP-negative reads. Figure 4 shows the fragment size distribution of the DeLoxer output (mate-paired, paired-end and LoxP-negative pairs). Table 1 shows the quality statistics for the DeLoxer output: Based on the Novoalign mapping and the resulting fragment size, we were able to determine the numbers of true mate-paired reads (fragment size 501–6500), true paired-end reads (fragment size 201–500) and short fragments resulting in overlapping reads (fragment size <200). For the LoxP-negative reads, Table 1 shows statistics for the reverse–forward mapping, as only a negligible number of reads mapped in the forward–reverse orientation (see Figure 4).
Figure 4.
Fragment size distribution of (a) mate-paired reads, (b) paired-end reads and (c) LoxP-negative reads.
Table 1.
DeLoxer output quality statistics
| Number of reads | Percentage of reads (%) | Fragment size (mean ± SD) | Size after trimming (mean ± SD) | |
|---|---|---|---|---|
| Total 2 × 100 reads | 78 607 373 | 100.00 | ||
| Mate-paired reads (LoxP positive) | 22 494 162 | 28.62 | 79 ± 22 | |
| Uniquely aligned pairs | 18 970 394 | 100.00 | ||
| True mate-paired reads | 18 951 404 | 99.90 | 2313 ± 812 | |
| Paired-end reads | 7216 | 0.04 | 400 ± 78 | |
| Short fragments | 1359 | 0.01 | 95 ± 66 | |
| Non-unique alignment | 2 488 148 | |||
| Unaligned | 1 035 620 | |||
| Duplicate reads | 5 518 899 | 29.09 | ||
| Paired-end reads (LoxP positive) | 22 424 011 | 28.53 | 78 ± 22 | |
| Uniquely aligned pairs | 17 963 827 | 100.00 | ||
| True paired-end reads | 12 677 878 | 70.57 | 256 ± 40 | |
| Mate-paired reads | 2663 | 0.01 | 3299 ± 2261 | |
| Short fragments | 5 283 278 | 29.41 | 168 ± 32 | |
| Non-unique alignment | 3 478 415 | |||
| Unaligned | 981 769 | |||
| Duplicate reads | 5 008 216 | 27.88 | ||
| LoxP negative, low quality | 5 517 705 | 7.02 | ||
| LoxP negative, quality filtered | 22 288 114 | 28.35 | 83 ± 18 | |
| Uniquely aligned pairs | 12 409 334 | 100.00 | ||
| Mate-paired reads | 11 567 200 | 93.21 | 2279 ± 813 | |
| Paired-end reads | 46 637 | 0.38 | 290 ± 64 | |
| Short fragments | 777 291 | 6.26 | 61 ± 17 | |
| Non-unique alignment | 1 613 963 | |||
| Unaligned | 8 264 817 | |||
| Duplicate reads | 3 243 252 | 26.14 | ||
| Single reads (LoxP positive) | 5 820 905 | 7.41 | ||
| Both reads too short (LoxP positive) | 62 476 | 0.08 |
Data generated by sequencing a 3-kb mate-paired S. cerevisiae DNA library (prepared using 14 cycles of PCR), sequenced in one Illumina HiSeq flowcell lane.
Picard tools program Markduplicates (http://picard.sourceforge.net) was used to determine the number of duplicate reads in each data set. Of the mapped reads in the mate-paired, paired-end and quality filtered LoxP-negative data sets, 29, 28 and 26% were marked as duplicate reads respectively. Samtools (3) was used to calculate the coverage depth of each base after the mate-paired, paired-end and the LoxP-negative read pairs were mapped against the S288C reference genome. The average coverage depth was 245×, 233× and 188× respectively, covering 98, 96 and 98% of the genome, respectively. The results from the Samtools depth tool are plotted in Supplementary Figure S3.
The yield and quality statistics output from the HiSeq 2000 pipeline can be found in Supplementary Figures S4–S6. Supplementary Figure S4 shows the cluster densities and the percentage of past filter clusters for each lane in the Illumina run, making it possible to compare the S. cerevisiae lane (lane 7) to the PhiX control lane (lane 8) and the other lanes which contain exome sequencing projects (not related to this study). Supplementary Figure S5 shows the percentage of the reads with a quality score >30 by cycle for the S. cerevisiae lane. Supplementary Figure S6 shows the error rate by cycle for the S. cerevisiae lane.
Library yield
The standard ‘Illumina Mate Pair Library v2 Sample Preparation Guide for 2–5 kb libraries’ requires 18 cycles of PCR amplification to yield enough library product. The method presented here was optimized to produce higher yield with fewer cycles of PCR amplification. The use of a Covaris S2 AFA instrument for fragmentation was a major factor in the optimization. Starting with 5 µg of genomic DNA, >1 µg of 3 kb fragmented DNA is available for input into the Cre-Lox recombination reaction. Table 2 shows the library yields with varying input amounts of fragmented DNA into the Cre-Lox recombination reaction and varying cycles of PCR amplification.
Table 2.
Library yields
| Input into Cre-Lox recombination reaction (ng) | Number of PCR cycles | Library yield (ng) |
|---|---|---|
| 400 | 18 | 1.08 |
| 600 | 16 | 2.49 |
| 1000 | 14 | 2.52 |
De novo assembly of S. cerevisiae
The output from DeLoxer was used to perform two de novo assemblies of the S. cerevisiae genome using SOAPdenovo version 1.05 (Beijing Genomics Institute) with default parameters. An optimal k-mer size of 45 was determined iteratively by reassembly and contig quality evaluation. Using only the paired-end reads both in the creation of contigs and in the scaffolding step, SOAPdenovo produced an assembly with a longest scaffold size of 454 266 bp, a scaffold N50 of 100 470 bp and 11 691 897 nt in scaffolds. Using the paired-end (contiging and scaffolding), mate-paired (scaffolding only) and LoxP-negative (scaffolding only) reads SOAPdenovo produced an assembly with a longest scaffold size of 849 572 bp, a scaffold N50 of 302 716 bp and 15 307 544 scaffolded nucleotides. Adding mate-paired data to the scaffolding step doubled the longest scaffold, tripled N50 and added 4 M nucleotides to the assembly.
To compare our results to the standard Illumina mate-paired protocol, we ran an alternative version of DeLoxer(BluntLoxer) to mimic a standard Illumina mate-paired data set from our data set. Bluntloxer only removes the LoxP sequence and concatenates the sequence to the left and right of the LoxP sequence, mimicking blunt-end ligation. BluntLoxer does not categorize the reads into paired-end, mate-paired and LoxP-negative reads. Using fastx_trimmer from the FASTX-Toolkit(hannonlab.cshl.edu/fastx_toolkit), the reads were trimmed to 36 bp. This generates a data set that is identical to our DeLoxer data set, except for the mimicked blunt-end ligation and the read length. The optimal k-mer size for this data set was empirically determined to be 35. Using the categorized paired-end reads from the original DeLoxer(contiging and scaffolding) with an additional scaffolding step using the uncategorized output from BluntLoxer, SOAPdenovo produced an assembly with a longest scaffold size of 631 169 bp, a scaffold N50 of 166 670 bp and 16 328 952 scaffolded nucleotides.
DISCUSSION
Mate-paired reads can be mapped to a reference using information about distance between the reads to resolve larger structural rearrangements including insertions, deletions and inversions. Distance information is useful in de novo assembly with short reads, enhancing scaffolding of contigs and enhancing assembly across repetitive regions. Illumina mate-paired libraries lack a recognizable sequence at the junction site between the two joined ends of the circularized DNA fragments used to create the mate-paired library (1). The presence of ‘unmarked’ junctions sites confounds de novo assembly efforts, possibly reducing assembly performance. Thus, when sequencing a mate-paired library, Illumina recommends a read length no longer than 36 bases to decrease the possibility that a sequence read will pass through the junction (1). To sequence Illumina mate-paired libraries with a read length >36 bp without running into the problem of a high percentage of unusable junction reads, we adapted the Illumina mate-paired protocol to use Cre-Lox recombination instead of blunt end circularization. This results in a LoxP sequence between both joined ends at the junction site, which allows reads to be split or trimmed at the junction and efficiently identifies mate-paired reads. We present a fast, multi-threaded bio-informatics tool (DeLoxer) for this purpose.
We tested this new method by preparing and sequencing a mate-paired library with an insert size of 3 kb from S. cerevisiae. The library preparation proved to be robust, yielding sufficient sequence-able library from 400 ng fragmented DNA when using 18 PCR cycles. When 1 µg of fragmented DNA is available, sufficient library can be produced using only 14 cycles. PCR can be a source of duplicate sequences and amplification bias, reducing the complexity of a library (4). So keeping the number of cycles of PCR amplification as low as possible is important. The protocol was evaluated with mate-paired libraries created from fragments up to 3 kb in length. Cre-Lox recombination will be less efficient using longer fragments, but library yields using 3 kb fragments were sufficiently high to suggest the protocol can be used successfully with longer fragments.
The quality statistics of the sequence data resulting from the S. cerevisiae library preparation show that our method generates a number of usable reads within the expected range of a successful HiSeq 2000 lane. The percentage of duplicate reads was ∼28% for the mapped LoxP-positive reads and is within the expected range of duplication when sequencing a 12-Mbp genome at 1300× coverage. The original read length of 100 bp was reduced to a mean read length of ∼80 bp after trimming LoxP sequences and quality filtering the LoxP-negative reads.
Nearly all reads in the LoxP-positive mate-paired category proved to be true mate-paired reads. The mean fragment size of 2.3 kb was lower than the targeted 3 kb fragment size, possibly because Cre-Lox recombination is more efficient with shorter fragments and is biased toward the shorter fragments in the fragmented DNA pool. A size selection step after DNA fragmentation could increase the mean mate-paired fragment size and reduce the standard deviation. An optional size selection step was added to the protocol available in Supplementary Methods. As illustrated in Figure 4, 71% of reads in the LoxP-positive paired-end category proved to be true, non-overlapping paired-end reads. The remaining 29% were from short DNA fragments smaller than the 2 × 100 bp read length. The reason for this bimodal fragment length distribution is unclear. The LoxP-negative reads nearly all mapped in the reverse–forward orientation (the orientation when the sequenced fragment contains a circularization junction) and proved to be true mate-paired reads. Similar to the LoxP-positive paired-end reads, this data set also contained a small subset of overlapping reads from short fragments. Altogether, 29% of reads were true mate-paired from long inserts, 28% were paired from short, fragment size inserts, 28% were LoxP-negative but proved to be mate-paired after mapping, and 15% were single reads, too short, or low quality after processing. The high proportion of mate-paired LoxP-negative reads indicates the biotin enrichment was highly successful.
It is beyond the scope of this study to directly compare the CreLox protocol with the standard Illumina mate-paired protocol and to prove that data resulting from the CreLox protocol performs better in de novo assembly. It is however safe to say that longer (5), categorized reads free of junction reads (1) are more useful for de novo assembly compared to shorter reads containing a mixture of mate-paired, paired-end and junction reads. Using only DeLoxer output from the S. cerevisiae mate-paired library preparation, we performed two de novo assemblies. We compared assembly performance using only the LoxP-positive paired-end reads to assembly performance using the same data complemented with a scaffolding step using the LoxP-positive mate-paired and the LoxP-negative reads. Results show that the additional scaffolding step doubled the size of the longest scaffold and tripled the N50 size of the assembly. Because the paired-end reads alone provided 370× coverage, the increase in assembly performance is not likely to be the result of additional coverage but due to the distance information contained in the mate-paired reads. Using only data obtained from the mate-paired library preparation, it was possible to produce a de novo assembly with a longest scaffold size of 849 572 bp (longest chromosome is 1.5 Mbp) and a scaffold N50 of 302 716 bp (N50 of the finished genome is 942 kbp). We also performed a de novo assembly using the an alternative version of DeLoxer (BluntLoxer) to mimic a standard Illumina mate-paired data set. BluntLoxer only removes the LoxP sequence and concatenates the sequence to the left and right of the LoxP sequence, mimicking blunt-end ligation. The reads were then trimmed to 36 bp, which is the maximum read length recommended by the standard Illumina mate-paired protocol. Reads from a standard Illumina mate-paired protocol, mimicked or otherwise, can not be used by themselves to perform a useful assembly because an initial contiging step using paired-end reads is required. These paired-end reads normally have to be generated by a separate paired-end protocol. Our DeLoxer output provides both paired-end and mate-paired reads from one protocol because the LoxPsequence makes it possible to categorize the reads. The paired-end reads from DeLoxer were used for contiging and the uncategorized reads from BluntLoxer were used for scaffolding. The results of this assembly show a scaffold N50 which is 50% smaller compared to the assembly using the original DeLoxer output.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods and Supplementary Figures 1–6.
FUNDING
Scientific Research-Flanders, Belgium (F.W.O.-Vlaanderen); National Institutes of Health (grant 1RC2EY02678-01, partial and grant U19 A1063603-06) and National Science Foundation (grant DBI0852081, partial). Funding for open access charge: The Scripps Research Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge the support of the Fund for Scientific Research-Flanders, Belgium (F.W.O.-Vlaanderen).
REFERENCES
- 1.Illumina. Mate Pair Library v2 Sample Preparation Guide For 2–5 kb Libraries. 2009. [Google Scholar]
- 2.Roche. GS FLX Paired End DNA Library Preparation Method Manual, GS FLX Titanium Series. 2009. [Google Scholar]
- 3.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schatz MC, Delcher AL, Salzberg SL. Assembly of large genomes using second-generation sequencing. Genome Res. 2010;20:1165–1173. doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.